- *Corresponding Author:
- Guangjun Wang
Department of Pediatrics, Shandong Provincial Third Hospital, Tianqiao, Shandong 250031, China
E-mail: guangjun2010@yeah.net
| Date of Received | 22 June 2022 |
| Date of Revision | 24 May 2023 |
| Date of Acceptance | 04 July 2024 |
| Indian J Pharm Sci 2024;86(3):1171-1175 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Studies have shown that natural flavone and flavonoids are molecules derived from the natural source and can selectively kill certain cancer cells. However, the exact process by which these bioactive substances affect acute myeloid leukemia is still unknown. To further understand about the medication resistance and find possible treatments for pediatric acute myeloid leukemia, we used network pharmacology methods. We specifically examined the pharmacological impacts of polyphenols, particularly kaempferol, catechin and quercetin on the treatment of acute myeloid leukemia. Using public databases including PubChem, Kyoto Encyclopedia of Genes and Genomes Mapper, Universal Protein Resource, DisGenet and cystoscope databases, we conducted a thorough search for the potential targets of polyphenols and acute myeloid leukemia. Consequently, we identified polyphenol target genes and acute myeloid leukemia disease targets. Among the 253 genes showed overlapping; however, some might be potential as polyphenol-based acute myeloid leukemia therapy targets. The specific method by which these bioactive substances exert their anti-acute myeloid leukemia effects has been elucidated through the use of protein-protein interaction network analysis, gene ontology analysis and Kyoto Encyclopaedia of Genes and Genomes pathway enrichment analysis. Further cytological examinations confirmed these results, illustrating the polyphenol's capacity to hinder the growth of acute myeloid leukemia cells through different pathways. Our study combines network pharmacology-based predictions with systematic review studies for polyphenolic-mediated therapy of acute myeloid leukemia. This provides new perspectives on therapeutic approaches to address drug resistance in pediatric acute myeloid leukemia.
Keywords
Acute myeloid leukemia, network pharmacology, flavanols, leukemogenesis, quercetin
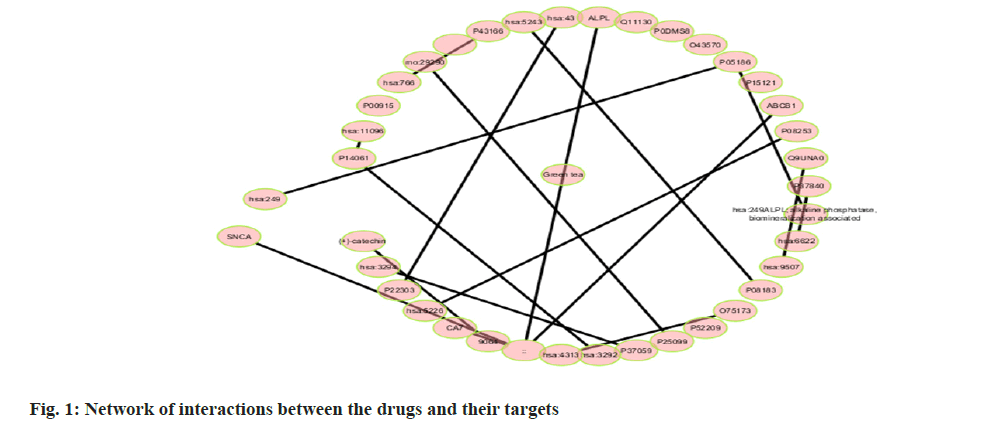
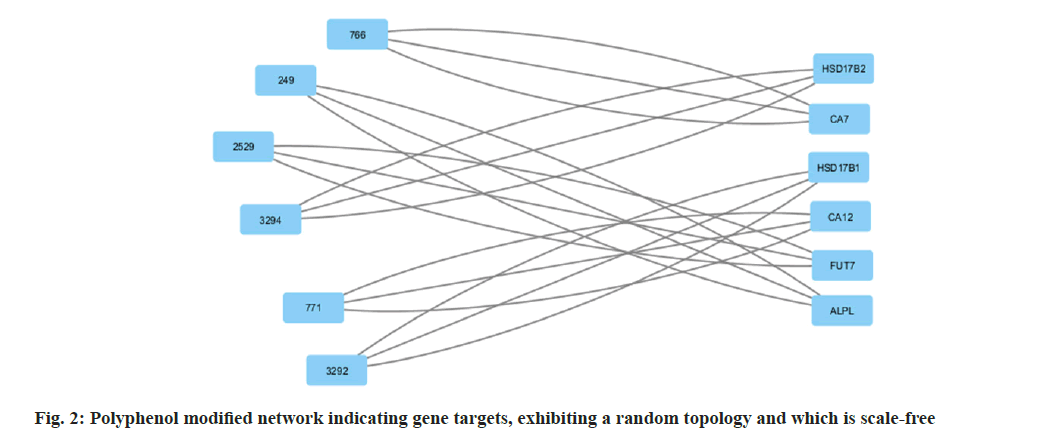
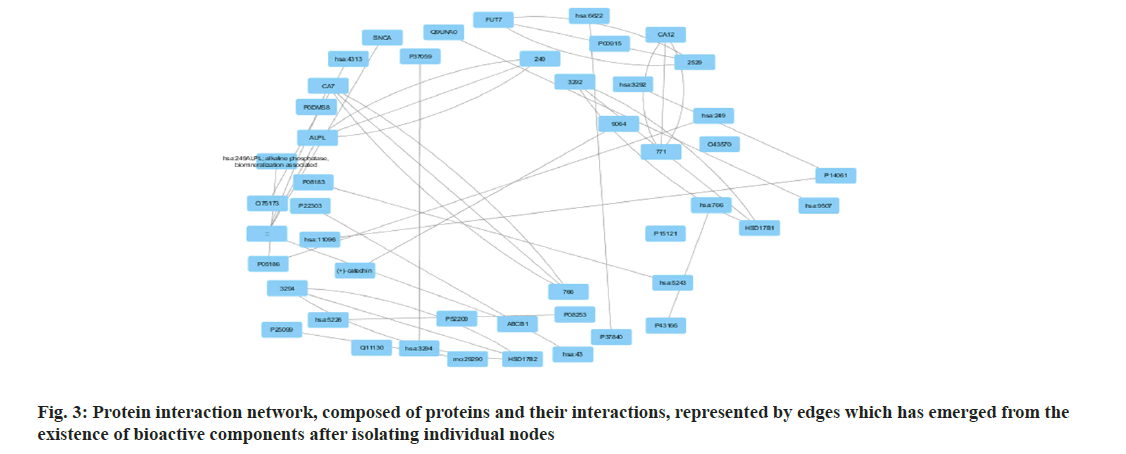
Among many different types of blood cancers, Acute Myeloid Leukaemia (AML) is characterised by undifferentiated growth of myeloid cells in the blood and bone marrow[1]. Especially for children with AML, medication resistance is a major obstacle despite advances in treatment methods such as targeted treatments, stem cell transplantation and intense chemotherapy[2]. There are numerous biological mechanisms that contribute to the emergence of drug resistance. These include changes in drug metabolism, upregulation of drug efflux pumps, improved Deoxyribonucleic Acid (DNA) repair mechanisms and dysregulation of apoptotic pathways[3]. The short non-coding Ribonucleic Acid (RNA) molecules known as microRNAs (miRNAs) target particular messenger RNAs (mRNAs) for the destruction or translational suppression, and they play an essential role in controlling gene expression[4]. There is mounting evidence that microRNAs are involved in a wide range of biological functions, such as drug resistance, cell differentiation, apoptosis and cell