- *Corresponding Author:
- Huilin Zheng
Department of Biological and Chemical Engineering, Zhejiang University, Yiwu 322000, China
E-mail: zhenghuilin@zust.edu.cn
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “54-66” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In this study, we investigated the gene expression changes of liver sinusoidal endothelial cell and kupffer cell in healthy and cirrhotic states of the liver using single-cell ribonucleic acid sequencing data from more than 60 000 hepatic cells from five human healthy liver samples and five human cirrhotic liver samples, and further explored the roles of liver sinusoidal endothelial cell and kupffer cell in cirrhotic livers. We constructed the relevant signaling pathways for the interaction between liver sinusoidal endothelial cell and kupffer cell in healthy and cirrhotic livers based on the CellChat algorithm and revealed the possible pathogenic mechanism of such interactions in cirrhosis. Results showed that the cell numbers of both liver sinusoidal endothelial cell and kupffer cell were significantly reduced in cirrhotic livers compared to healthy livers. Genes associated with antigen processing and presentation and immune response (CTSL, CTSB, LGMN, HSPA1A, JUN, and CLEC1B, etc.) were down-regulated in liver sinusoidal endothelial cell from cirrhotic livers, suggesting that the immune function of liver sinusoidal endothelial cell from cirrhotic livers was impaired; genes associated with cellular senescence, apoptosis, programmed necrosis and tumor necrosis factor signaling pathway (CTSH, EDN1, IL1R1, CXCL2 and IGFBP3, etc.) were up-regulated in liver sinusoidal endothelial cell from cirrhotic livers, suggesting that liver sinusoidal endothelial cell was impaired, which resulted in a significant reduction in its cellular population. This study provides a new perspective for a deeper understanding of the roles and interactions between liver sinusoidal endothelial cell and kupffer cell in cirrhosis, and offers potential targets for the development of new therapeutic approaches.
Keywords
RNA sequencing, cirrhosis, liver sinusoidal endothelial cell, kupffer cell, hypertension
Cirrhosis is a serious chronic liver disease characterized by the replacement of normal liver tissue by extensive fibrotic tissue and liver nodules, leading to severe impairment of liver function[1-9]. Cirrhosis is one of the major causes of death worldwide, with approximately 2 million deaths per year due to cirrhosis and its complications, such as portal hypertension, liver ascites and liver cancer[2]. Cirrhosis is triggered by a variety of causes, including viral liver disease, alcoholic liver disease, non-alcoholic fatty liver disease and autoimmune liver disease[3].
Although the pathogenesis of cirrhosis is not fully understood, it is known that Liver Sinusoidal Endothelial Cells (LSEC) and liver macrophages play an important role in the liver tissue microenvironment[4]. LSEC are the largest population of endothelial cells in the liver, they form the inner layer of the liver sinusoids and are in direct contact with platelets, leukocytes and erythrocytes in the blood and are involved in blood flow regulation, immunomodulation, angiogenesis and liver regeneration[5]. In cirrhosis, the structure and function of LSEC are altered, leading to increased vascular permeability, which may accelerate inflammatory cell infiltration and liver fibrosis[6,7]. Kupffer Cells (KC) are an intrinsic macrophage population in the liver, predominantly in the liver sinusoids and in close proximity to LSEC[8]. KC has the function of phagocytosis and removal of pathogens from the blood, secretion of inflammatory factors and promotion of the immune response[4]. In cirrhosis, activation of KC and overproduction of pro-inflammatory factors are thought to be important factors driving liver fibrosis[9]. Research has indicated that LSEC and KC can play an important role in maintaining normal liver function and metabolic homeostasis, either through direct contact or indirect signaling between them[10]. During cirrhosis, LSEC and KC may experience phenotypic shifts and functional alterations, and the interactions between these two cell types may have an important impact on disease progression. Therefore, delving into the pathogenesis of cirrhosis and discovering the molecular regulatory networks between cells in the liver and searching for new therapeutic targets are important topics in the current field of hepatology.
Single cell RNA sequencing (scRNA-seq) technology can reveal the heterogeneity of different cell types or subtypes in terms of gene expression, functional status and interactions at the level of individual cells[11], which provides new methods and perspectives for in-depth investigation of the roles and interactions of LSEC and KC in cirrhosis as well as pathological mechanisms.
In this study, we investigated the changes in gene expression and cellular functions of LSEC and KC in cirrhotic livers compared with those of LSEC and KC in healthy livers based on scRNA-seq analysis, and deeply explored the roles of LSEC and KC in cirrhotic livers as well as the interactions between these two types of cells, thus revealing their potential pathological mechanisms in liver cirrhosis.
Materials and Methods
Human liver scRNA-seq data availability and analysis:
The data source for this study was the GSE136103 (scRNA-seq) dataset from the Gene Expression Omnibus (GEO) database, which includes scRNA-seq data from five human healthy liver samples (male=4, female=1) and five human cirrhotic liver samples (male=3, female=2).
The gene expression matrix of scRNA-seq data (GSE136103) was preprocessed and analyzed using the Seurat package (version 4.1.1) in R (version 4.1.2), retaining genes expressed in at least 3 cells, cells expressing 300-6000 genes, and removing cells with >30 % mitochondrial content. After quality control, a total of 25 526 genes and 60 879 cells were included in the subsequent analysis. Next, the data after QC were normalized using the normalize data function, and after normalization, the top 2000 Hypervariable Genes (HVGs) out of 25 526 genes were identified using the find variable gene function with default parameters. The gene expression of HVGs was linearly transformed using the scale data function to normalize the expression values, thus removing the effect of highly expressed genes. The Run Principal Components Analysis (PCA) function was used to downsize the data, and the number of principal components for downsizing was determined based on the JackStraw function. Then, the Harmony algorithm was used to remove the batch effect generated during the multi-sample merging process, and the K-Nearest Neighbor (KNN) of each cell was calculated using the find neighbors function based on the KNN and Shared Nearest Neighbors (SNN) methods. The find clusters function was used to identify different cell clusters based on SNN, where the resolution was set to 0.4-1.2, and the cluster function was used to observe the effect of cell clustering under different resolution parameters. Finally, the clusters of cells were projected and visualized based on the t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm using the run t-SNE function.
Identification of liver cell types:
Differentially expressed genes between different cell clusters were identified using the find all markers function, and further screening of differentially expressed genes between clusters was performed according to p<0.05, log2 (fold-change) >0.8 to obtain significantly differentially expressed genes for each cell cluster. Finally, liver cell types were identified based on the manually sorted liver major cell marker genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis:
Differently expressed genes of LSEC and KC from healthy and cirrhotic livers were analyzed by KEGG for exploring the roles and associations of the differentially expressed genes in metabolic pathways and biological processes[12], so as to unravel the functional characteristics and biological significance of the gene sets. The clusterProfiler R package (version 4.2.2) was used to visualize KEGG data.
Constructing Protein-Protein Interaction (PPI) networks:
Protein interaction analysis of differentially expressed gene sets were performed using the String database (https://string-db.org)[13]. The PPI network graph was constructed using Cytoscape (version 3.9) and the core genes in the gene set were identified by sequencing the differentially expressed genes according to the degree algorithm using the cytoHubba plugin in Cystoscope.
Cell communication analysis:
CellChat (version 1.5.0) was used for cell communication analysis, and Seurat objects were exported as input to CellChat[14]. Using the create CellChat function to construct CellChat objects for LSEC and KC in healthy and cirrhotic livers, respectively, and download the human ligand-receptor interaction database. Cellular communication probabilities between LSEC and KC are calculated using the compute commune probe function with default parameters.
