- *Corresponding Author:
- V. S. Kawade
CSIR- Unit for Research and Development of Information Products, Tapovan, NCL Estate, Pashan Road, Pune-411 008, India
E-mail: vikram.kawade@urdip.res.in
| Date of Submission | 09 February 2017 |
| Date of Revision | 27 November 2017 |
| Date of Acceptance | 23 June 2018 |
| Indian J Pharm Sci 2018;80(4):702-708 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cell signalling mechanism plays a vital role in cell functioning. Imbalance and disregulation between these signals, such as phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin signalling pathway; may lead to uncontrolled cell proliferation. Generation of drug resistance is the hurdle in current cancer treatment. Designing an effective combination therapy for inhibition of two or more proteins of a given pathway might help to overcome the drug resistance and side effect related issues in cancer treatment. In this regard, application of computational tools firstly to predict newer combinations against phosphatidylinositol-3-kinase, protein kinase b and mammalian target of rapamycin involved in a single pathway have been proposed. The results obtained using the computational tools were shortlisted based on Glide score and binding interactions of the drugs to the receptors of the pathway. One of the predicted combinations was further subjected to biological evaluation using the Western blot assay. The experimental results revealed synergistic effects that supported the predictions. The study also provided insights for the application and development of computational tools to predict newer combinations in a given network pharmacology.
Keywords
PI3K/Akt/mTOR pathway, anticancer activity, molecular docking, combination therapy, synergistic effect
According to WHO’s global health repository data, cancer is one of the major causes of mortality[1]. In 2012, there were 14.1 million of new cancer cases diagnosed in world and around 8.2 million deaths were caused by cancer alone[2]. The recent advances in targeted immunotherapies and cytotoxic chemotherapy have led to an improved survival rate[3]. However till date the treatment of cancer is a major challenge for the reasons associated with the complexity of cell signalling pathways, heterogeneity of tumor cell, development of resistance to current therapy[4] and inevitable side effects[5]. Hence a need for the development of newer and more effective therapies to treat cancer and reduce side effects continues to exist.
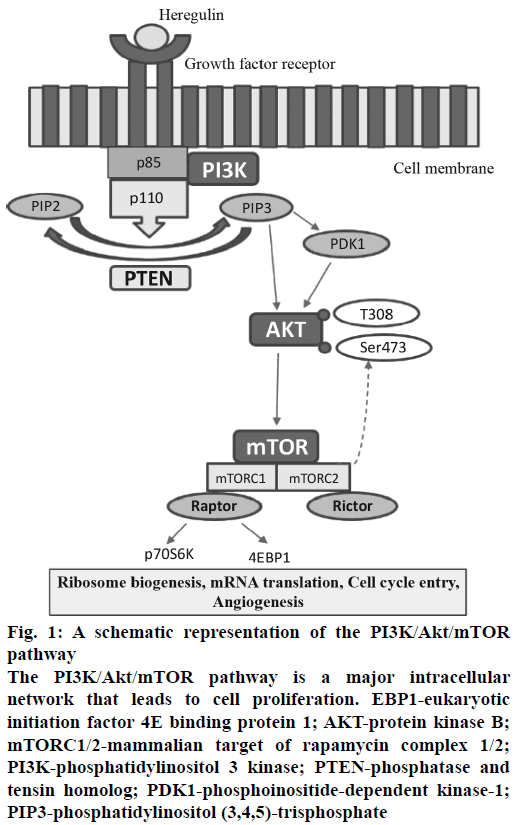
In cancer, during the process of tumorigenesis, alterations occur in cell signalling pathway and cellular machinery. It induces cell to acquire capacity to proliferate independently and invade other tissues and metastasize to distant locations. This intricacy of cellular signalling networks has major implications for the understanding of tumour cell behaviour and also helpful to design a novel cancer therapy[6]. The phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) is a major signalling pathway, which involved in cell cycle regulation as well as progression[7,8].
