- *Corresponding Author:
- C. Tang
Department of Pharmacology, Southwest Medical University, Luzhou, Sichuan 646099, China
E-mail: 576680355@qq.com
| This article was originally published in a special issue, “Role of Biomedicine in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(2) Spl Issue “146-158” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A common clinical treatment for hyperlipidemia in China is hawthorn, a traditional Chinese herb. However, few researches have been done on its active ingredients and their possible mechanisms for treating hyperlipidemia. The active chemical constituents of hawthorn were obtained from the encyclopedia of traditional Chinese medicine, the bioavailability and druglikeness properties of each active ingredient of hawthorn were analyzed using SwissADME and their possible targets of action were predicted using SwissTargetPrediction and PharmMapper. Then, we used the Gene Expression Omnibus-related gene microarray data to mine the differentially expressed genes for hyperlipidemia. Meanwhile, we collected relevant targets of hyperlipidemia through the DrugBank, Online Mendelian Inheritance in Man, GeneCards, DisGeNET and Therapeutic Target Database. Based on this data, a component-target-disease pathway and a protein-protein interaction networks were constructed using search tool for the retrieval of interacting genes/proteins. Finally, Metascape was systematically used to analyze the targets of hawthorn intervention in hyperlipidemia. A total of 8 active components and 312 hawthorn targets were gathered and 343 prospective targets associated with hyperlipidemia were predicted, 25 of which shared targets with hawthorn-related targets. Gene ontology enrichment analysis revealed that the mechanism of action of hawthorn in intervention of hyperlipidemia lies in the regulation of lipid metabolism, nutrition level, hormone level and cholesterol transport process. The Kyoto encyclopedia of genes and genomes enrichment analysis revealed that the peroxisome proliferator activated receptor signaling pathway contained the majority of the core targets. Molecular docking verified that hawthorn could bind closely to the core targets. According to the findings of network pharmacology and enrichment analysis, we predicted that the main active components of hawthorn to intervene in hyperlipidemia are epicatechin, officinalic acid and quercetin, which mainly intervene in hyperlipidemia through the peroxisome proliferator activated receptor signaling pathway.
Keywords
Network pharmacology, hawthorn, hyperlipidemia, gene expression omnibus database
Cardiovascular Disease (CVD) is the major cause of morbidity and death in the world today. In a global survey of CVD, it is reported that the number of people with CVD worldwide increased rapidly from 1.271 billion to 523 million between 1990 and 2019, and the number of deaths increased from 12.1 million to 18.6 million, with 9.6 million men and 8.9 million women[1]. Metabolic CVD is a class of diseases in which disorders of multiple nutrient metabolites impair the heart and blood vessel function. It is closely related to CVD development and mainly includes abnormalities in blood lipids, blood glucose, uric acid and obesity[2]. A prospective cohort study of 6255 subjects found that patients with metabolic syndrome had a prominently increased risk of death from CVD than healthy subjects and patients with diabetes combined with CVD were at higher risk[3]. Hyperlipidemia is a common metabolic syndrome characterized by elevated levels of Total Cholesterol (TC), Triglycerides (TGs), Low-Density Lipoprotein Cholesterol (LDL-C) and decreased High-Density Lipoprotein Cholesterol (HDL-C)[4]. It is a significant risk factor for coronary artery disease[5]. Furthermore, lowering TC, TGs and LDL-C levels in blood can considerably lower the incidence of coronary artery disease and the risk of death[6]. Therefore, effective lipid control is essential for the treatment of CVD and for improving the quality of life of patients. At present, hyperlipidemia cannot be cured temporarily and requires long-term oral drug treatment. Atorvastatin calcium, rosuvastatin calcium, simvastatin and other Hydroxymethylglutaryl-Coenzyme A (HMG-CoA) reductase inhibitors and some other lipid-lowering drugs are commonly used in the first line of clinical practice, and long-term clinical observation has found that they have good lipid-lowering effects[7]. However, there are certain adverse effects such as muscle damage, liver function impairment, allergy, etc.[8]. Therefore, the development of effective drugs with less toxic side effects is an inevitable clinical requirement.
Most interventions in Chinese medicine for hyperlipidemia use methods to strengthen the spleen and resolve phlegm, and hawthorn is one of the most used single herbs with a long history[9]. Metabolomic analysis shows that hawthorn exerts its activity by regulating lipid metabolism, energy metabolism, oxidative stress and amino acid metabolism[10]. In addition, hawthorn extract can effectively lower serum cholesterol, TGs, LDL-C and increase HDL-C levels, reducing a series of inflammatory factors, inhibits progression and protects vascular endothelial cells[11]. Meanwhile, Shatoor et al. found that hawthorn can effectively reduce blood lipids and reverse atherosclerosis in hyperlipidemic rats through an antioxidant mechanism and regulation of oxidized LDL-C (ox-LDL-C) levels[12]. It has potential value in the treatment of hyperlipidemia. However, the specific pharmacological mechanism of hawthorn intervention in hyperlipidemia is still unclear. This study comprehensively elucidated the potential therapeutic targets of hawthorn intervention in hyperlipidemia using network pharmacology, Gene Expression Omnibus (GEO) and molecular docking to provide a strong theoretical basis for clinical research and drug use.
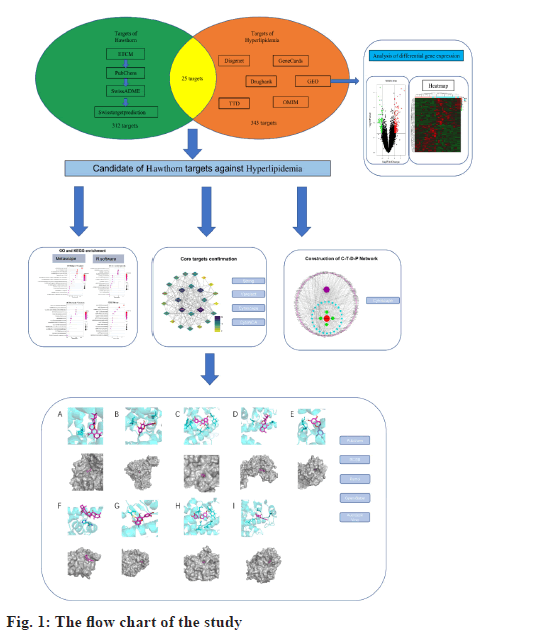
Network pharmacology is a systematic analysis method based on the interaction of disease, drugs, active components, target gene and target protein[13]. It emphasizes the analysis of potential targets of drug interventions on diseases from a systemic level and the holistic perspective of network data, which coincides with the holistic concept of Chinese medicine and is important for studying the role of Traditional Chinese Medicine at the micro level[14]. The GEO database is a publicly accessible archive of data from a wide range of high-throughput genetic studies, including single and dual-channel microarraybased assays that detect messenger Ribonucleic Acid (mRNA), genomic Deoxyribonucleic Acid (DNA), and protein abundance, as well as non-array technologies[15,16]. Combining network pharmacology and GEO could reveal the disease mechanisms and molecular targets for drug therapy[17]. Therefore, this study will systematically use network pharmacology combined with GEO to analyze the mechanism of hawthorn intervention on hyperlipidemia (fig. 1). First, we obtained its composition and potential targets to intervene in hyperlipidemia. Protein- Protein Interaction (PPI) models of potential targets for intervention in hyperlipidemia were constructed using the potential targets. Following that, a comprehensive examination of possible targets and biofunctional modules was carried out. Meanwhile, molecular docking was used to validate the interactions between active components and key targets.