Results and Discussion
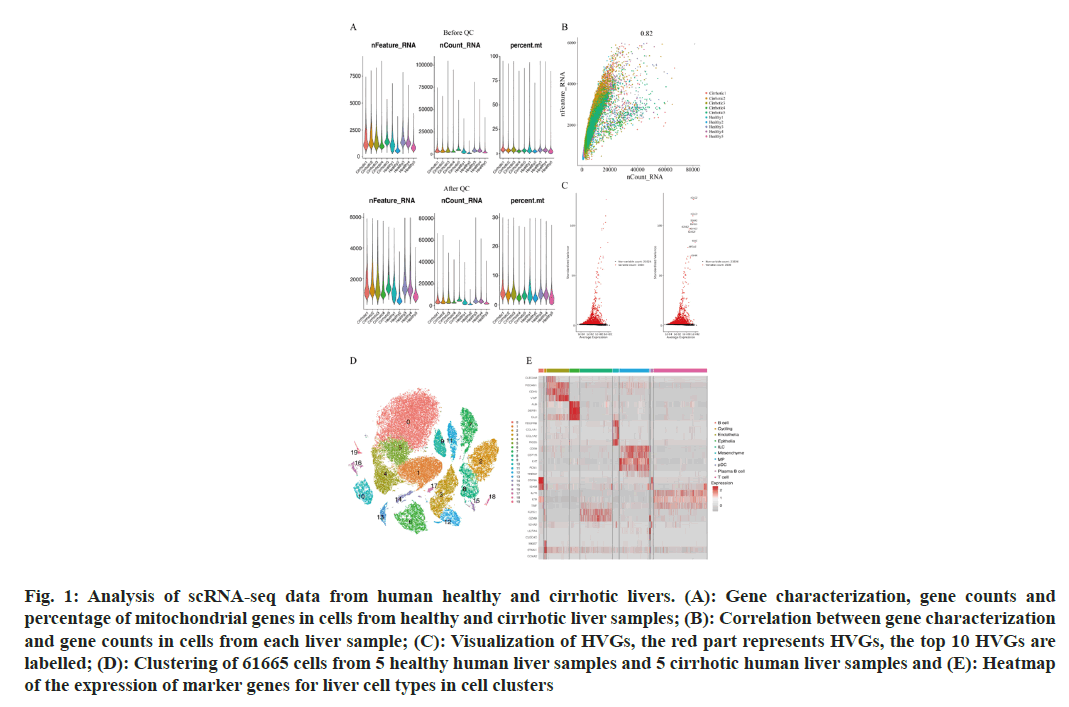
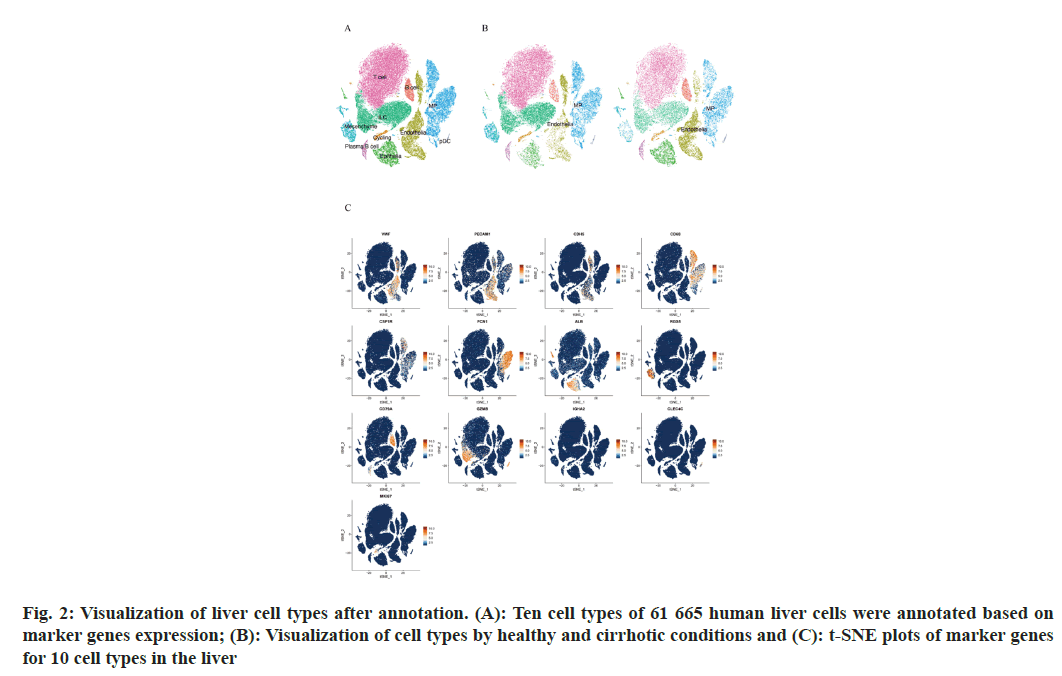
To describe the major cellular composition of healthy and cirrhotic liver tissues, we analyzed the scRNA-seq data (GSE136103) from the GEO database, including 35 365 cells from 5 human healthy livers and 26 300 cells from 5 human cirrhotic livers. After quality control, 25 812 cells from cirrhotic liver tissue and 35 067 cells from healthy liver tissue were retained, and the gene expression profiles, gene counts and mitochondrial gene expression for each sample before and after QC are shown here (fig. 1A). The nCount_RNA, which represents the number of Unique Molecular Identifiers (UMIs), and nFeature_ RNA, which represents the number of genes, were positively correlated and had a correlation coefficient of 0.82 (fig. 1B). We identified 2000 HVGs and labeled the top 10 HVGs (fig. 1C), most of which were involved in the activation of immune responses and were associated with the regulation of antigen-binding activity and immunoglobulin receptor-binding activity. We carried out principal component analysis and identified 20 principal components with p<0.05 based on the JackStraw function. 20 cell clusters were identified using the FindNeighbors function and FindClusters function (fig. 1D), and examined the expression of marker genes in cell clusters based on liver cell type marker genes obtained from previous studies[15]. Marker genes of different cell types showed high expression in the corresponding cell lines, indicating that a good clustering effect has been achieved (fig. 1E). A total of 10 liver cell types were identified in the 20 cell clusters, which correspond to endothelial cell (CLE+Platelet Endothelial Cell Adhesion Molecule 1 (PECAM-1)+Cadherin 5 (CDH5)+Von Willebrand Factor (VWF)+Intercellular Adhesion Molecule (ICAM) 2+); epithelial cell (Albumin (ALB)+Defensin Beta 1 (DEFB1)+Concentration of Clusterin (CLU)+); mesenchymal cell (Platelet Derived Growth Factor Receptor Beta (PDGFRB)+ Collagen Type I Alpha 1 (COL1A1)+COL1A2+Regulator of G Protein Signaling 5 (RGS5)+); Mononuclear Phagocyte (MP, CD68+Integrin Subunit Alpha M (ITGAM)+Colony Stimulating Factor 1 Receptor (CSF1R)+Lysozyme (LYZ)+Ficolin 1 (FCN1)+Triggering Receptor Expressed on Myeloid cells 2 (TREM2)+); B cell (CD79A+CD19+Immunoglobulin Heavy Constant Mu (IGHM)+); T cells (Interleukin 7 Receptor (IL7R)+Lymphotoxin Beta (LTB)+CD3 Epsilon subunit of T-cell receptor complex (CD3E)+Tumor Necrosis Factor (TNF)); Innate Lymphocyte (ILC, Granzyme A (GZMA)+Natural Killer cell Granule protein 7 (NKG7)+Killer Cell Lectin Like Receptor C1 (KLRC1)+Granzyme B (GZMB)); Plasma-like B cell (CD79A+Immunoglobulin Heavy Constant Alpha 2 (IGHA2)+); plasmacytoid Dendritic Cell (pDC), Leukocyte Immunoglobulin Like Receptor A4 (LILRA4)+C-Type Lectin Domain Family 4 Member C (CLEC4C)+GZMB+); and Cycling cell (Marker of proliferation Ki-67 (MKI67)+Stathmin 1 (STMN1)+Cyclin A2 (CCNA2)+) (fig. 2A), we can directly observe that most of the independent cell clusters are annotated as different liver cell types, while most of the adjacent cell clusters are annotated as the same cell type.