proliferation. Imbalanced miRNA expression profiles in AML have been associated with leukemogenesis, progression of the disease and medication resistance[5,6]. Some microRNAs have been linked to AML, and one of these, miR-186, may be a tumour suppressor that controls cell growth, death and resistance to drugs[7]. Multiple investigations have shown that miR-186 is downregulated in AML samples and cell lines, which may indicate that it plays an important role in leukemogenesis and the development of the illness[7]. With this study, we hope to learn more about miR-186 function in childhood AML and whether it can be used as a target for therapy to combat medication resistance. To understand how miR-186 regulates various processes, this study will use a holistic strategy that integrates experimental methods with network pharmacology. Notably, this study aims to explore the regulatory pathways and targets associated with miR-186, with a particular focus on the potential interaction between miR-186 and B-lymphoma Moloney murine leukemia virus Insertion region-1 (BMI1), a key regulator of the stem cell self-renewal and is implicated in leukemogenesis. By leveraging bioinformatics tools and molecular biology techniques, this study seeks to identify and validate the downstream targets of miR-186 and elucidate the molecular mechanisms underlying its tumor-suppressive functions in AML. The integration of network pharmacology approaches in this study holds the potential to uncover novel therapeutic strategies by identifying drug-target interactions and predicting the efficacy of existing or novel compounds in modulating the miR-186-mediated pathways implicated in drug resistance. Overall, our systematic analysis and therapeutic use of network pharmacology explored the mechanisms of treatment resistance and the involvement of miR-186 in pediatric AML, providing a comprehensive and multidisciplinary strategy to address the key clinical challenge. By elucidating the molecular underpinnings of miR-186 dysregulation and its impact on drug resistance, this study aims to lay the foundation for developing targeted therapeutic interventions and improving treatment outcomes for paediatric AML patients. Polyphenols, which comprise flavonoids and flavanols, are thought to mediate many of the effects. Flavanols, which are also known as catechins, including (-) Epiafzelechin (EZ), (+)-Catechin (C), (-)-Epicatechin (EC), (+)-Gallocatechin (GC), (-)-Epigallocatechin (EGC), their respective 3-Gallate Esters (-)-EZG, (+)-CG (11), (-)-Epicatechin-3-Gallate (ECG), Gallocatechin Gallate (GCG), and (-)-EGCG, and two 3-(3-O-methyl) gallate esters (-)-ECMG and (-)-EGCMG. Beyond that, three flavonoids namely kaempferol, quercetin and myricetin-have been identified[6]. There are wide range of bioactivities that have spurred extensive study, through several studies detailing these findings. Nonetheless, EGCG which is a primary catechin was the primary focus of the majority of these investigations due to its potential anticancer effects. The many bioactivities are not supported by such biased research. Kaempferol and quercetin are the only two examples of the many different Green Tea Polyphenolics (GTPs) that have demonstrated diverse bioactivities; these compounds may have anti-inflammatory properties[8]. In addition, GTPs frequently bind to several proteins. One such example is EGCG, which binds to numerous