Materials and Methods
Hawthorn target acquisition:
Firstly, using the Encyclopedia of Traditional Chinese Medicine (ETCM)[18] and published literature the individual active components of hawthorn are collected. Then, all Structure Data Format (SDF) files were obtained from PubChem[19]. Next, the obtained SDF files were imported into SwissADME[20] for absorption prediction. The criteria for setting the potential core compounds were, Pharmacokinetics- Gastro Intestinal (GI) absorption was set to “High”, indicating good oral bioavailability of the ingredient; Druglikeness was set to 5 categories of pharmacophore prediction (Lipinski, Ghose, Veber, Egan, Muegge) with two or more “Yes” results[21,22]. Finally, the screened core chemical components were uploaded to SwissTargetPrediction[23] and PharmMapper[24] to predict potential targets and the screened targets were combined and de-duplicated.
Therapy targets of hyperlipidemia:
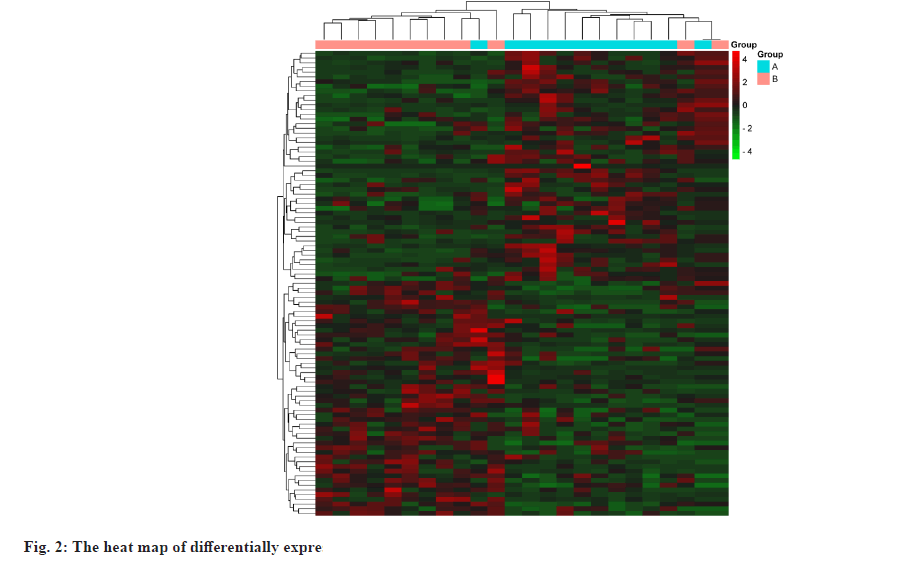
The datasets were retrieved using the keyword “hyperlipidemia” in the GEO database and downloaded the retrieved gene chip related files. Converting probe IDs to gene names using the R software limma package[25], obtaining the expression matrix and ultimately screening for differential genes with Fold Change (FC)>2 and p value<0.05 as the threshold, using R software pheatmap package[26] to draw heat map. Utilizing ggplot2[27], we created a volcano plot and differentially expressed gene sets were normalized to the UniProt protein database[28].
Online Mendelian Inheritance in Man (OMIM) [29], DrugBank[30], GeneCards[31], DisGeNET[32] and Therapeutic Target Database (TTD)[33] can provide the latest data on human genes and free information on currently known disease-related targets in which hyperlipidemia was used as a keyword with the condition “human” or “Homo sapiens.” The relevant hyperlipidemia targets were gathered in the aforementioned database, merged and de-duplicated with the differentially expressed gene set obtained from GEO analysis and all targets were standardized using UniProt.
Component-Target-Disease-Pathway (C-T-D-P) network construction:
Based on the data gathered, we constructed the C-TD- P network using Cytoscape 3.8.0[34] to demonstrate the correlation between bioactive compounds, potential targets and pathways. Potential targets were used for subsequent protein interaction and functional enrichment analysis.
PPI analysis:
We extracted intersect of targets in hyperlipidemia and hawthorn and defined them as potential targets for hawthorn intervention in hyperlipidemia. PPI[35] refers to studying correlations between compounds and disease-related proteins from the perspective of biochemistry, signal transduction pathways and genetic networks. PPI analysis of disease-associated proteins allows better presentation of their relevance to drug compounds. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a web-based online tool[36] that provides online analysis of PPIs by uploading potential intervention targets to STRING to obtain PPI network relationships. The results are imported into Cytoscape for further visualization and analysis. CytoNCA plugin[37] of cytoscape was used to analyze and evaluate PPI networks and Molecular Complex Detection (MCODE)[38] was used to obtain potential protein functional modules and analyze related functions. In addition, in order to identify the core of the potential targets more accurately, we imported the potential targets of hawthorn intervention in hyperlipidemia into the VarElect dataset[39] for correlation analysis of potential targets with hyperlipidemia.
Biology enrichment analysis:
Functional enrichment analysis (Gene Ontology (GO)[40] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[41]) was performed on potential targets to facilitate mechanistic understanding of hawthorn intervention in hyperlipidemia. Metascape is a web-based tool that enables researchers thorough gene list annotation and functional enrichment[42]. Potential targets were uploaded to Metascape for GO Biological Process (BP), Cell Component (CC), Molecular Function (MF) and KEGG pathway enrichment analysis, and entries with p value<0.01 are considered significantly enriched. Finally, the top 15 eligible GO enrichment analysis results and KEGG pathways were selected for further research.
Molecular docking:
Import the core compound “SDF” file of Hawthorn into Open Babel 3.1.1[43] to convert the file format to “mol2”. Protein Data Bank (PDB)[44] collects the structures of the core targets and saves them as “mol2” file format, using PyMOL software[45] to separate modified ligands and remove water. AutoDockTools[46] was used to add polar hydrogens and assign Gasteiger charges for proteins and ligands, and the resulting structure was saved in Protein Data Bank, Partial Charge (Q), and Atom Type (T) (PDBQT) format. Finally, in order to determine whether the ligand and proteins selected after this procedure would bind well, dockings were performed with AutoDock Vina[47], where a docking score affinity<-4.25 kcal/mol can suggest binding activity between ligand and proteins, score<-5.0 kcal/mol indicates improved binding activity and score<-7.0 kcal/mol indicates substantial docking activity between ligand and proteins[48].
Results and Discussion
Hawthorn targets were shown here. Active components of hawthorn were obtained from ETCM and their structures were downloaded through PubChem after uploading the “SDF” files to SwissADME to assess the GI absorption and druglikeness. A total of 8 potential core compounds were obtained and shown in Table 1. 209 potential hawthorn targets were collected after importing them into SwissTargetPrediction and 369 potential targets of hawthorn were collected after prediction by PharmMapper. After removing duplicates, a total of 312 potential targets of hawthorn were obtained.
| Compound | PubChem ID | Molecular formula |
|---|---|---|
| Acetic acid | 176 | C2H4O2 |
| Epicatechin | 72 276 | C15H14O6 |
| Malic acid | 525 | C4H6O5 |
| Officinalic acid | 10 577 334 | C30H44O6 |
| Oxalic acid | 971 | C2H2O4 |
| Propionic acid | 1032 | C3H6O2 |
| Quercetin | 5 280 343 | C15H10O7 |
| Succinic acid | 1110 | C4H6O4 |
Table 1: Potential Core Compounds of Hawthorn
Targets of hyperlipidemia were shown here. After searching from the GEO datasets, the original dataset of the GSE1010 chip and its annotation file GPL96 were downloaded and analyzed using R software. There were 253 differentially expressed genes which were obtained, of which 164 had their increased expression and 89 were downregulated. Following this, a heat map for hyperlipidemia target was created using the pheatmap R package (fig. 2). Finally, a volcano diagram was developed using the R package ggplot2 (fig. 3). We searched OMIM, DrugBank, GeneCards, DisGeNET and TTD. A total of 141 hyperlipidemia-associated targets were obtained. After pooling and de-duplication with GEO differential expressed genes, 343 hyperlipidemiaassociated genes were collected.