Fig. 1: Analysis of scRNA-seq data from human healthy and cirrhotic livers. (A): Gene characterization, gene counts and percentage of mitochondrial genes in cells from healthy and cirrhotic liver samples; (B): Correlation between gene characterization and gene counts in cells from each liver sample; (C): Visualization of HVGs, the red part represents HVGs, the top 10 HVGs are labelled; (D): Clustering of 61665 cells from 5 healthy human liver samples and 5 cirrhotic human liver samples and (E): Heatmap of the expression of marker genes for liver cell types in cell clusters
We focused on the liver endothelial cells and liver macrophages, and all 60 879 cells were labeled according to healthy or cirrhotic condition (fig. 2B and fig. 2C). The distribution of liver endothelial cell populations and liver macrophage populations in cirrhotic livers appeared to be more distinctly different compared with healthy livers, with part of endothelial cell populations (cluster 11) and part of macrophage populations (clusters 7, 2 and 15) having a considerably lower distribution density in the cirrhotic group, whereas the other part of the endothelial cell populations (clusters 3, 12, and 17) and macrophage populations (cluster 8) were more dense in the cirrhotic group, suggesting that subtypes of endothelial cell populations and macrophage populations may be altered in some ways in cirrhotic livers.
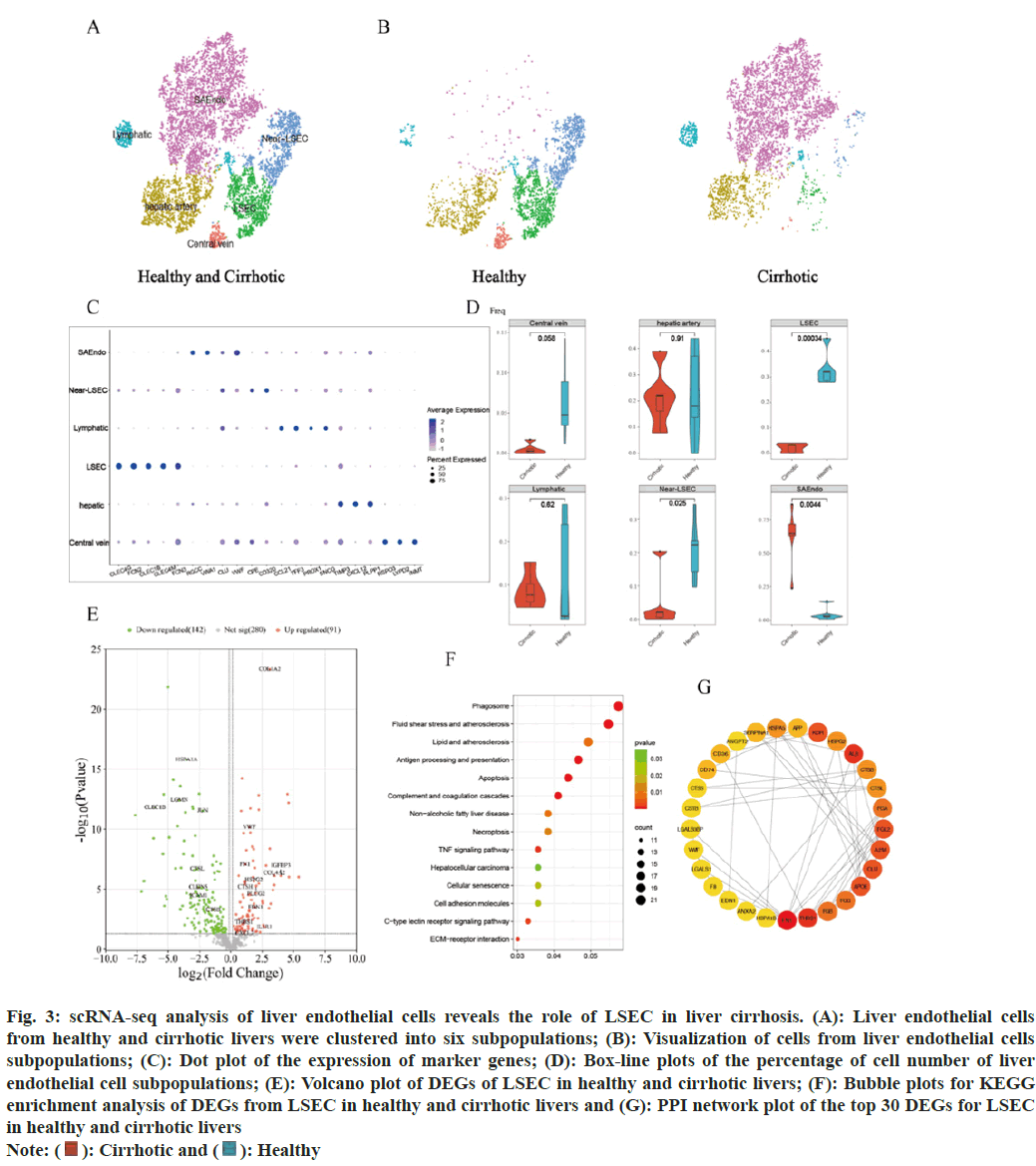
The distribution of some subsets of the endothelial cells differs more markedly in healthy and cirrhotic livers. In order to reveal the cellular composition of subpopulations of endothelial cells in healthy and cirrhotic livers and to further investigate the effect of LSEC in cirrhosis, endothelial cell subsets (cluster 3, 11, 12 and 17, a total of 8202 cells) were extracted for subpopulation clustering analysis and subpopulation cell type annotation. Re-clustering of endothelial cells identified 12 cell clusters and 6 endothelial cell subtypes were identified. Cell clusters were annotated as LSEC (cluster 2, CLEC4G+FCN2+CLEC1B+CLEC4M+FCN3+); scar-associated endothelial cells (SAEndo, cluster 0, 1, 8, 4, Regulator of Cell Cycle (RGCC)+Von Willebrand Factor A Domain Containing 1 (VWA1)+CLU+VWF+); lymphoid endothelial cells (cluster 9, 11, C-C Motif Chemokine 21 (CCL21)+Trefoil Factor 3 (TFF3)+Prospero Homeobox 1 (PROX1)+Synuclein Gamma (SNCG)+); hepatic artery endothelial cells (cluster 3, 5, TIMP Metallopeptidase Inhibitor 3 (TIMP3)+C-X-C Motif Chemokine Ligand 12 (CXCL12)+Phospholipid Phosphatase 1 (PLPP1)+) and central vein endothelial cells (cluster 10, R-Spondin 3 (RSPO3)+LYPD2+Indolethylamine N-Methyltransferase (INMT)+). Cluster 6 and 7 are closely adjacent to LSEC, and the DEGs of cluster 6 and 7 are almost the same as those of the LSECs, but those 2 clusters did not express the marker genes of LSEC. We annotated cluster 6 and 7 as near Liver Sinusoidal Endothelial Cell (near-LSEC) (fig. 3A). We used the t-SNE algorithm to visualize the endothelial cell subpopulations in the healthy and cirrhotic groups, respectively, and found that the cellular distribution of the endothelial cell subpopulations in the cirrhotic group was significantly different compared with that in the healthy group (fig. 3B). Expression of endothelial cell subpopulation cell type marker genes was visualized using dot plots (fig. 3C), marker genes showed high expression only in the endothelial cell subpopulation to which it corresponded. We counted the proportion of each endothelial cell subtype in each liver sample of the healthy group and the cirrhosis group. Compared with the healthy group, the proportion of LSEC and near-LSEC in the endothelial cell group of the cirrhosis group was significantly lower than that of the healthy group (p=0.00034 and p=0.025), while the proportion of SAEndo in the endothelial cell group of the liver cirrhosis group was significantly higher than that of the healthy group (p=0.0044) (fig. 3D). We focused on LSEC, which had a remarkably lower cell number and percentage in cirrhotic livers. We hypothesized that LSEC were severely damaged in the disease state, resulting in a diminished ability to survive and proliferate, which may also be accompanied by phenotypic and functional alterations that further exacerbate cirrhosis.