PI3K is a primary signalling target from PI3K/Akt/ mTOR pathway and involved in diverse functions. The PI3Ks phosphorylate phosphatidylinositol-4,5- bisphosphate (PIP2) to PIP3, which leads to the phosphorylation of Akt, which has an impact on cancer cell survival, growth and cell cycle[9]. Phosphatase and tensin homolog (PTEN) is a tumor suppressor and PTEN has the opposite action, which dephosphorylates PIP3 into PIP2[10]. Akt-independent activation of the PI3K pathway and that PIK3CA mutations can lead to cancer[11,12]. The protein mTOR is a serine/threonine protein kinase and has a downstream substrate of PI3K and Akt. mTOR has two different complexes mTORC1 and mTORC2, both of which have a different mechanism of action. mTORC1 is better characterized than the mTORC2, and it has also been observed that mTORC2 is also inhibited by mTORC1 inhibitors at specific doses and it affects cell metabolism and leads to cancer cell growth[13,14].
The pathway specific target Akt catalyses the activation of other targets, including mTOR and initiates cell proliferation by the protein synthesis machinery[15,16]. Another approach is dual inhibition of PI3K/mTOR using suitable PI3K/mTOR inhibitors that target the common p110 subunit. The advantage of dual PI3K/mTOR inhibitors is the complete inhibition of the PI3K/Akt/mTOR signalling pathway (Figure 1), which improve the clinical efficacy[15]. Several studies have demonstrated the deregulation of PI3K/Akt/ mTOR pathway which is linked to around 12 types of human cancers namely: breast cancer, ovarian cancer, endometrial cancer[17], prostate cancer, lung cancer, gastric cancer, melanoma, pancreatic cancer[18], glioblastoma multiform and acute myelogenous leukaemia.
Figure 1: A schematic representation of the PI3K/Akt/mTOR pathway
The PI3K/Akt/mTOR pathway is a major intracellular network that leads to cell proliferation. EBP1-eukaryotic initiation factor 4E binding protein 1; AKT-protein kinase B; mTORC1/2-mammalian target of rapamycin complex 1/2; PI3K-phosphatidylinositol 3 kinase; PTEN-phosphatase and tensin homolog; PDK1-phosphoinositide-dependent kinase-1; PIP3-phosphatidylinositol (3,4,5)-trisphosphate
Combination therapy is of recent focus for treating cancer[19] and other multifaceted disease condition such as infectious diseases[20] and acquired immunodeficiency syndrome (AIDS)[21]. Clinically approved combination therapies are mainly used to increase the therapeutic efficacy, to minimize the dose-dependent side effects and prevent development of drug resistance. The evaluation of combination therapies are mainly carried out using the traditional experimental drug discovery approaches and recently include pathway or network-based mathematical approach and integrated approach[22].
Conventional laboratory-based cancer research revolves around in vitro assays and in vivo trials. These are expensive and time consuming methods[23]. Therefore, pharmaceutical industries incorporate computer-aided drug design tools to mainly identify the hits and optimize them to the leads, which are then subjected to in vitro screening against the receptors for the therapeutic area under consideration[23,24].
In this paper, we illustrate the application of the known computational drug discovery tools mainly to predict which of the known anticancer drugs can modulate the activity of two receptors of the PI3K/Akt/mTOR pathway in order to yield a combination therapy. The predicted combinations were further subjected to in vitro evaluation to confirm the predicted biological activity of the combination.
Materials and Methods
Virtual library generation
A diverse set of 1803 FDA-approved drugs were collated to generate a virtual library of drugs. The approved drugs were considered, as the drugs have been already evaluated for their safety concerns. Additionally the approved drugs also have the optimal ADME parameters.
Protein and ligand preparation
The co-crystal structure of the proteins, PI3K, Akt and mTOR (PDB ID: 4JPS, 3MV5 and 3QAR) were used for carrying out the molecular docking studies. The protein structures were downloaded from the RCSB protein data bank[25] and were prepared using the Protein Preparation Wizard in Maestro version 10.2, Schrödinger, LLC, New York, 2015. Each of the proteins under consideration was initially optimized from its raw state with OPLS 2005 force field using the Protein Preparation Wizard. The water molecules beyond the 5 Å distances from the native ligands were removed. This step was followed by bond order assignment, hydrogen atom addition, determination of atom type and correction of the charge and protonation states.