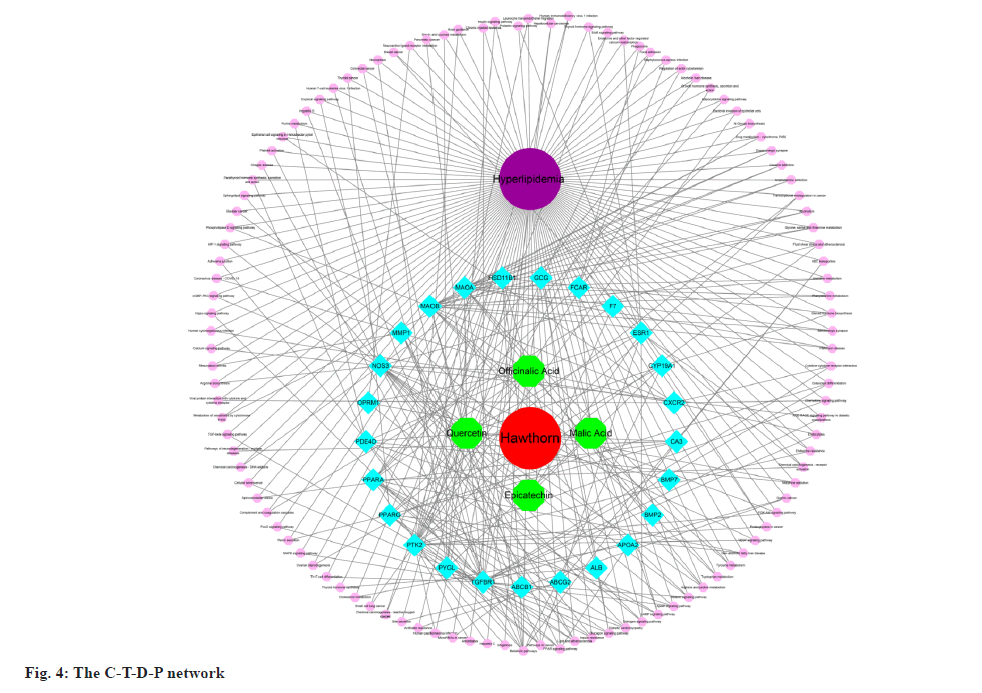
Using Cytoscape, we created a C-T-D-P network based on the data presented above. The network consisted of 142 nodes, including four active components of hawthorn, hyperlipidemia and 25 common targets, with 321 signaling pathways connected between the nodes (fig. 4). This network has initially revealed a complex mechanism of multiple targets and signaling pathways for compounds of hawthorn to intervene in hyperlipidemia.
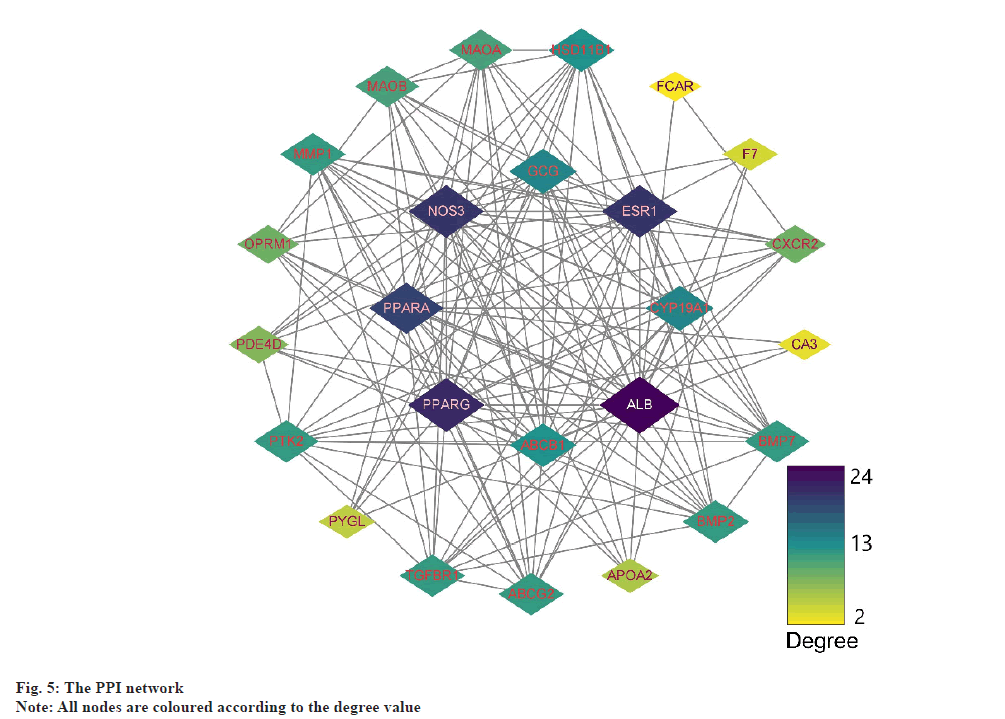
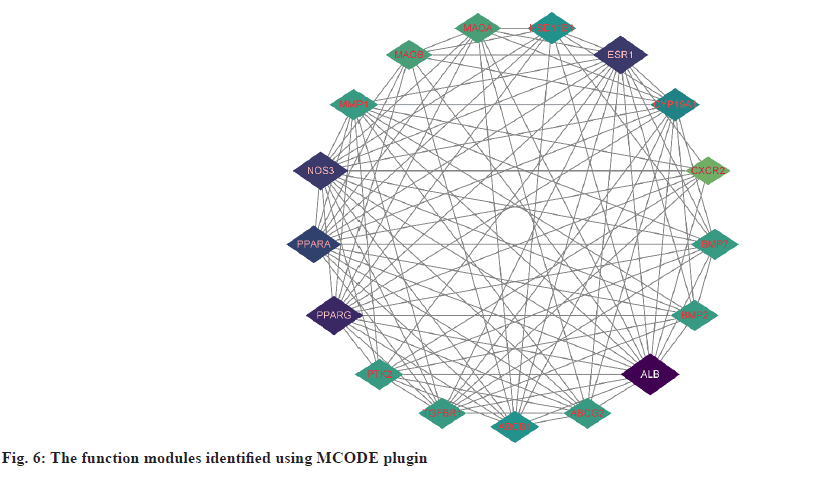
PPI network analysis was explained here. The further analysis used, STRING to analyze PPI for all 25 common targets. The data was loaded into Cytoscape and degree values of each gene were determined with the CytoNCA plugin. Meanwhile, PPI networks were constructed based on the STRING database (fig. 5). The targets with top 8 degree values were defined as hub targets. The names and functions of these hub genes are shown in Table 2. Using the MCODE plugin, a working module was also identified (fig. 6). In order to determine the target of the hub genes more accurately, we used VarElect to analyze the correlation. According to the findings, 17 targets were directly associated to hyperlipidemia, whereas 8 were indirectly related to hyperlipidemia (Table 3). After comparing with the hub target, it was found that Albumin (ALB), Peroxisome Proliferator Activated Receptor Alpha (PPARA) and Peroxisome Proliferator Activated Receptor Gamma (PPARG) had the highest score of correlation. Each active component of hawthorn will be molecularly docked to 3 hub targets.