Fig. 3: scRNA-seq analysis of liver endothelial cells reveals the role of LSEC in liver cirrhosis. (A): Liver endothelial cells
from healthy and cirrhotic livers were clustered into six subpopulations; (B): Visualization of cells from liver endothelial cells
subpopulations; (C): Dot plot of the expression of marker genes; (D): Box-line plots of the percentage of cell number of liver
endothelial cell subpopulations; (E): Volcano plot of DEGs of LSEC in healthy and cirrhotic livers; (F): Bubble plots for KEGG
enrichment analysis of DEGs from LSEC in healthy and cirrhotic livers and (G): PPI network plot of the top 30 DEGs for LSEC in healthy and cirrhotic livers
Note:  Healthy
Healthy
To further investigate whether the gene expression characteristics and functions of LSEC in cirrhotic livers were altered compared to healthy livers, DEGs of LSEC in the healthy and cirrhotic groups were identified using the find markers function, setting p<0.05 as the criterion, and a total of 233 DEGs were identified, including 91 genes up regulated in LSEC of cirrhosis and 142 genes down regulated in LSEC of cirrhosis (fig. 3E). KEGG enrichment analysis of these DEGs showed that the DEGs of LSEC were mainly enriched in biological pathways associated with cellular senescence, apoptosis, programmed necrosis, Extracellular Matrix (ECM) receptor interactions, antigen processing and presentation, complement and coagulation cascades, atherosclerosis, cell adhesion molecules, C-type lectin receptor signaling pathway, and TNF signaling pathway (fig. 3F). Among the DEGs of LSEC, some genes associated with cellular senescence, apoptosis, programmed necrosis, atherosclerosis, and TNF signaling pathways (CTSH, EDN1, IL1R1, CXCL2 and IGFBP3), which were up-regulated in LSEC of cirrhotic liver, may be an important contributor to the reduced number of cirrhotic LSEC. Some genes associated with ECM receptor interactions (VWF, COL1A2, COL4A2, FN1, ITGB5, HSPG2, LAMC1, and THBS1), were similarly up-regulated in LSEC of cirrhotic liver. These genes are critical for signal transduction and communication between LSEC and other cells in the hepatic sinusoids, which may reflect the fact that in cirrhotic liver enhanced interactions between LSEC and other cells, and plays an important role in driving disease progression. Some genes related to cell adhesion molecules (CLDN5, ICAM1), were down-regulated in LSEC of cirrhotic liver, which may suggest that the adhesion between LSEC and leukocytes (such as macrophages) decreases in liver cirrhosis, resulting in the migration of leukocytes across endothelial cells affected[16]. Some genes associated with antigen processing and presentation and immune response (CTSL, CTSB, LGMN, HSPA1A, JUN, and CLEC1B), were down-regulated in LSEC of cirrhotic liver, which reflected the impaired normal immunity of LSEC in cirrhotic livers[17]. In addition, we selected the DEGs of LSEC to construct the PPI network and selected the top 30 DEGs as hub genes according to the degree method (fig. 3G). Among these hub genes, FN1 and THBS1 were the core genes of the PPI network, interacting with a variety of other genes, and both core genes were up regulated in cirrhotic LSEC. FN1 encodes fibronectin, which is involved in cell adhesion and wound healing[18]. THBS1 encodes an adhesive glycoprotein that mediating interactions between cell-cell and cell-matrix[19]. This protein binds to fibronectin and has been shown to play an important role in inflammatory response, angiogenesis, cellular senescence, apoptosis and tumorigenesis[20,21]. We hypothesized that the up regulation of FN1 and THBS1 in cirrhotic LSEC might lead to senescence or even apoptosis of LSEC on the one hand, and exacerbate the inflammatory response and accelerate hepatic fibrosis on the other hand.
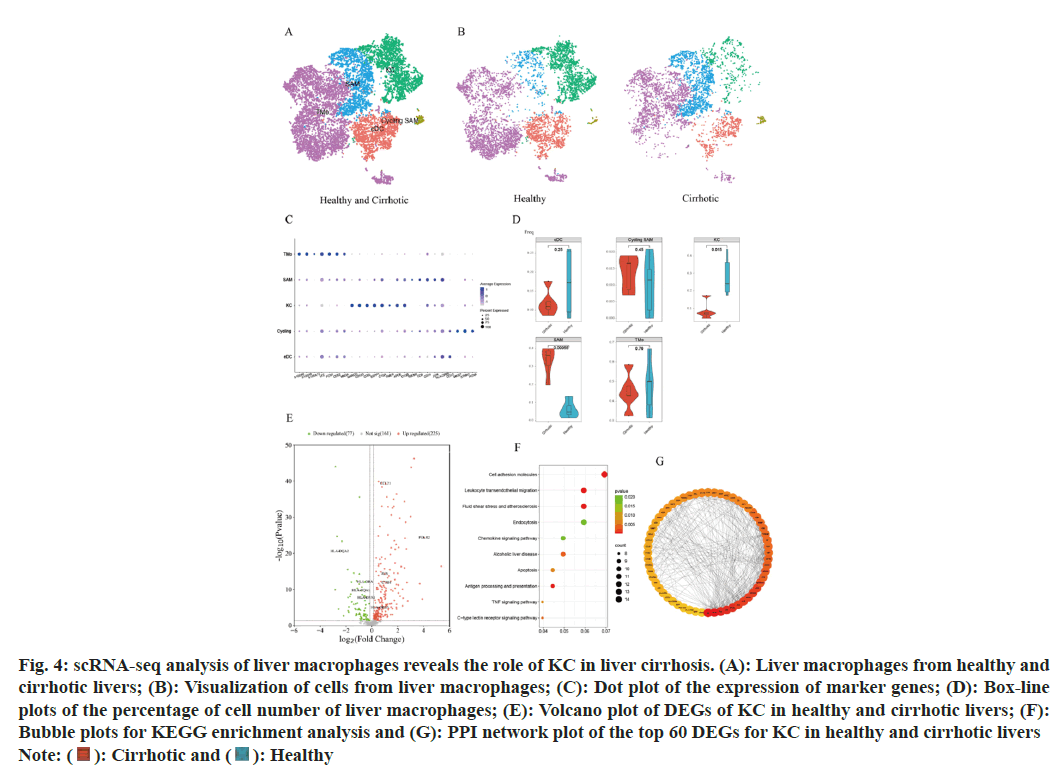
Significant differences in the distribution of certain subsets of macrophage populations are observed between healthy and cirrhotic livers. To reveal the cellular composition of subpopulations of macrophage populations in healthy and cirrhotic livers and to further explore the effect of KC in cirrhosis, a subset of macrophage populations (cluster 2, 7, 8, and 15, a total of 10 958 cells) was extracted for subpopulation clustering analysis and subpopulation cell type annotation. The re-clustering of the macrophage population identified 12 cell clusters and 5 macrophage subtypes were identified (fig. 4A). Cell clusters were annotated as Tissue Monocytes (TMo), (cluster 0, 2, 6, 8, 9, 10, S100A8+S100A9+S100A12+LYZ+FCN1 +CD52+MNDA+); KC (cluster 4,5, MARCO+CD163+CD5L +SE PP1+C1QA +TIMD4+); Scar-Associated Macrophages (SAM, cluster 3,7, APOE+C1QB+TREM2+CTSD+CD9+); conventional Dendritic Cells (cDC), cluster 1, CD1C+); and cycling Scar-Associated Macrophages (cycling SAM, cluster 11, MKI67+STMN1+PCNA+).