sites in cancer cells and is thus able to mediate various signal pathways[9]. Further, data mining was carried out. By utilising search in PubMed and Google Scholar, we were able to gather all of the research on GTP targets. Among the biological test results, only the confirmed and active targets were chosen. To find the ligands and receptors, researchers used the web server PubChem[9], based on the three Dimensional (3D) similarity procedure; to predict powerful targets, we used Keggs-Mapper, which is based on the similar property principle, which states that molecules with similar structures should have the same bioactivities[10]. Higher hybrid score indicates that the query and target molecules are more closely aligned in terms of form and chemotype identities[11]. An innovative technique called reverse docking can fit a chemical with a recognised biological function into the binding sites of every 3D structure in a specific protein database. The Mapper server is an open-source web application that uses pharmacophore mapping to find possible binding sites for small molecule probes, such as medicines, natural products, or newly found compounds without known binding site[12]. Supporting this is a database that has a wide variety of pharmacophores retrieved from all of the targets in Target Bank, DrugBank, BindingDB and Potential Drug Target Database (PDTD). As far as human protein targets go, PharmMapper has over 7000 receptor-based pharmacophore models stored which are accessible. Users can submit molecules and the programme will determine the optimal mapping postures for them. Network construction and analysis was carried. Three kinds of visual networks namely, a network of interactions between active polyphenolic compounds and their potential targets, known as the Compound-Target (C-T) network; potential targets and their associated pathways make up the Target-Pathway (T-P) network and based on the known therapeutic targets and the probable targets of polyphenols, Target-Target (T-T) network was constructed[13]. Universal Protein Resource (UniProt) database was utilised to construct the unique interaction networks for every protein. This database integrates both known and anticipated protein interactions[14]. The chosen network interactions were based on network and cytoscape (http://www. cytoscape.org/) software version 2.8.3. From the outcomes of the pathway enrichment, the interactions that made up the "target-pathway" network were chosen. The degree of a node was proportional to its size. The bioactive molecule depicted in fig. 1 was sought using PubMed, ChemDraw and other databases such as binding database Kyoto Encyclopedia of Genes and Genomes (KEGG) Mapper, for creating a network. The finding of targets for diverse components was made possible by selecting just the functional and confirmed targets from the PubMed search results. The ChemDraw employs a 3D similarity search to offer several targets, whereas the binding database provides targets together with a fit score value. Due to the presence of a common structural framework, polyphenols frequently have shared targets. Following the elimination of redundant and non-Homo sapiens targets, only Homo sapiens targets were chosen for further study. Interactions between drugs and their target molecules was studied. The intricate web of interactions between medications and their targets is illustrated in fig. 