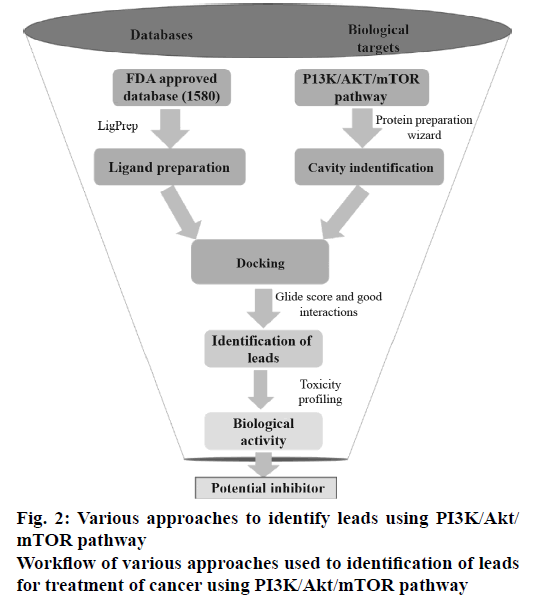
All the FDA-approved drugs were downloaded from the Approved drugs (Drug Bank Release Version 5.0.1). The tautomers, ionization states, ring conformations and stereochemistry for the drugs under consideration were calculated using default parameters. The OPLS_2005 force field was used for energy minimization of all legends using LigPrep v2.5 module (Schrödinger LLC, New York, 2015, version 10.2). These prepared ligands were used for molecular docking study mainly to identify which of the drugs bind to the active site of PI3K, Akt and mTOR proteins. The work flow diagram (Figure 2) depicts the steps, which were carried out to determine the best combinations of the known anticancer drugs for the PI3K/Akt/mTOR pathway under consideration.
Molecular docking studies
Molecular docking module of the proprietary software used in the present analysis incorporates an automated computer algorithm to determine the interaction between a ligand with various amino acids present in the active site of the protein under analysis[26,27]. The most promising ligands that can primarily bind to the amino acids in the active site of the proteins under consideration were identified using the molecular docking tool using a structure-based drug design approach[28]. The molecular docking step also helped to identify the orientation of the ligand towards the protein, based on which a docking score for each of the protein-ligand interaction was generated[29,30]. It also predicted the free molecular binding energy of ligandreceptor complex as well as binding affinity of ligand with the amino acid residues of the protein[31].
Each of the 1805 FDA-approved drugs was subjected to molecular docking against the three proteins under consideration. The Glide module of the proprietary software used in the current analysis provides three different levels of docking precision; high throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP).
In this current study, the ligands from the SP docking level were shortlisted based on the docking score and were further subjected to docking into each of the three proteins under consideration by XP docking protocol. The final GlideScore was ranked and the binding pattern of each drug onto the active pocket of the proteins was manually identified. The top ranked drugs based on the docking score for each of the proteins are enlisted in Table 1. The drugs which had a greater or comparable GlideScore to that of the reference ligand were selected as the most promising combination.
| Sr. no. | PI3K | Akt | mTOR | |||
|---|---|---|---|---|---|---|
| Compound name | GlideScore | Compound name | GlideScore | Compound name |
GlideScore | |
| 1 | Wortmannin | –10.220 | Mitoxantrone | –9.455 | 17β,18-Norestradiol | –9.854 |
| 2 | LY294002 | –10.038 | Vemurafenib | –9.178 | Riboflavin | –9.522 |
| 3 | Quinostatin | –10.025 | MK-2206 | –9.211 | Rapamycin | –9.319 |
| 4 | Sirolimus | –9.921 | Theophylline | –9.150 | 5’AMP | –9.238 |
| 5 | Pilaralisib | –9.590 | ATP | –9.039 | Everolimus | –9.124 |
| Reference ligand | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro- 2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol- 2-yl}pyrrolidine-1,2-dicarboxamide | –9.133 | 1-(4-amino-6-methyl-1,3,5-triazin-2-yl)-N-(1H-pyrazol-3-yl)-1H-benzimidazol-2-amine | –8.975 | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin- 4-yl)pyrrolidin-3-amine | –8.586 |
Table 1: Top Five Drugs Based on the Glidescore for PI3k, Akt and mTOR Receptors
Cell lysates preparation and western blot analysis