Fig. 4: scRNA-seq analysis of liver macrophages reveals the role of KC in liver cirrhosis. (A): Liver macrophages from healthy and
cirrhotic livers; (B): Visualization of cells from liver macrophages; (C): Dot plot of the expression of marker genes; (D): Box-line
plots of the percentage of cell number of liver macrophages; (E): Volcano plot of DEGs of KC in healthy and cirrhotic livers; (F):
Bubble plots for KEGG enrichment analysis and (G): PPI network plot of the top 60 DEGs for KC in healthy and cirrhotic livers
Note:  Healthy
Healthy
The t-SNE algorithm was utilized to visualize the macrophage subpopulations within both the healthy and cirrhotic groups, revealing a distinct cell distribution in the cirrhotic group macrophage subpopulations compared to the healthy group (fig. 4B). Dot plots were used to visualize the expression of macrophage subpopulation cell type marker genes (fig. 4C), showing high expression of marker genes exclusively within their corresponding macrophage subpopulations. We quantified the proportion of each macrophage subtype within each liver sample for both the healthy and cirrhotic groups. Compared to the healthy group, the proportion of KC significantly decreased within the macrophage population of the cirrhotic group (p=0.015), while SAM were predominantly found in cirrhotic livers (p=0.00055) (fig. 4D). We focused on KC, which significantly decreased in number and proportion within cirrhotic livers, suggests that the local immune microenvironment of cirrhotic livers is altered, limiting the normal functions of KC such as pathogen clearance and cytokine production, thereby exacerbating liver inflammation and fibrosis.
To further investigate whether the gene expression characteristics and functions of KC in cirrhotic livers have altered compared to healthy livers, DEGs of KC in the healthy and cirrhotic groups were identified using the find markers function. Setting p<0.05 as the criterion, a total of 302 DEGs were identified, including 225 up regulated and 77 down regulated genes in cirrhotic KC (fig. 4E). The KEGG enrichment analysis of these DEGs revealed a high degree of similarity with the KEGG results of DEGs in LSEC, primarily enriching in pathways related to apoptosis, antigen processing and presentation, complement and coagulation cascades, atherosclerosis, cell adhesion molecules, leukocyte transendothelial migration, C-type lectin receptor signaling pathway, TNF signaling pathway, chemokine signaling pathway, and phagocytosis (fig. 4F). Among the DEGs in KC, genes associated with apoptosis, atherosclerosis, and the TNF signaling pathway (JUN, CTSH, TUBA1A, JUNB, and CYBA) were up-regulated in cirrhotic KC, potentially contributing to the reduction in KC numbers in cirrhosis. Genes related to leukocyte transendothelial migration (ITGB2, CXCR4, NCF4, CD99, ITGAM, and CYBA) were also up-regulated in cirrhotic KC, mediating the migration of leukocytes (such as TMo) to the site of liver injury post-damage. This suggests that KC in cirrhosis may facilitate the recruitment of immune cells into the liver, replenishing the immune cell numbers and thereby maintaining the immune microenvironment stability to some extent. Genes associated with cell adhesion molecules (ITGB2, HLA-DRB5, and CD99) were up-regulated in cirrhotic KC, whereas HLA-DRA, HLA-DPA1, HLA-DQA1, and HLA-DQA2 were down-regulated. Genes related to immune and inflammatory responses (CXCR4, CCL21, FOLR2, HLA-A, HLA-F, JUN, PYCARD, and LSP1) saw increased expression in cirrhotic KC. The upregulation of these genes in cirrhosis could further enhance the inflammatory response of KC, releasing more inflammatory factors, thereby exacerbating liver inflammation and damage, even leading to KC apoptosis and accelerating liver fibrosis. We constructed the PPI network using the DEGs of KC and selected the top 60 genes as hub genes based on the degree method (fig. 4G). Among these hub genes, ALB, TNF, APOE, IL1B, APOA1, APP, CLU, CSF1R, SERPINA1, and APOH were the top 10 hub genes, all of which were up-regulated in cirrhotic KC and interacted with various other genes. ALB (Albumin), APOE (Apolipoprotein E), APOH (Apolipoprotein H), and APOA1 (Apolipoprotein A1) are genes related to lipid metabolism and transport, and their upregulation suggests involvement of KC in the dysregulation of lipid metabolism in cirrhosis, leading to liver lipid metabolic disorder[22]. Tumor Necrosis Factor (TNF) and Interleukin-1 Beta (IL-1β) are inflammation-related genes, and their upregulation indicates that cirrhotic liver KCs may play a significant role in inflammatory responses[21]. Amyloid β Precursor Protein (APP) and Clusterin (CLU) are associated with neural system function and apoptosis pathways, and their upregulation may reflect an enhanced response of KC to neurotransmitters and apoptosis signals in cirrhosis, leading to KC apoptosis. Colony Stimulating Factor 1 Receptor (CSF1R) and Serpin Family Member A1 (SERPINA1) are also related to cell signaling and inflammation, and their upregulation might suggest that KC are involved in more intense immune responses, which on one hand fight disease and on the other hand may lead to KC damage or even apoptosis[23]. In summary, these upregulated core genes reveal multiple significant roles that KC may play in the progression of cirrhosis, including regulating lipid metabolism, participating in inflammatory responses, affecting neural system function, and influencing apoptosis pathways.
Based on scRNA-seq data analysis, we have independently explored the differences in cell distribution, cell proportion, gene expression, and cellular functions between LSEC and KC in both healthy and cirrhotic livers, further investigating the effects and potential pathogenic mechanisms of LSEC and KC in cirrhosis.
However, the aforementioned studies treated these two cell types separately. Within the liver sinusoids, LSEC and KC are in close proximity, suggesting complex interactions between them that could significantly influence disease progression in cirrhosis. To explore the potential cellular communications and their differences between LSEC and KC in healthy and cirrhotic livers, we utilized the CellChat algorithm to analyze the scRNA-seq data of LSEC and KC from healthy and cirrhotic livers (fig. 5A)[24]. This analysis aims to identify potential interaction modes between LSEC and KC and the differences present in healthy and disease states, thereby revealing the molecular mechanisms of LSEC and KC interactions in the pathogenesis of cirrhosis at the molecular level.