2. Proteins of interest were classified according to the diseases with which they were related, as found in the Disease and Gene Annotations (DGA) database[15]. All the targets identified were associated with cancer, which aligns with prior research findings that indicate this compound has anti-cancer properties[16]. In addition, there was a correlation between the remaining targets and diseases like diabetes, heart disease, Alzheimer's, muscle disease, mental illness and inflammation. The degree directly correlates to the size of the nodes. According to the results, most of the GTPs had multiple targets, such as 13 (kaempferol), 14 (quercetin), 6 ((-)-EGCG), and 15 (quercetin). It is possible for many polyphenols to act on the majority of targets at once. The degree of each node is a property which can be used to distinguish scale-free network topologies from random ones. The degree of distribution of nodes in a scale-free network is not uniform, in contrast to a random network where it usually follows a Poisson distribution. A scale-free structure is characteristic of most biologically-based network models. The polyphenol-modified network had a power-law distribution of node degrees. It appears that the network does not exhibit a random topology and seems to be scale-free. Pathway enrichment analysis was studied. We investigated the possible pathways because many bioactivities are affected by >1 pathway. To assess the likely pathways affected by the polyphenols, this study used pathway enrichment with the hypergeometric test; KEGG pathways were located at last. Making the protein-gene-disease pathway network confirmed the findings. The pathways, which include hsa05200, hsa05215, hsa05214, hsa04510 and hsa05218 are shown in fig. 3. In addition, alternative pathways pertinent to these diseases were selected for AML because of the historical links between green tea and cancer, diabetes, cardiovascular disease, neurological disease, muscle disease, inflammation and diabetes. Additional pathways pertinent to these disorders were selected for AML, in addition to the correlations of green tea to cancer, diabetes, cardiovascular disease, neurological conditions, muscular disease and inflammation. The anticancer properties of green tea are among its most talked-about effects. A number of studies have shown that green tea may have chemotherapeutic effects against several cancers, including myeloid leukaemia. Overall, the unique pharmacological properties of green tea, including its ability to combat myeloid leukaemia, are highly intriguing. We employed a network pharmacology methodology to analyse the complex pathways by which the polyphenolic flavonoids found in green tea exert their many biological effects. Our investigation discovered a cooperative and mutually beneficial interaction among different polyphenols, which is consistent with the comprehensive approach of network pharmacology. Through the process of mapping the network of interactions between drugs, targets, pathways and diseases, we have obtained the information on how the components of green tea can influence various targets in complicated diseases. This understanding has led to positive therapeutic result. This study not only improves our comprehension of the mechanics of green tea, but also offers vital data for future research on creating powerful medicinal drugs that target specific signalling pathways.