| Gene symbol | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|
| ALB | 24 | 99.3894 | 1 |
| PPARG | 21 | 46.5841 | 0.88889 |
| NOS3 | 20 | 31.1672 | 0.85714 |
| ESR1 | 20 | 29.3894 | 0.85714 |
| PPARA | 19 | 36.4794 | 0.82759 |
| CYP19A1 | 14 | 5.18066 | 0.70588 |
| ABCB1 | 13 | 4.07669 | 0.68571 |
| HSD11B1 | 13 | 6.72114 | 0.68571 |
Table 2: Names and Topological Parameters of the Hub Targets
| Symbol | Description | Scores |
|---|---|---|
| APOA2 | Apolipoprotein A2 | 16.74 |
| ALB | Albumin | 11.15 |
| PPARA | Peroxisome Proliferator Activated Receptor Alpha | 11.06 |
| PPARG | Peroxisome Proliferator Activated Receptor Gamma | 8.9 |
| NOS3 | Nitric Oxide Synthase 3 | 3.81 |
| PYGL | Glycogen Phosphorylase L | 3.59 |
| CYP19A1 | Cytochrome P450 Family 19 Subfamily A Member 1 | 3.05 |
| ESR1 | Estrogen Receptor 1 | 2.97 |
| F7 | Coagulation Factor VII | 2.63 |
| GCG | Glucagon | 2.2 |
| ABCB1 | ATP Binding Cassette Subfamily B Member 1 | 0.9 |
| HSD11B1 | Hydroxysteroid 11-Beta Dehydrogenase 1 | 0.5 |
| PTK2 | Protein Tyrosine Kinase 2 | 0.49 |
| MMP1 | Matrix Metallopeptidase 1 | 0.38 |
| BMP2 | Bone Morphogenetic Protein 2 | 0.22 |
| PDE4D | Phosphodiesterase 4D | 0.12 |
| ABCG2 | ATP Binding Cassette Subfamily G Member 2 (Junior Blood Group) | 0.12 |
Table 3: The Information of Targets Genes Directly Associated to Hyperlipidemia
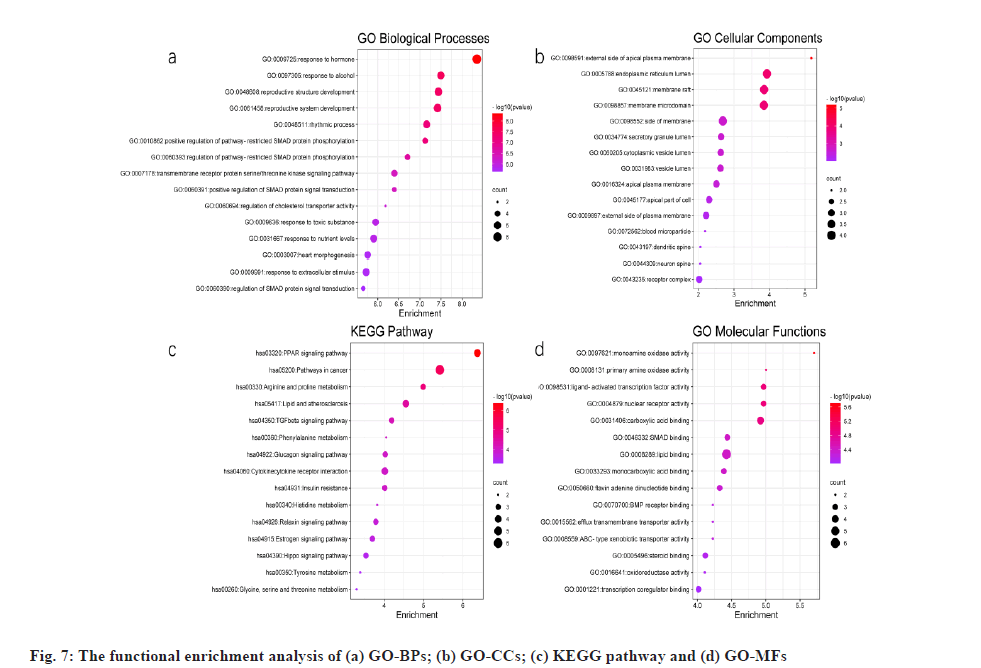
Functional enrichment analysis and its results were explained here. 25 potential targets were uploaded to Metascape for GO enrichment analysis: BP, CC, MF and KEGG pathways. The results of p<0.01 were derived and the bubble diagram of enrichment analysis was drawn using the ggplot package in R language (fig. 7).
Potential targets in GO BPs were mainly involved in nutrient level regulation, hormone levels, growth, heart rate, cholesterol transport and other processes (fig. 7a). GO CCs are involved in the apical plasma membrane, endoplasmic reticulum, plasma membrane rafts, vesicle lumen and secretory granule lumen (fig. 7b). The KEGG pathway focuses on PPAR signaling pathway, lipid and atherosclerosis, Transforming Growth Factor beta (TGF-β) signaling pathway, glucagon signaling pathway, insulin resistance and cytokine receptor interactions (fig. 7c). GO MFs are mainly involved in monoamine oxidase activity, nuclear receptor activity, lipid binding, transcription factor activity, steroid binding, carboxylic acidbinding, etc. (fig. 7d).
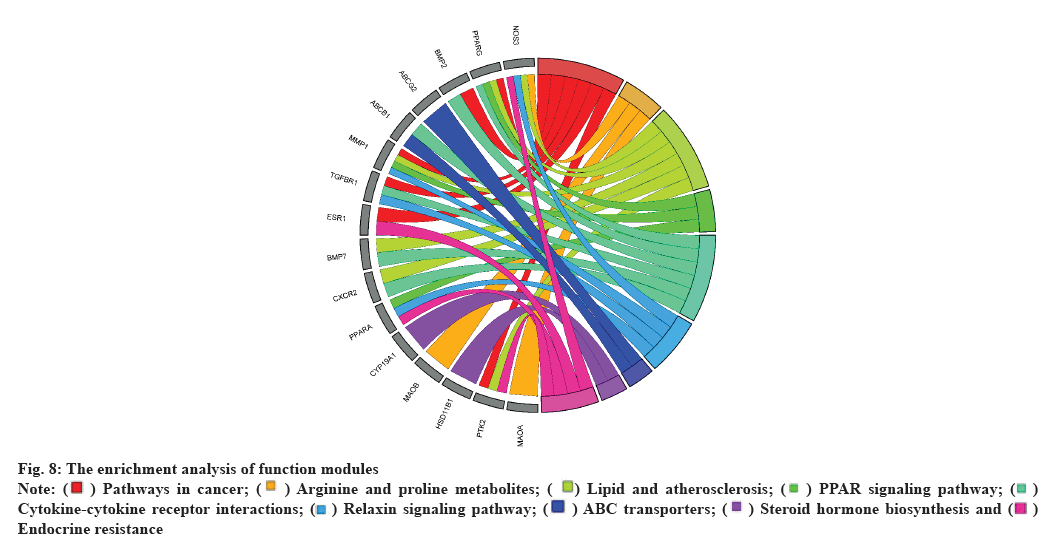
The functional modules were subjected to GO and KEGG enrichment analyses and their findings were shown in R using the ggplot package (fig. 8). The enrichment analysis showed that the modules could be involved in cancer pathways, lipid binding, transport, steroid hormones bioregulation, TGF- β-Suppressor of Mothers against Decapentaplegic (TGF-β-SMAD) signaling pathway regulation, redox, etc.