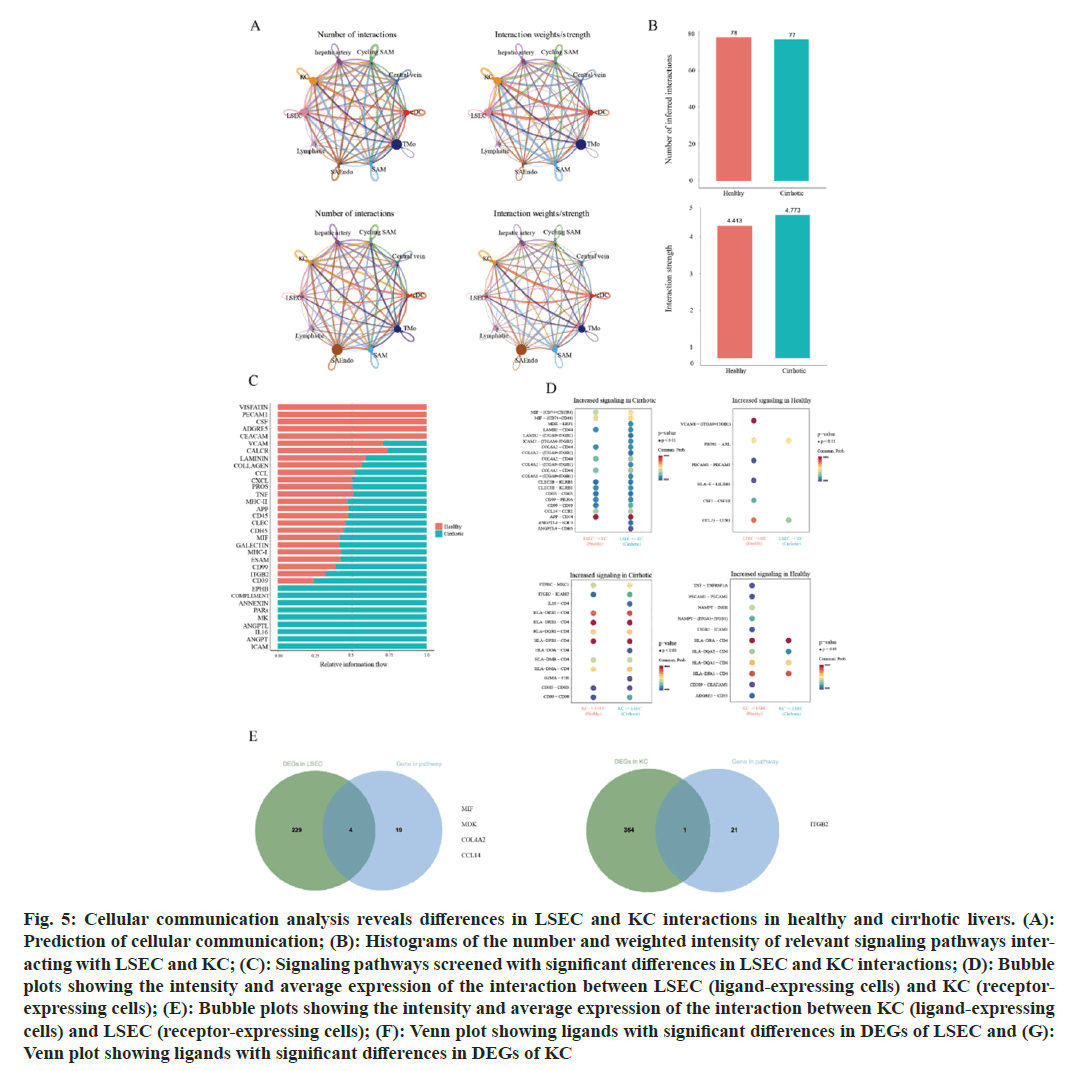
Fig. 5: Cellular communication analysis reveals differences in LSEC and KC interactions in healthy and cirrhotic livers. (A): Prediction of cellular communication; (B): Histograms of the number and weighted intensity of relevant signaling pathways interacting with LSEC and KC; (C): Signaling pathways screened with significant differences in LSEC and KC interactions; (D): Bubble plots showing the intensity and average expression of the interaction between LSEC (ligand-expressing cells) and KC (receptor-expressing cells); (E): Bubble plots showing the intensity and average expression of the interaction between KC (ligand-expressing cells) and LSEC (receptor-expressing cells); (F): Venn plot showing ligands with significant differences in DEGs of LSEC and (G): Venn plot showing ligands with significant differences in DEGs of KC
Given the significant differences in cell quantity, cellular functions, and gene expression between these two cell types in both the cirrhotic and healthy groups, we hypothesized that the cell communication between LSEC and KC might also differ significantly, with the interaction intensity between LSEC and KC in cirrhotic livers being considerably lower than in healthy livers. However, in predicting the communication pathways between LSEC and KC, we identified 155 interaction modes, with 78 present in the healthy group and 77 in the cirrhotic group, indicating a comparable number of cellular communication pathways between the two groups. The weighted cell communication strength of the LSEC-KC communication pathways in the healthy group was 4.413, which is lower than that in the cirrhotic group at 4.773 (fig. 5B). This result contradicts our initial assumption, suggesting that despite a significant reduction in the numbers of LSEC and KC in cirrhotic livers compared to healthy ones, the quantity of cell communication pathways and the weighted cell communication strength between the two groups are nearly equivalent. We believe that the communication mechanisms between LSEC and KC in cirrhotic livers might have changed, leading to an adjustment in the modes of interaction and signaling pathways between these two cell types during the progression of cirrhosis, making the number and strength of interactions in the cirrhotic group comparable to those in the healthy group.
To validate our hypothesis, we extracted signaling pathways from the LSEC-KC communication channels in both the healthy and cirrhotic groups that exhibited significant differences (fig. 5C). A total of 34 signaling pathways showed significant differences in communication strength between the two groups. Among these, pathways such as VISFATIN, PECAM1, CSF, ADGRE, and CEACAM were exclusively enriched in the healthy group. However, 21 pathways were notably more enriched in the cirrhotic group than in the healthy group, especially the EPHB, COMPLEMENT, ANNEXIN, PARs, MK, ANGPTL, IL16, ANGPT, and ICAM pathways, which were exclusively enriched in the cirrhotic group. Since each signaling pathway for cellular communication contains a ligand expressed in one cell type and a receptor expressed in another, we predicted the strength and expression of LSEC-KC communication at the level of ligand-receptor pairs (fig. 5D). The results showed that when LSEC act as the ligand-expressing cell, the number and communication strength of ligand-receptor pairs related to LSEC-KC cell communication in the cirrhotic group were significantly higher than in the healthy group, for instance, MIF-(CD74+CXCR4), MDK-LRP1, COL4A2-CD44, CD99-CD99, CCL14-CCR1, and APP-CD74. Conversely, pairs like VCAM1-(ITGA9+ITGB1), PROS1-AXL, HLA-F-LILRB1, PECAM1-PECAM1, CSF1-CSF1R, and CCL23-CCR1 showed significantly weakened intensity in the cirrhotic group. When KC acted as ligand-expressing cells, ligand-receptor pairs with higher interaction strength in the cirrhotic group were also found, such as PTPRC-MRC1, HLA-DRB5-CD4, ITGB2-ICAM2, HLA-DRB1-CD4, HLA-DMA-CD4, and CD99-CD99. Nonetheless, a considerable number of interactions showed significantly reduced strength or were even absent in the cirrhotic group, for example, TNF-TNFRSF1A, NAMPT-INSR, NAMPT-(ITGA5+ITGB1), ITGB2-ICAM1, HLA-DQA2-CD4, HLA-DQA1-CD4, and ADGRE5-CD55 (fig. 5E). We screened ligand-receptor pairs expressed in DEGs of LSEC and DEGs of KC, encompassing five signaling pathways:; MIF, MK, COLLAGEN, CCL, and ITGB2. MIF, MDK, COL4A2 and CCL14, as ligands expressed by LSEC, were upregulated in LSEC in the cirrhotic group. ITGB2, as a ligand expressed by KC, was upregulated in the cirrhotic group. These signaling pathways that are upregulated in cirrhotic livers and their corresponding ligands in the corresponding cell types may accelerate cirrhosis by some mechanism.