Based on the commercial availability of the compounds from the top 5 combinations as predicted by the molecular docking method, the combination vemurafenib and riboflavin were considered for further evaluation using in vitro assay models. Human malignant melanoma A375 cells were treated with vemurafenib and riboflavin at 1 and 50 μM concentrations, respectively for 48 h time period. Cells were collected by centrifugation at 400×g at 4°, washed with phosphate buffered saline (PBS) twice and cell pellet collected were lysed with cold cell lysis buffer (RIPA, Sigma) having protease inhibitors NaF (50 mM), sodium orthovanadate (NaVO4; 0.5 mM), phenylmethylsulfonyl fluoride (PMSF; 2 mM) and protease inhibitor cocktail (1 % v/v) for 40 min and centrifuged at 12 000 g for 15 min at 4°. After that, supernatant was collected and proceed for Western blot analysis. Protein measurement was done by using Bio-Rad protein assay kit and equal amount of protein lysates (50-70 mg) were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis. Proteins were electro-transferred to PVDF membrane for 1.40 h at 4° at 100 V using Bio-Rad electrode assembly. After transfer of proteins, membrane was blocked by incubation with 5 % non-fat milk in Trisbuffered saline containing 0.1 % Tween-20 (TBST) for 50 min at room temperature to prevent any nonspecific binding of desired antibody. Blots were incubated with respective primary antibodies for 4 h at room temperature. They are washed two times with TBST and were incubated with horseradish peroxidase conjugated secondary antibodies for another 1 h. Membranes were washed three times with 15 min interval with TBST buffer and signals were detected using ECL plus chemiluminescence's kit on X-ray film.
Results and Discussion
Recent drug discovery approaches for combating cancer consist of combination therapy mainly to reduce the side effects and drug resistance of a single drug[32]. Amongst the pathways which play a role in the tumor initiation and progression of cancer[33,34], the PI3K/ Akt/mTOR is one of the major signalling pathway associated with the proliferation of tumor in many human cancer[35]. The proteins of this pathway are hence considered as one of the important therapeutic targets for the discovery of anticancer drugs.
The role of molecular modelling and cheminformatics in the development, research and drug design has been proved to be significant in terms of developing alternative methodologies. The computational techniques such as pharmacophore-based screening[36,37], e-pharmacophore-based screening[38,39] have been successfully incorporated into the drug discovery process to firstly identify the hits followed by the optimization step to discover the leads. However none of the reported studies discuss the application of computational techniques leading to the prediction and discovery of drugs, which can be used in combination therapy.
In the current study, known molecular docking tools were evaluated to identify drugs, which showed a potential binding interaction towards multiple proteins involved in the PI3K/Akt/mTOR pathway. For the study, the FDA-approved drugs were considered as the data set to evaluate the binding interactions towards each of the protein in the pathway under consideration. The molecular docking studies resulted into around 19 drugs binding to PI3K protein, 31 to Akt while 27 to mTOR protein. Shortlisting the most potent drugs for the pathway under consideration was done using multiple filtration approaches such as, molecular docking-based scoring, ADME property-based screening and major toxicity endpoints. The top hits obtained by molecular docking were further docked into the active binding sites of the protein using XP protocol of the computational tool.
The drugs which were well reported in the prior art with respect to binding interaction to any of the three proteins under consideration were excluded from further in vitro evaluation. This step ensured the consideration of only those drugs, which were reported as anticancer drugs, but were not yet reported for modulating the enzymes in the PI3K/Akt/mTOR pathway. The molecular docking studies of these drugs using the XP docking protocol resulted into top five combinations, which showed possible molecular interactions with the amino acids in the active site of the proteins under consideration.