Fig. 8: The enrichment analysis of function modules
Note: ( ) Pathways in cancer; (
) Pathways in cancer; (  ) Arginine and proline metabolites; (
) Arginine and proline metabolites; ( ) Lipid and atherosclerosis; (
) Lipid and atherosclerosis; ( ) PPAR signaling pathway; (
) PPAR signaling pathway; ( )
Cytokine-cytokine receptor interactions; (
)
Cytokine-cytokine receptor interactions; ( ) Relaxin signaling pathway; (
) Relaxin signaling pathway; ( ) ABC transporters; (
) ABC transporters; ( ) Steroid hormone biosynthesis and (
) Steroid hormone biosynthesis and ( )
Endocrine resistance
)
Endocrine resistance
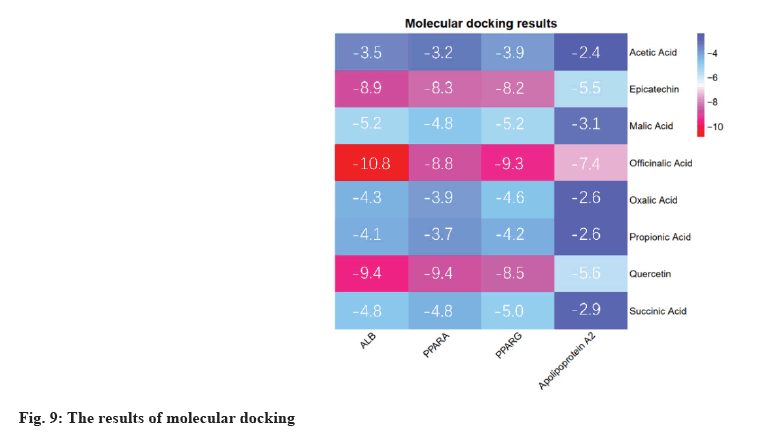
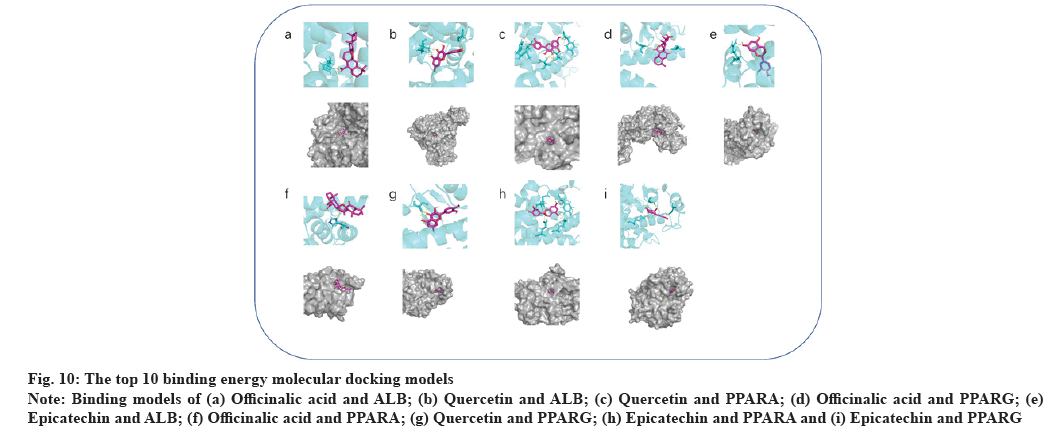
Molecular docking results were explained here.The protein PDB files (ALB: 4L8U, PPARA: 1I7G and PPARG: 1FM6) were obtained from the PDB database. In further analysis, they were preprocessed with ligands for molecular docking using AutoDock Vina and results were exported (fig. 9). The results were visualized using PyMOL (fig. 10). As a result, 24 receptor-ligand docking results were obtained, which depicted that, the docking model becomes more stable if the binding energy is less. Finally, we included 9 groups with the least binding energy. In the binding model of officinalic acid to ALB, the officinalic acid core can form hydrogen bonds with residues H242 (fig. 10a). In the binding model of quercetin to ALB, the quercetin core can form hydrogen bonds with residues K199, A210 (fig. 10b). In the binding model of quercetin to PPARA, the quercetin core can form hydrogen bonds with residues Y214, M320, T283, T279 and Y334 (fig. 10c). In the binding model of officinalic acid to PPARG, the officinalic acid core can form hydrogen bonds with residues L476, K457 (fig. 10d). In the binding model of epicatechin to ALB, the epicatechin core can form hydrogen bonds with residues R10 (fig. 10e). In the binding model of officinalic acid to ALB, the officinalic acid core can form hydrogen bonds with residues H411 (fig. 10f).
Fig. 10: The top 10 binding energy molecular docking models
Note: Binding models of (a) Officinalic acid and ALB; (b) Quercetin and ALB; (c) Quercetin and PPARA; (d) Officinalic acid and PPARG; (e)
Epicatechin and ALB; (f) Officinalic acid and PPARA; (g) Quercetin and PPARG; (h) Epicatechin and PPARA and (i) Epicatechin and PPARG
In the binding model of quercetin to PPARG, the quercetin core can form hydrogen bonds with residues S342 (fig. 10g). In the binding model of epicatechin to PPARA, the epicatechin core can form hydrogen bonds with residues M220, E282, E286 and Y334 (fig. 10h). In the binding model of epicatechin to PPARG, epicatechin core can form hydrogen bonds with residues E259, E291, S342 (fig. 10i).
Hyperlipidemia belongs to “lipid turbidity” in traditional Chinese medicine. Most medical practitioners believe that the method of removing stagnation and strengthening the spleen is to resolve phlegm which is crucial in intervening in hyperlipidemia[9]. Modern clinical studies have shown that almost patients with hyperlipidemia have disorders of lipid metabolism due to decreased activity of lipoprotein degrading enzymes and increased rate of lipoprotein synthesis, or abnormal lipoprotein structure and lipoprotein receptor structure, ultimately leading to excessive accumulation of lipids in the body[49]. Several studies have shown that hawthorn can regulate lipid metabolism, effectively reducing cholesterol and TG content in hyperlipidemic rats[10- 12]. Based on the network pharmacological screening, 3 active compounds (quercetin, epicatechin and officinalic acid) of hawthorn had a potential role in the intervention in hyperlipidemia. Quercetin is a flavonol compound with various biological activities. In the pharmacological effect study of quercetin, it was found that quercetin could significantly improve lipid metabolism in rats, which includes inhibiting the activity of HMG-CoA reductase, down-regulating the HMG-COA expression, reduction in TC, TGs and LDL levels[50]. In addition, quercetin also induces TC and HDL binding through activation of the PPARG/Liver X Receptor-α (LXRα) pathway to increase HDL-C selective lipid uptake and promotes cholesterol transportation to the liver, thereby reducing lipid accumulation[51,52]. Meanwhile, Cheng et al. showed that epicatechin could effectively reduce TC, TGs and LDL by regulating the Insulin- Induced Gene 1 protein (INSIG1)-Sterol Regulatory Element-Binding Protein (SREBP)-SREBP Cleavage-Activating Protein (SCAP) pathway and other genes related to lipid metabolism (including LXRα, Fas Cell Surface death receptor (FAS) and Sirtuin 1 (SIRT1)), which could increase the amount of HDL in serum, significantly improving lipid metabolism[53]. Currently, clinical experiments have confirmed that epicatechin can reduce the level of membrane lipid peroxidation, which can improve the membrane fluidity and lipid transport in patients with hypercholesterolemia and healthy donors[54]. It is expected that these active components will be used in potential directions for hawthorn intervention in hyperlipidemia.