LSEC and KC are two crucial non-parenchymal cells within liver sinusoids, playing pivotal roles in both physiological functions and pathological processes of the liver. Their interaction significantly influences the development of liver diseases. Normally, signal exchange between LSEC and KC contributes to maintaining liver homeostasis and immune tolerance. However, in conditions like cirrhosis, this signaling may become disrupted, resulting in immune dysfunction and increased fibrosis. Our findings indicate that upregulated ligand-receptor pairs in cirrhotic LSEC and KC may exacerbate liver fibrosis, suggesting their significant involvement in cirrhosis pathology.
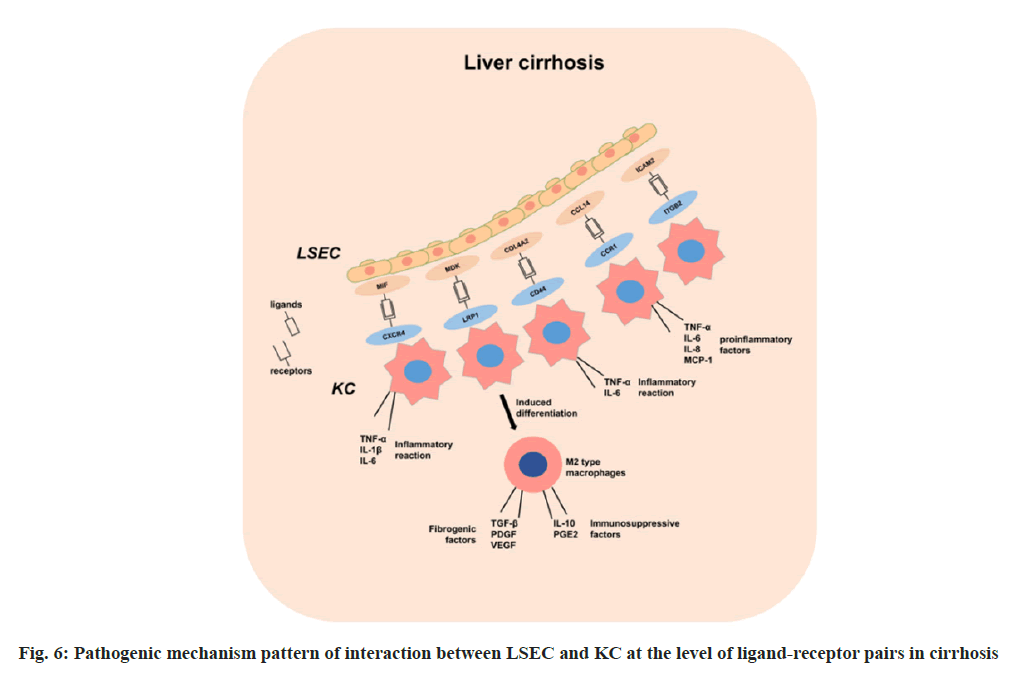
MIF, a macrophage migration inhibitory factor, acts as an immune regulatory factor by activating various signaling pathways, such as ERK, PI3K/AKT, JNK, upon binding to the CXCR4 receptor. This activation regulates cell proliferation, migration, differentiation, and apoptosis[25]. In cirrhosis, the upregulation of the MIF-CXCR4 signaling pathway promotes the release of pro-inflammatory factors from endothelial cells LSEC and macrophages KC, triggering a heightened inflammatory response, which exacerbates liver damage and necrosis[26]. These pro-inflammatory factors further stimulate LSECs and other cells to secrete MIF, perpetuating a cycle of sustained liver inflammation.
Previous studies indicate that the MDK-LRP1 signaling pathway, known for its role in gallbladder cancer, also influences immune escape and fibrosis in cirrhosis[27]. In cirrhotic livers, increased MDK expression in LSEC binds to LRP1 expressed by KC, potentially affecting KC polarization and function, leading to the differentiation of KC into M2 macrophages. These M2 macrophages secrete profibrotic factors, such as TGF-β, PDGF and VEGF, promoting hepatic stellate cell activation and collagen deposition, thereby exacerbating liver fibrosis[27]. Additionally, M2 macrophages secrete immunosuppressive factors like IL-10 and PGE2, inhibiting immune effector cell activity and promoting immune escape in cirrhosis[28,29]. This signaling pathway may be a novel target for the treatment of cirrhosis and deserves further investigation and validation.
The COL4A2-CD44 signaling pathway is implicated in promoting liver fibrosis progression. COL4A2 promotes LSEC capillarization, reducing barrier function and increasing vascular permeability, while CD44 regulates KC inflammatory response and immune function, affecting liver damage repair and fibrosis control[30]. Studies suggest that COL4A2 induces KC polarization toward the M1 type, releasing pro-inflammatory factors such as TNF-α and IL-6, enhancing liver inflammation and fibrosis, whereas CD44 inhibits M2 polarization, reducing anti-inflammatory factor (IL-10, TGF-β) secretion and impairing liver repair.
The CCL14-CCR1 signaling pathway acts as a chemokine-receptor axis, involved in regulating inflammatory and immune responses, plays a role in cirrhosis by regulating immune cell migration and activation[31]. CCL14 binding to CCR1 activates LSEC and KC, inducing inflammatory factor and chemokine secretion (TNF-α, IL-6, IL-8 and MCP-1), exacerbating liver inflammation. These cytokines can further recruit and activate other immune cells such as neutrophils, monocytes, and T cells, exacerbating the inflammatory response in the liver[32]. In addition, on the one hand, CCL14 can induce LSEC apoptosis via CCR1, leading to endothelial barrier dysfunction in the hepatic sinusoids and increasing the sensitivity of the liver to toxins and foreign antigens. On the other hand, CCL14 can promote M1-type polarization of KCs via CCR1, enhancing their secretion of pro-fibrotic factors such as TGF-β, PDGF and CTGF[33]. These factors can stimulate the activation and proliferation of hepatic stellate cells and promote collagen synthesis and deposition.
ITGB2 is part of the integrin family, which plays an important role in the immune response, and defects in the ITGB2 gene result in defective leukocyte adhesion. ICAM2 is a member of the family of intercellular adhesion molecules, a group of proteins that play a key role in cell adhesion and signaling, and are involved in cell adhesion and migration, as well as in inflammatory responses through interactions with integrin’s[34]. Upregulation of the ITGB2-ICAM2 pathway in cirrhotic livers may promote migration of inflammatory cells and exacerbate the inflammatory response in the liver, and this pathway may also inhibit hepatic vascular remodeling, affecting hepatic blood flow. We mapped the pathogenic mechanism pattern of interaction between LSEC and KC at the level of ligand-receptor pairs in cirrhosis (fig. 6).
In this study, we examined scRNA-seq data of LSEC and KC in healthy and cirrhotic human livers using scRNA-seq data analysis technology, and investigated the cellular distribution, number ratio, gene expression and cellular function of LSEC and KC in both conditions. The aim was to elucidate the roles and potential pathogenic factors of LSEC and KC in liver cirrhosis. Using the CellChat algorithm, we performed cellular communication analysis of LSEC and KC in healthy and cirrhotic livers and identified potential interactions between LSEC and KC at the level of signaling pathway-associated ligand-receptor pairs. We further explored the pathogenic mechanisms of LSEC and KC interactions in cirrhosis. Reduction in the number of LSEC and KC may be a key indicator of the severity and prognosis of cirrhosis. Therefore, the reduction of LSEC and KC may be a potential therapeutic target that has the potential to stop or reverse cirrhosis by protecting or restoring the number and function of LSEC and KC. In addition, exploring the use of signaling pathways and ligand-receptor pairs that mediate communication between LSEC and KC to intervene or reverse cirrhosis is also a promising direction for future research.