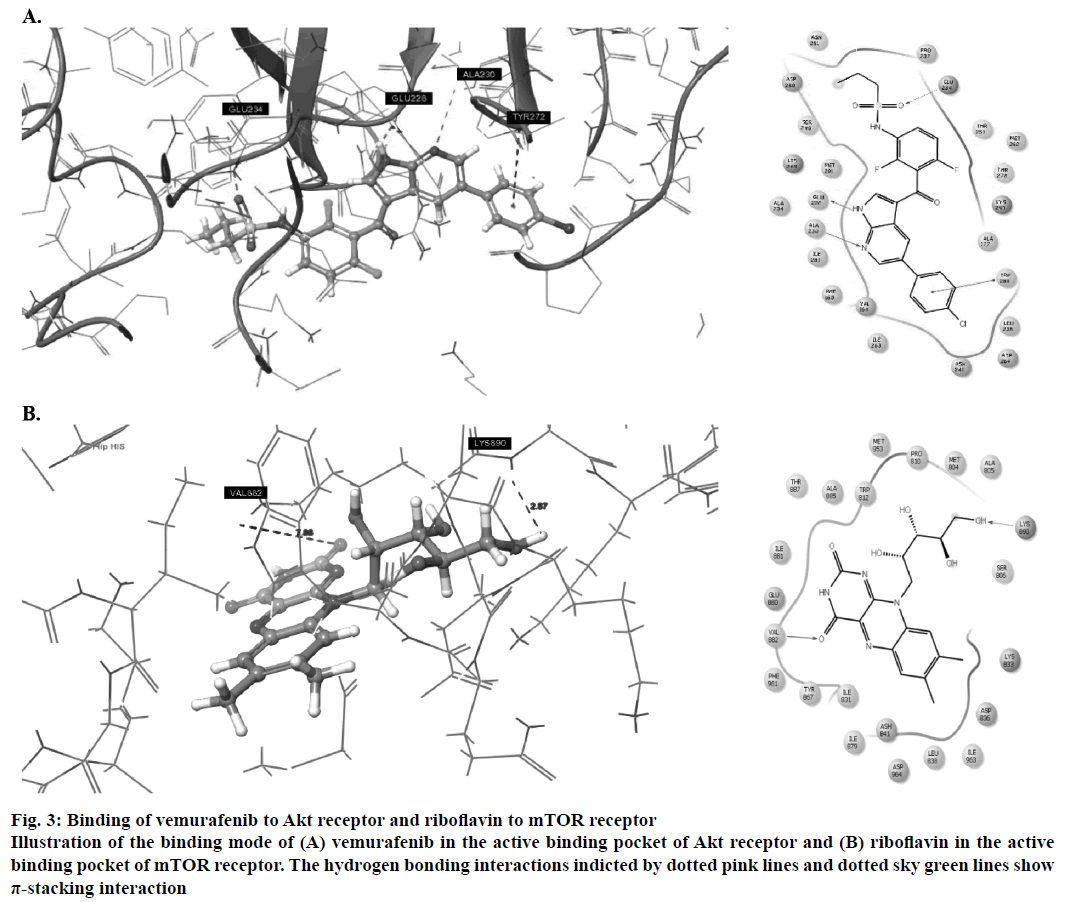
Amongst the predicted combinations, vemurafenib demonstrated good interactions with the binding domain of Akt and revealed three hydrogen bond interactions with the amino acid residues ALA230 (O-H…N: 2.35Å), GLU226 (N…N-H: 1.58Å) and GLU234 (N-H…O: 2.09Å) and one π-stacking interaction with TYR272 at a distance of 4.76Å (Figure 3A). Other amino acid residues such as VAL164, ALA177, ILU800, THR291 and MET281 showed good hydrophobic interactions. Riboflavin showed good GlideScore as well as docking interactions with mTOR receptor. It showed two hydrogen bonding interactions (Figure 3B) with active site amino acid residues VAL882 and LYS890 at 1.82 Å and 2.07 Å, respectively.