After screening the related datasets, a total of 25 potential intervention targets were collected. Based on this target, a PPI network was constructed to evaluate its association and a C-T-D-P diagram was drawn to visually express the relevant targets and pathways of hawthorn components intervening in hyperlipidemia. Meanwhile, potential functional modules were identified through the MCODE plugin. The potential targets in GO BPs are mainly involved in regulating nutrient levels, hormone levels, growth, heart rate, cholesterol transport and other processes. The KEGG pathway focuses on PPAR signaling pathway, lipid and atherosclerosis, TGF-β signaling pathway, glucagon signaling pathway, insulin resistance, and cytokine receptor interactions. The enrichment analysis showed that the modules could be involved in cancer pathways, lipid binding, transport, steroid hormone bioregulation, TGF-β- SMAD signaling pathway regulation and redox regulation.
Impaired lipid metabolism is a significant factor in the development of hyperlipidemia and PPAR signaling was the most enriched KEGG pathway in the analysis of the potential target. Furthermore, the PPAR signaling is closely related to the lipid metabolism pathway. Therefore, we hypothesize that hawthorn intervenes in hyperlipidemia by modulating the PPAR pathway to improve lipid metabolism. The PPAR signaling pathway can be involved in regulating a variety of cellular functions. Nan et al. found that up-regulating the expression of Lipoprotein Lipase (LPL) through the PPAR signaling pathway and inhibiting the expression of Fatty Acid Translocase (FAT)/Cluster of Differentiation 36 (CD36) signaling pathway could effectively improve lipid metabolism in hyperlipidemic rats and correct the lipid metabolism and lipid deposition imbalances in the liver[55]. LPL, a critical enzyme involved in lipid metabolism, plays an essential role in lipoprotein metabolism and lipid partitioning[56]. Mutations in the human LPL gene cause various abnormalities in lipid metabolism and lead to familial hyperlipidemia[57]. CD36 is a membrane receptor known to play a role in lipid metabolism by promoting the uptake of longchain fatty acids[58]. Meanwhile, TGF-β-SMAD is an important signaling pathway related to the regulation of cardiovascular function. It plays an important role in BPs such as atherosclerosis, myocardial fibrosis, regulation of cardiomyocyte apoptosis and vascular remodeling[59]. TGF-β-SMAD is also closely related to lipid metabolism. Lu et al. found that calcitriol could inhibit the activation of Interleukin-1 beta (IL-1β) and Tumor Necrosis Factor alpha (TNF-α), effectively reducing TC, TGs and LDL-C through the TGF-β-SMAD signaling pathway, which can effectively improve lipid metabolism and atherosclerosis in hyperlipidemic rats[60].
Finally, PPI and disease association analysis was performed and the core targets (ALB, PPARA and PPARG) were identified. ALB has the highest degree of value, association and centrality, making it a primary target not just for PPI networks but also for possible functional modules. In addition, disease association analysis using VarElect showed that ALB was strongly associated with hyperlipidemia. It can be concluded that ALB is the main target of hawthorn to intervene in hyperlipidemia. ALB, the most abundant protein in human blood, plays a key role in lipid and fatty acid metabolism, binding and transport[61]. In addition, studies on congenital hypoalbuminemia have found that congenital hypoalbuminemia significantly affects lipid metabolism, leading to severe hypoalbuminemia and elevated levels of LDL-C, leading to the occurrence of hyperlipidemia[62]. It has also been noted that people suffering from severe anaplasmosis are often accompanied by severe hypercholesterolemia and elevated LDL. These aberrant metabolisms might be the product of a compensation mechanism for severe ALB deficit and they give evidence for a putative function of serum ALB in the regulation of lipoprotein metabolism[63]. Faingold et al. found that the ALBbound nitrosyl iron complex can inhibit oxidative stress-induced lipid peroxidation[64]. PPARA is a key regulator of lipid metabolism, which can regulate the β-oxidation of fatty acids and accelerates the bioavailability of lipids. A further study by Eurlings et al. verified that PPARA was identified as a gene that regulates intra and extracellular lipid metabolism, such as lipolysis and mitochondrial β-oxidation. A change in these factors, in turn, leads to decreased expression of Fatty Acid Binding Protein (FABP) and Fatty Acid Transport Protein (FATP). Interestingly, the absorption of Free Fatty Acids (FFA) requires the mediation of FABP and FATP, leading to blocked FFA entry into hepatocytes for β-oxidation. These changes could increase the TG synthesis from FFA and Very- Low-Density Lipoprotein (VLDL) production[65-67]. Meanwhile, experiments conducted by Stec et al. confirmed that PPARA mutant mice acquire obesity in a sexually dimorphic way, showing that PPARA plays an important role in the prevention of hyperlipidemia and fatty liver disease[68]. PPARG, a key pathway of the PPAR signaling pathway, has a positive role in regulating lipids[69]. Ruotolo et al. found that PPARG can significantly improve dyslipidemia by modulating insulin resistance[70]. Recently, an animal study suggests that PPARG can regulate adipocyte differentiation to suppress hyperlipidemia[71]. Further mechanism investigations have revealed that PPARG mutation and lipid metabolism related to PPARG are important in the progression of hyperlipidemia, which is one of the cores of the development of hyperlipidemia[72,73]. In conclusion, based on the results of network pharmacology and enrichment analysis, we predicted that the main active components of hawthorn to intervene in hyperlipidemia are epicatechin, officinalic acid and quercetin, which mainly intervene in hyperlipidemia through the PPAR signaling pathway.
Funding:
This work was funded by General Project of Sichuan Provincial Administration of Traditional Chinese Medicine (2021MS114).
Author’s contributions:
Xuejing Song, Hong Li and Chao Tang conceived, designed and planned the study. Hong Li, Yi Liang and Xuejing Song acquired and analysed the data. Hong Li and Jun Chen interpreted the results. Hong Li and Chao Tang drafted the manuscript. Yi Liang and Xuejing Song contributed to the critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
Conflicts of interest:
The authors declare that there is no conflict of interest.
References
- Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global burden of cardiovascular diseases and risk factors, 1990-2019: Update from the GBD 2019 study. J Am Coll Cardiol 2020;76(25):2982-3021.
[Crossref] [Google Scholar] [PubMed]
- Ralston J, Nugent R. Toward a broader response to cardiometabolic disease. Nat Med 2019;25(11):1644-6.
[Crossref] [Google Scholar] [PubMed]
- Malik S, Wong ND, Franklin SS, Kamath TV, L’Italien GJ, Pio JR, et al. Impact of the metabolic syndrome on mortality from coronary heart disease, cardiovascular disease and all causes in United States adults. Circulation 2004;110(10):1245-50.
[Crossref] [Google Scholar] [PubMed]
- Stewart J, McCallin T, Martinez J, Chacko S, Yusuf S. Hyperlipidemia. Pediatr Rev 2020;41(8):393-402.
[Crossref] [Google Scholar] [PubMed]
- Brown JC, Gerhardt TE, Kwon E. Risk factors for coronary artery disease. Treasure Island (FL): StatPearls Publishing LLC; 2022.