Acknowledgements:
This study was funded by grants from the Zhejiang Province Key Research and Development Project (Grant No: 2020C03057, 2020C03071 and 2021C03145); pioneer and Leading Goose R&D Program of Zhejiang (grant number 2022C03043); National Science and Technology Project (grant number 2022CSJGG1000/2023ZY1068).
Conflict of interests:
The authors declared no conflict of interests.
References
- Marcellin P, Kutala BK. Liver diseases: A major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int 2018;38(1):2-6.
[Crossref] [Google Scholar] [PubMed]
- de Smet V, Eysackers N, Merens V, Kazemzadeh DM, Halder G, Verhulst S, et al. Initiation of hepatic stellate cell activation extends into chronic liver disease. Cell Death Dis 2021;12(12):1110.
[Crossref] [Google Scholar] [PubMed]
- Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol 2019;70(1):151-71.
[Crossref] [Google Scholar] [PubMed]
- Matsuda M, Seki E. The liver fibrosis niche: Novel insights into the interplay between fibrosis-composing mesenchymal cells, immune cells, endothelial cells, and extracellular matrix. Food Chem Toxicol 2020;143:111556.
[Crossref] [Google Scholar] [PubMed]
- Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, et al. Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J Hepatol 2017;66(1):212-27.
[Crossref] [Google Scholar] [PubMed]
- Deleve LD. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 2015;61(5):1740-6.
[Crossref] [Google Scholar] [PubMed]
- Tanaka M, Miyajima A. Liver regeneration and fibrosis after inflammation. Inflamm Regen 2016;36:1-6.
[Crossref] [Google Scholar] [PubMed]
- Smedsrød B, de Bleser PJ, Braet F, Lovisetti P, Vanderkerken K, Wisse E, et al. Cell biology of liver endothelial and Kupffer cells. Gut 1994;35(11):1509.
[Crossref] [Google Scholar] [PubMed]
- Verreck FA, de Boer T, Langenberg DM, van der Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-γ-and CD40L-mediated costimulation. J Leukocyte Biol 2006;79(2):285-93.
[Crossref] [Google Scholar] [PubMed]
- Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain kupffer cell identity. Immunity 2019;51(4):655-70.
[Crossref] [Google Scholar] [PubMed]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell 2017;65(4):631-43.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia Of Genes And Genomes. Nucl Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucl Acids Res 2021;49(D1):D605-12.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Sun D, Wang C. Evaluation of cell-cell interaction methods by integrating single-cell RNA sequencing data with spatial information. Genome Biol 2022;23(1):218.
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BE, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575(7783):512-8.
[Crossref] [Google Scholar] [PubMed]
- Duan Y, Pan X, Luo J, Xiao X, Li J, Bestman PL, et al. Association of inflammatory cytokines with non-alcoholic fatty liver disease. Front Immunol 2022;13:880298.
[Crossref] [Google Scholar] [PubMed]
- Lalor PF, Herbert J, Bicknell R, Adams DH. Hepatic sinusoidal endothelium avidly binds platelets in an integrin-dependent manner, leading to platelet and endothelial activation and leukocyte recruitment. Am J Physiol Gastroint Liver Physiol 2013;304(5):G469-78.
[Crossref] [Google Scholar] [PubMed]
- Rostagno A, Williams MJ, Baron M, Campbell ID, Gold LI. Further characterization of the NH2-terminal fibrin-binding site on fibronectin. J Biol Chem 1994;269(50):31938-45.
[Google Scholar] [PubMed]
- Adams JC, Bentley AA, Kvansakul M, Hatherley D, Hohenester E. Extracellular matrix retention of thrombospondin 1 is controlled by its conserved C-terminal region. J Cell Sci 2008;121(6):784-95.
[Crossref] [Google Scholar] [PubMed]
- Meijles DN, Sahoo S, Al Ghouleh I, Amaral JH, Bienes-Martinez R, Knupp HE, et al. The matricellular protein TSP1 promotes human and mouse endothelial cell senescence through CD47 and Nox1. Sci Signal 2017;10(501):eaaj1784.
[Crossref] [Google Scholar] [PubMed]
- Lumeng CN, deYoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 2007;56(1):16-23.
- Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: From pathogenesis to novel therapeutic strategies. Cell Mol Immunol 2016;13(3):316-27.
[Crossref] [Google Scholar] [PubMed]
- Jung K, Kang M, Park C, Hyun CY, Jeon Y, Park SH, et al. Protective role of V-set and immunoglobulin domain‐containing 4 expressed on kupffer cells during immune-mediated liver injury by inducing tolerance of liver T-and natural killer T-cells. Hepatology 2012;56(5):1838-48.
[Crossref] [Google Scholar] [PubMed]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12(1):1088.
[Crossref] [Google Scholar] [PubMed]
- Assis DN, Leng L, Du X, Zhang CK, Grieb G, Merk M, et al. The role of macrophage migration inhibitory factor in autoimmune liver disease. Hepatology 2014;59(2):580-91.
[Crossref] [Google Scholar] [PubMed]
- Hughes CE, Nibbs RJ. A guide to chemokines and their receptors. FEBS J 2018;285(16):2944-71.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Zuo C, Liu L, Hu Y, Yang B, Qiu S, et al. Single-cell RNA-sequencing atlas reveals an MDK-dependent immunosuppressive environment in ErbB pathway-mutated gallbladder cancer. J Hepatol 2021;75(5):1128-41.
[Crossref] [Google Scholar] [PubMed]
- N Zekri AR, El Kassas M, Salam ES, Hassan RM, Mohanad M, Gabr RM, et al. The possible role of Dickkopf-1, Golgi protein-73 and Midkine as predictors of hepatocarcinogenesis: A review and an Egyptian study. Sci Rep 2020;10(1):5156.
[Crossref] [Google Scholar] [PubMed]
- Sorrelle N, Dominguez AT, Brekken RA. From top to bottom: Midkine and pleiotrophin as emerging players in immune regulation. J Leukoc Boil 2017;102(2):277-86.
[Crossref] [Google Scholar] [PubMed]
- Senbanjo LT, Chellaiah MA. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front Cell Devel Biol 2017;5:18.
[Crossref] [Google Scholar] [PubMed]
- Märkl F, Huynh D, Endres S, Kobold S. Utilizing chemokines in cancer immunotherapy. Trends Cancer 2022;8(8):670-82.
[Crossref] [Google Scholar] [PubMed]
- Zhu M, Xu W, Wei C, Huang J, Xu J, Zhang Y, et al. CCL14 serves as a novel prognostic factor and tumor suppressor of HCC by modulating cell cycle and promoting apoptosis. Cell Death Dis 2019;10(11):796.
[Crossref] [Google Scholar] [PubMed]
- Omwenga I, Aboge GO, Mitema ES, Obiero G, Ngaywa C, Ngwili N, et al. Antimicrobial usage and detection of multidrug-resistant Staphylococcus aureus, including methicillin-resistant strains in raw milk of livestock from Northern Kenya. Microbial Drug Resist 2021;27(6):843-54.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Zhao Q, Kong M, Mao L, Yang Y, Xu Y. Myocardin-related transcription factor A regulates integrin beta 2 transcription to promote macrophage infiltration and cardiac hypertrophy in mice. Cardiovasc Res 2022;118(3):844-58.
[Crossref] [Google Scholar] [PubMed]