Figure 3: Binding of vemurafenib to Akt receptor and riboflavin to mTOR receptor
Illustration of the binding mode of (A) vemurafenib in the active binding pocket of Akt receptor and (B) riboflavin in the active binding pocket of mTOR receptor. The hydrogen bonding interactions indicted by dotted pink lines and dotted sky green lines show π-stacking interaction
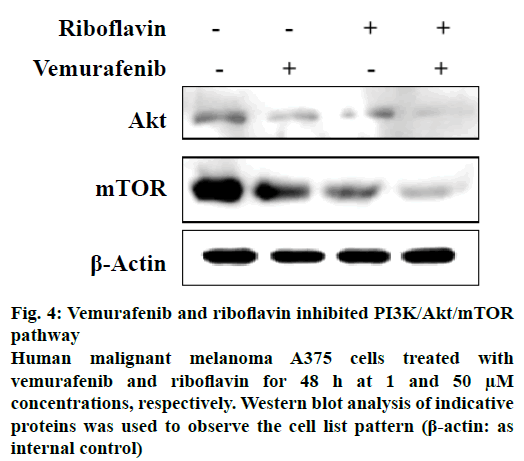
Other drugs which showed interactions to two or more proteins of the pathway included wotmannin and MK-2206, LY-294002, mitoxantrone and rapamycin. Based on the predicted binding interactions, commercial availability, cost parameters and the availability of the in vitro assay under consideration the authors chose to experimentally evaluate the effect of the combination of vemurafenib and riboflavin from the predicted result set. The anticancer activity of vemurafenib and riboflavin was tested against PI3K/Akt/mTOR pathway using an enzyme inhibition assay. The selected combination showed good PI3K/Akt/mTOR inhibitory activity (Figure 4). Combination of vemurafenib (1 μM) and riboflavin (50 μM) showed high biological activity in human malignant melanoma A375 cells at 48 h time period. It is also well-known that PI3K/Akt/mTOR pathway plays an important role in the cell proliferation and survival of human malignant melanoma A375 cells. The in vitro experimental results thus support that the predicted combination of vemurafenib and riboflavin are effective in suppressing the expression of tumours by targeting the Akt and mTOR proteins of the pathway under consideration. The experimental evaluation studies also reveal that vemurafenib was effective in regulating Akt at lower concentrations (1 μM), however the effect of the second therapeutic compound riboflavin was at higher concentrations. This provides further scope for research in terms of optimising the predicted combination.
Figure 4: Vemurafenib and riboflavin inhibited PI3K/Akt/mTOR pathway
Human malignant melanoma A375 cells treated with vemurafenib and riboflavin for 48 h at 1 and 50 μM concentrations, respectively. Western blot analysis of indicative proteins was used to observe the cell list pattern (β-actin: as internal control)
The literature survey for the PI3K/Akt/mTOR pathway and its role in proliferating tumor in human cancers lead to the finding that the pathway and its functioning play an important role in the development and progression of skin, breast, ovary, endometrial, prostate, lung, gastric, melanoma, and acute myelogenous leukaemia. Hence as the predicted combinations has provided key insights about their biological activity in modulating two of the proteins of the pathway, the findings can be further utilized to design and optimise dosage of each of the identified drugs for a combination therapy.
The present work thus illustrated the application of the well-known computational drug discovery tools as an alternative method to identify potential new combinations for a therapeutic area under consideration. The scientific literature survey along with experimental evaluation using an enzyme assay supported the predictions obtained using the computational methods and provided key insights to evaluate new combination of drugs by targeting two or more proteins forming a network pharmacology.
Acknowledgements
Authors are thankful to the CSIR-Unit for Research and Development of Information Products, Pune for providing the required financial and technical support for carrying out the analysis.
Conflicts of interest
There are no conflicts of interest.
References
- Global Health Observatory data repository. World Health Organization: WHO. Available from: http://apps.who.int/gho/data/node.main.CODWORLD?lang=en.
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86.
- Urruticoechea A, Alemany R, Balart J, Villanueva A, Vinals F, Capella G. Recent advances in cancer therapy: an overview. Curr Pharm Des 2010;16:3-10.
- Gottesman MM, Lavi O, Hall MD, Gillet JP. Toward a better understanding of the complexity of cancer drug resistance. Annu Rev Pharmacol Toxicol 2016;56:85-102.
- Gerber DE. Targeted therapies: a new generation of cancer treatments. Am Fam Physician 2008;77:311-9.
- Martin GS. Cell signalling and cancer. Cancer cell 2003;4:167-74.
- Polivka J Jr, Janku F. Molecular targets for cancer therapy in the PI3K/Akt/mTOR pathway. Pharmacol Ther 2014;142:164-75.
- Owonikoko TK, Khuri FR. Targeting the PI3K/Akt/mTOR pathway: biomarkers of success and tribulation. Am Soc Clin Oncol Educ Book 2013;2013:395-401.
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110 alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA 2008;105:2652-7.
- Maehama T, Dixon JE. The tumour suppressor, PTEN/MMOLAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 1998;273:13375-8.
- Zhang W, Haines BB, Efferson C, Zhu J, Ware C, Kunii K, et al. Evidence of mTOR activation by an Akt-independent mechanism provides support for the combined treatment of PTEN-deficient prostate tumours with mTOR and AKT inhibitors. Transl Oncol 2012;5:422-9.