- Ibi D, Boot M, Dollé ME, Jukema JW, Rosendaal FR, Christodoulides C, et al. Apolipoprotein AV is a potential target for treating coronary artery disease: Evidence from genetic and metabolomic analyses. J Lipid Res 2022;63(5):1-9.
[Crossref] [Google Scholar] [PubMed]
- Nußbaumer B, Glechner A, Kaminski-Hartenthaler A, Mahlknecht P, Gartlehner G. Ezetimibe-statin combination therapy: Efficacy and safety as compared with statin monotherapy-a systematic review. Dtsch Arztebl Int 2016;113(26):445.
[Crossref] [Google Scholar] [PubMed]
- Alonso R, Cuevas A, Cafferata A. Diagnosis and management of statin intolerance. J Atheroscler Thromb 2019;26(3):207-215.
[Crossref] [Google Scholar] [PubMed]
- Qi XJ, Zhou RJ, Liu HP, Gao S, Xie YY, Hou JX, et al. Analysis on regularity of traditional Chinese patent medicine in treating hyperlipidemia. Zhongguo Zhong Yao Za Zhi 2019;44(19):4277-84.
[Crossref] [Google Scholar] [PubMed]
- Zeng L, Luo L, Xue Q, He Q, Chen X, Meng J, et al. LC-MS based plasma metabolomics study of the intervention effect of different polar parts of Hawthorn on hyperlipidemia rats. J Sep Sci 2021;44(5):963-72.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Liang R, Wang L, Yan R, Hou R, Gao S, et al. Effects of an aqueous extract of Crataegus pinnatifida Bge. var. major NE Br. fruit on experimental atherosclerosis in rats. J Ethnopharmacol 2013;148(2):563-9.
[Crossref] [Google Scholar] [PubMed]
- Shatoor AS, Al Humayed S, Alkhateeb MA, Shatoor KA, Aldera H, Alassiri M, et al. Crataegus aronia protects and reverses vascular inflammation in a high fat diet rat model by an antioxidant mechanism and modulating serum levels of oxidized low-density lipoprotein. Pharm Biol 2019;57(1):37-47.
[Crossref] [Google Scholar] [PubMed]
- Shao LI, Zhang B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin J Nat Med 2013;11(2):110-20.
[Crossref] [Google Scholar] [PubMed]
- Wu CW, Lu L, Liang SW, Chen C, Wang SM. Application of drug-target prediction technology in network pharmacology of traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi 2016;41(3):377-82.
[Crossref] [Google Scholar] [PubMed]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37:D885-90.
[Crossref] [Google Scholar] [PubMed]
- Li J, Yin Y, Zhang M, Cui J, Zhang Z, Zhang Z, et al. GsmPlot: A web server to visualize epigenome data in NCBI. BMC Bioinformatics 2020;21:1-7.
[Crossref] [Google Scholar] [PubMed]
- Gu S, Xue Y, Gao Y, Shen S, Zhang Y, Chen K, et al. Mechanisms of indigo naturalis on treating ulcerative colitis explored by GEO gene chips combined with network pharmacology and molecular docking. Sci Rep 2020;10(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 2019;47(D1):D976-82.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res 2016;44(1):D1202-13.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem 2001;44(12):1841-6.
[Crossref] [Google Scholar] [PubMed]
- Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem 2012;4(2):90-8.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(1):W357-64.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 2017;45(W1):W356-60.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Wang Z, Zhu R, Wang F, Cheng Y, Liu Y. Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2. J Vis Exp 2021 (175):e62528.
[Crossref] [Google Scholar] [PubMed]
- Hu K. Become competent in generating RNA-seq heat maps in one day for novices without prior R experience. Methods Mol Biol 2021:269-303.
[Crossref] [Google Scholar] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol 2013;2(10):1-6.
[Crossref] [Google Scholar] [PubMed]
- UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res 2021;49(1):D480-9.
[Crossref] [Google Scholar] [PubMed]
- Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics 2017;58(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46(1):1074-82.
[Crossref] [Google Scholar] [PubMed]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54(1):1-30.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48(1):D845-55.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res 2020;48(1):D1031-41.
[Crossref] [Google Scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Rabbani G, Baig MH, Ahmad K, Choi I. Protein-protein interactions and their role in various diseases and their prediction techniques. Curr Protein Pept Sci 2018;19(10):948-57.
[Crossref] [Google Scholar] [PubMed]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network analysis and visualization of proteomics data. J Proteome Res 2018;18(2):623-32.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
[Crossref] [Google Scholar] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4(1):1-27.
[Crossref] [Google Scholar] [PubMed]
- Stelzer G, Plaschkes I, Oz-Levi D, Alkelai A, Olender T, Zimmerman S, et al. VarElect: The phenotype-based variation prioritizer of the GeneCards Suite. BMC Genomics 2016;17(2):195-206.
[Crossref] [Google Scholar] [PubMed]
- Gene ontology consortium. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res 2019;47(1):D330-8.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1523.
[Crossref] [Google Scholar] [PubMed]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform 2011;3(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Goodsell DS, Zardecki C, di Costanzo L, Duarte JM, Hudson BP, Persikova I, et al. RCSB protein data bank: Enabling biomedical research and drug discovery. Protein Sci 2020;29(1):52-65.
[Crossref] [Google Scholar] [PubMed]
- Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ 2016;44(5):433-7.
[Crossref] [Google Scholar] [PubMed]
- Morris GM, Huey R, Olson AJ. Using AutoDock for ligand‐receptor docking. Curr Protoc Bioinformatics 2008;24(1):8-14.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One 2013;8(12):e83922.
[Crossref] [Google Scholar] [PubMed]
- Klop B, Elte JW, Cabezas MC. Dyslipidemia in obesity: Mechanisms and potential targets. Nutrients 2013;5(4):1218-40.
[Crossref] [Google Scholar] [PubMed]
- Khamis AA, Salama AF, Kenawy ME, Mohamed TM. Regulation of hepatic hydroxy methyl glutarate-CoA reductase for controlling hypercholesterolemia in rats. Biomed Pharmacother 2017;95:1242-50.
[Crossref] [Google Scholar] [PubMed]
- Cui Y, Hou P, Li F, Liu Q, Qin S, Zhou G, et al. Quercetin improves macrophage reverse cholesterol transport in apolipoprotein E-deficient mice fed a high-fat diet. Lipids Health Dis 2017;16:1-7.
[Crossref] [Google Scholar] [PubMed]
- Ren K, Jiang T, Zhao GJ. Quercetin induces the selective uptake of HDL-cholesterol via promoting SR-BI expression and the activation of the PPARγ/LXRα pathway. Food Funct 2018;9(1):624-35.
[Crossref] [Google Scholar] [PubMed]
- Cheng H, Xu N, Zhao W, Su J, Liang M, Xie Z, et al. (‐)‐Epicatechin regulates blood lipids and attenuates hepatic steatosis in rats fed high‐fat diet. Mol Nutr Food Res 2017;61(11):1700303.
[Crossref] [Google Scholar] [PubMed]
- Franiak-Pietryga I, Koter-Michalak M, Broncel M, Duchnowicz P, Chojnowska-Jezierska J. Anti-inflammatory and hypolipemic effects in vitro of simvastatin comparing to epicatechin in patients with type-2 hypercholesterolemia. Food Chem Toxicol 2009;47(2):393-7.
[Crossref] [Google Scholar] [PubMed]
- Hu N, Chen C, Wang J, Huang J, Yao D, Li C. Atorvastatin ester regulates lipid metabolism in hyperlipidemia ratsvia the PPAR-signaling pathway and HMGCR expression in the liver. Int J Mol Sci 2021;22(20):11107.