Funding:
This study was supported by the Shandong Province Medical and Health Technology Development plan project (Project name: The study of miR-186's effect and mechanism on AML cell resistance through targeted regulation of BMI 1 expression; No: 2019WSB35012).
Conflict of interests:
The authors declared no conflict of interests.
References
- Usman MD, Garba N. Diagnosis of acute myeloid leukemia: A Review. Bayero J Med Lab Sci 2018;3(2):171-82.
- Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD, et al. Advances in the treatment of acute myeloid leukemia: New drugs and new challenges. Cancer Discov 2020;10(4):506-25.
[Crossref] [Google Scholar] [PubMed]
- Colmegna B, Morosi L, D’Incalci M. Molecular and pharmacological mechanisms of drug resistance: An evolving paradigm. Handb Exp Pharmacol 2018;249:1-12.
[Crossref] [Google Scholar] [PubMed]
- Srijyothi L, Ponne S, Prathama T, Ashok C, Baluchamy S. Roles of non-coding RNAs in transcriptional regulation. Transcriptional and Post-transcriptional Regulation 2018;55.
- di Stefano C, Mirone G, Perna S, Marfe G. The roles of microRNAs in the pathogenesis and drug resistance of chronic myelogenous leukemia. Oncol Rep 2016;35(2):614-24.
[Crossref] [Google Scholar] [PubMed]
- Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N. MicroRNAs in cancer drug resistance: Basic evidence and clinical applications. J Cell Physiol 2019;234(3);2152-68.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Sha HH, Li HJ. Functions and mechanisms of miR-186 in human cancer. Biomed Pharmacother 2019;119:1-10.
[Crossref] [Google Scholar] [PubMed]
- Hamalainen M, Nieminen R, Vuorela P, Heinonen M, Moilanen E. Anti-inflammatory effects of flavonoids: Genistein, kaempferol, quercetin, and daidzein inhibit STAT-1 and NF-κB activations, whereas flavone, isorhamnetin, naringenin, and pelargonidin inhibit only NF-κB activation along with their inhibitory effect on iNOS expression and NO production in activated macrophages. Mediators Inflamm 2007;1-12.
[Crossref] [Google Scholar] [PubMed]
- Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (-)-epigallocatechin-3-gallate. Cancer Res 2006;66(5):2500-5.
[Crossref] [Google Scholar] [PubMed]
- MA Johnson, Maggiora GM. Concepts and applications of molecular similarity. 1990
- Lu W, Liu X, Cao X, Xue M, Liu K, Zhao Z, et al. SHAFTS: A hybrid approach for 3D molecular similarity calculation. 2. Prospective case study in the discovery of diverse p90 ribosomal S6 protein kinase 2 inhibitors to suppress cell migration. J Med Chem 2011;54(10):3564-74.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, et al. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 2010;38:609-14.
[Crossref] [Google Scholar] [PubMed]
- Li H, Liu L, Liu C, Zhuang J, Zhou C, Yang J, et al. Deciphering key pharmacological pathways of Qingdai acting on chronic myeloid leukemia using a network pharmacology-based strategy. Med Sci Monit 2018;24:5668-88.
[Crossref] [Google Scholar] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2012;41:808-15.
[Crossref] [Google Scholar] [PubMed]
- Peng K, Xu W, Zheng J, Huang K, Wang H, Tong J, et al. The Disease and Gene Annotations (DGA): An annotation resource for human disease. Nucleic Acids Res 2012;41:553-60.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Zhang DY, Zhang W, Zhao X, Yuan C, Ye F. The effect of green tea extract and EGCG on the signaling network in squamous cell carcinoma. Nutr Cancer 2011;63(3):466-75.
[Crossref] [Google Scholar] [PubMed]