- Bruhn MA, Pearson RB, Hannan RD, Sheppard KE. Akt-independent PI3-K signalling in cancer - emerging role for SGK3. Cancer Manag Res 2013;5:281-92.
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006;22:159-68.
- Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest 2011;121:1231-41.
- Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-Akt-mTOR signalling network in cancer. Chin J Cancer 2013;32:253-65.
- Manning BD, Cantley LC. Akt/PKB signalling: navigating downstream. Cell 2007;129:1261-74.
- Janku F, Wheler J, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/Akt/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol 2012;30:777-82.
- Wong MH, Xue A, Baxter RC, Pavlakis N, Smith RC. Upstream and downstream co-inhibition of mitogen-activated protein kinase and PI3K/Akt/mTOR pathways in pancreatic ductal adenocarcinoma. Neoplasia 2016;18:425-35.
- Li F, Zhao C, Wang L. Molecular‐targeted agent’s combination therapy for cancer: Developments and potentials. Int J Cancer 2014;134:1257-69.
- Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 2013;31:177-84.
- Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, et al. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 2016;22:839-50.
- Bulusu KC, Guha R, Mason DJ, Lewis RP, Muratov E, Motamedi YK, et al. Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives. Drug Discov Today 2016;21:225-38.
- Trisilowati, Mallet DG. In silico experimental modelling of cancer treatment. ISRN Oncol 2012;2012:828701.
- Das B, Samanta S. Design, docking studies of chalcone peptide hybrid compound as anti-inflammolatory and anti-arthritic agent. Int J Pharm Res 2015;7:59.
- A Structural View of Biology. RCSB PDB (Protein Data Bank). Available from: http://www.rcsb.org/pdb/home/home.do.
- Koska J, Spassov VZ, Maynard AJ, Yan L, Austin N, Flook PK, et al. Fully automated molecular mechanics based induced fit protein-ligand docking method. J Chem Inf Model 2008;48:1965-73.
- Yusuf D, Davis AM, Kleywegt GJ, Schmitt S. An alternative method for the evaluation of docking performance: RSR vs RMSD. J Chem Inf Model 2008;48:1411-22.
- Scior T, Bender A, Tresadern G, Medina-Franco JL, Martínez-Mayorga K, Langer T, et al. Recognizing pitfalls in virtual screening: A critical review. J Chem Inf Model 2012;52(4):867-81.
- Onodera K, Satou K, Hirota H. Evaluations of molecular docking programs for virtual screening. J Chem Inf Model 2007;47(4):1609-18.
- Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of several molecular docking programs: Pose prediction and virtual screening accuracy. J Chem Inf Model 2009;49(6):1455-74.
- Krovat EM, Steindl T, Langer T. Recent advances in docking and scoring. Curr Comput Aided Drug Des 2005;1:93-102.
- Fan W, Johnson KR, Miller MC 3rd. In vitro evaluation of combination chemotherapy against human tumor cells (Review). Oncol Rep 1998;5(5):1035-42.
- McAuliffe PF, Meric-Bernstam F, Mills GB, Gonzalez-Angulo AM. Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin Breast Cancer 2010;10:S59-65.
- Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol 2001;75(9):4283-96.
- Wu J, Chen C, Zhao KN. Phosphatidylinositol 3-kinase signaling as a therapeutic target for cervical cancer. Curr Cancer Drug Targets 2013;13(2):143-56.
- Yang SY. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today 2010;15(11-12):444-50.
- Chen Z, Tian G, Wang Z, Jiang H, Shen J, Zhu W. Multiple pharmacophore models combined with molecular docking: a reliable way for efficiently identifying novel PDE4 inhibitors with high structural diversity. J Chem Inf Model 2010;50:615-625.
- Muthusamy K, Singh KhD, Chinnasamy S, Nagamani S, Krishnasamy G, Thiyagarajan C, et al. High throughput virtual screening and E-pharmacophore filtering in the discovery of new BACE-1 inhibitors. Interdiscip Sci 2013;5(2):119-26.
- Veeramachaneni GK, Raj KK, Chalasani LM, Bondili JS, Talluri VR. High-throughput virtual screening with e-pharmacophore and molecular simulations study in the designing of pancreatic lipase inhibitors. Drug Des Devel Ther 2015;9:4397-412.