[Crossref] [Google Scholar] [PubMed]
- Wu SA, Kersten S, Qi L. Lipoprotein lipase and its regulators: An unfolding story. Trends Endocrinol Metab 2021;32(1):48-61.
[Crossref] [Google Scholar] [PubMed]
- Dron JS, Hegele RA. Genetics of hypertriglyceridemia. Front Endocrinol 2020;11:455.
[Crossref] [Google Scholar] [PubMed]
- Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr 2014;34:281-303.
[Crossref] [Google Scholar] [PubMed]
- Xu HM, Sui FH, Sun MH, Guo GL. Retracted: Downregulated microRNA‐224 aggravates vulnerable atherosclerotic plaques and vascular remodeling in acute coronary syndrome through activation of the TGF‐β/Smad pathway. J Cell Physiol 2019;234(3):2537-51.
[Crossref] [Google Scholar] [PubMed]
- Lu C, Yin Y, Cui Y, Wang L, Bai Y, Li J, et al. 1, 25 (OH) 2D3 improves blood lipid metabolism, liver function and atherosclerosis by constraining the TGF-beta/Smad signaling pathway in rats with hyperlipidemia. Cell Cycle 2019;18(22):3111-24.
[Crossref] [Google Scholar] [PubMed]
- Nishi K, Yamasaki K, Otagiri M. Serum albumin, lipid and drug binding. Subcell Biochem 2020;94:383-97.
[Crossref] [Google Scholar] [PubMed]
- Minchiotti L, Caridi G, Campagnoli M, Lugani F, Galliano M, Kragh-Hansen U. Diagnosis, phenotype and molecular genetics of congenital analbuminemia. Front Genet 2019;10:336.
[Crossref] [Google Scholar] [PubMed]
- Crook MA. Analbuminaemia: Clinical features and associated hypercholesterolaemia. Ann Clin Biochem 2016;53(5):525-6.
[Crossref] [Google Scholar] [PubMed]
- Faingold II, Poletaeva DA, Soldatova YV, Smolina AV, Pokidova OV, Kulikov AV, et al. Effects of albumin-bound nitrosyl iron complex with thiosulfate ligands on lipid peroxidation and activities of mitochondrial enzymes in vitro. Nitric Oxide 2021;117:46-52.
[Crossref] [Google Scholar] [PubMed]
- Eurlings PM, van der Kallen CJ, Geurts JM, Flavell DM, de Bruin TW. Identification of the PPARA locus on chromosome 22q13. 3 as a modifier gene in familial combined hyperlipidemia. Mol Genet Metab 2002;77(4):274-81.
[Crossref] [Google Scholar] [PubMed]
- Canbay A, Bechmann L, Gerken G. Lipid metabolism in the liver. Z Gastroenterol 2007;45(01):35-41.
[Crossref] [Google Scholar] [PubMed]
- Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of PPAR-α in lipid metabolism and inflammation. Endocr Rev 2018;39(5):760-802.
[Crossref] [Google Scholar] [PubMed]
- Stec DE, Gordon DM, Hipp JA, Hong S, Mitchell ZL, Franco NR, et al. Loss of hepatic PPARα promotes inflammation and serum hyperlipidemia in diet-induced obesity. Am J Physiol Regul Integr Comp Physiol 2019;317(5):R733-45.
[Crossref] [Google Scholar] [PubMed]
- Cataldi S, Costa V, Ciccodicola A, Aprile M. PPARγ and diabetes: Beyond the genome and towards personalized medicine. Curr Diab Rep 2021;21:1-5.
[Crossref] [Google Scholar] [PubMed]
- Ruotolo G, Howard BV. Dyslipidemia of the metabolic syndrome. Curr Cardiol Rep 2002;4(6):494-500.
[Crossref] [Google Scholar] [PubMed]
- Zhang X, Chen X, Qi T, Kong Q, Cheng H, Cao X, et al. HSPA12A is required for adipocyte differentiation and diet-induced obesity through a positive feedback regulation with PPARγ. Cell Death Differ 2019;26(11):2253-67.
[Crossref] [Google Scholar] [PubMed]
- Padova G, Prudente S, Vinciguerra F, Sudano D, Baratta R, Bellacchio E, et al. The novel loss of function Ile354Val mutation in PPARG causes familial partial lipodystrophy. Acta Diabetol 2020;57:589-96.
[Crossref] [Google Scholar] [PubMed]
- Glodowski M, Christen S, Saxon DR, Hegele RA, Eckel RH. Novel PPARG mutation in multiple family members with chylomicronemia. J Clin Lipidol 2021;15(3):431-4.
[Crossref] [Google Scholar] [PubMed]
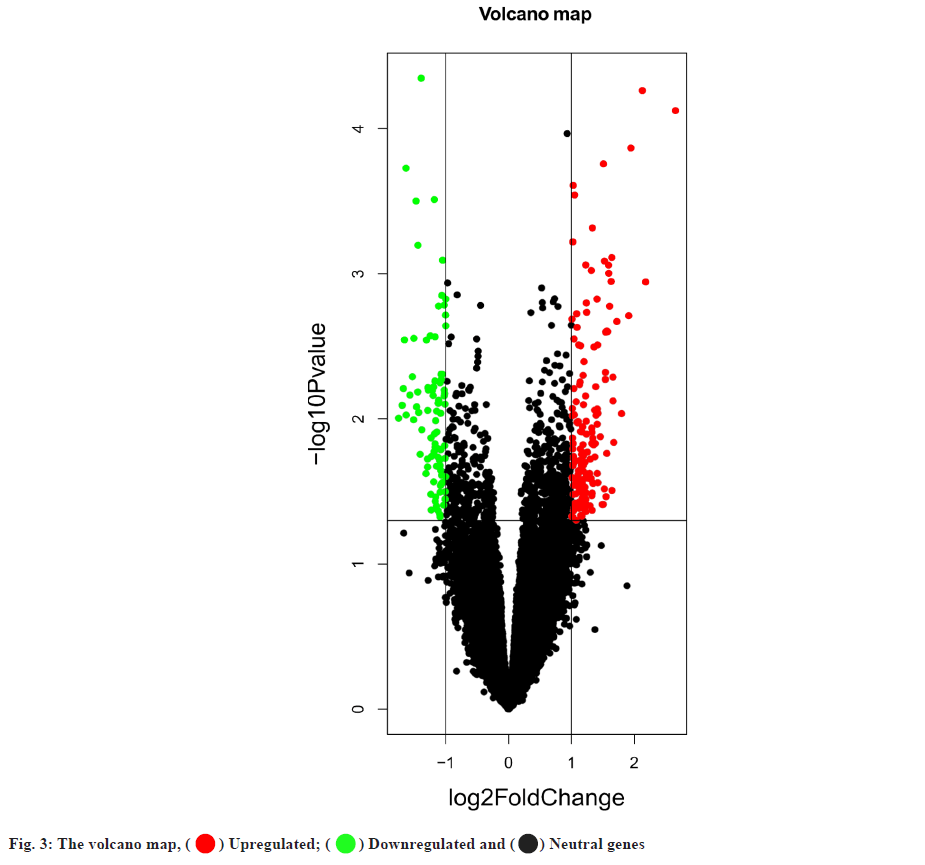
 ) Upregulated; (
) Upregulated; (  ) Downregulated and (
) Downregulated and ( ) Neutral genes
) Neutral genes