- *Corresponding Author:
- D. Pal
Department of Pharmaceutical Sciences, Guru Ghasidas Vishwavidyalaya (A Central University), Koni, Bilaspur-495 009, India
E-mail: drdilip2003@yahoo.co.in
| Date of Submission | 14 November 2019 |
| Date of Revision | 23 August 2019 |
| Date of Acceptance | 14 November 2019 |
| Indian J Pharm Sci 2020;82(1):41-53 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Synthesis and investigation of antimicrobial activity of 6 novel 1,8-naphthyridine-3-carboxylic acid derivatives are presented. Among the derivatives, compounds 4a–5b2 showed a broad-spectrum antimicrobial activity against all reference Gram-positive, Gram-negative bacteria and fungi. These compounds exhibited remarkable bactericidal activity against Staphylococcus and Bacillus sps. The tested substances 4a–5b2 were found also found to be active against E. coli, Salmonella and Shigella sps. The chlorine substituted compounds 4a and 5a2 were found to be the most active towards the tested microorganisms. Compounds 4a–5b2 were found to be fungicidal against Candida sp. with a MIC values in the range of 400-2000 μg/ml. Docking studies of these compounds with Salmonella typhi OmpF complexed with ciprofloxacin using PDB- 4KRA revealed that the compounds acted as covalent crosslinker on the DNA gyrase B of the former and intercalate the latter both with higher C score values. Thus, the antibacterial activity against tested strains suggested 1,8-naphthyridine-3-carboxylic acid derivatives warrant further evaluation as potential novel antiinfective agents. The antifungal activity of thse compounds was comparable to that of griseofulvin. The drug-likeness data of synthesized compounds made them promising leads for the future development as antifungal agents.
Keywords
1,8-Naphthyridine, antimicrobial activities, antifungal action, ADMET, molecular docking
Microbes are causative agents for various types of diseases like pneumonia, amoebiasis, typhoid, malaria, common cold, cough and other infections[1]. Infectious diseases are a major health problem in the hird world countries. These diseases are treated with drugs of synthetic to natural origin including antibacterial, antiquorum sensing, antiviral, antifungal and antiparasitic agents[2,3]. Infectious diseases caused by bacterial pathogens have become the main public health problem due to extensive occurrence of drug resistance. Resistance to antimicrobial agents has increased health concerns and resulted in mortality and morbidity from treatment failures[4]. Quinolones are potent antibacterial agents used in the treatment of a wide range of bacterial infections. The history of quinolones started with the discovery of nalidixic acid in 1962. Later, first-generation quinolones such as enoxacin, lomefloxacin and norfloxacin, were discovered[5]. The second, third, and fourth-generation quinolones are called fluoroquinolones since they have a fluorine atom attached to the central ring system[6]. Although fluoroquinolones are extremely successful antibacterial agents, due to their extensive usage, fluoroquinoloneresistant bacteria have inevitably emerged. According to WHO report 2014, high rates of resistance observed in bacteria that cause common health-care-associated and community-acquired infections such as the urinary tract infection and pneumonia in all WHO regions.
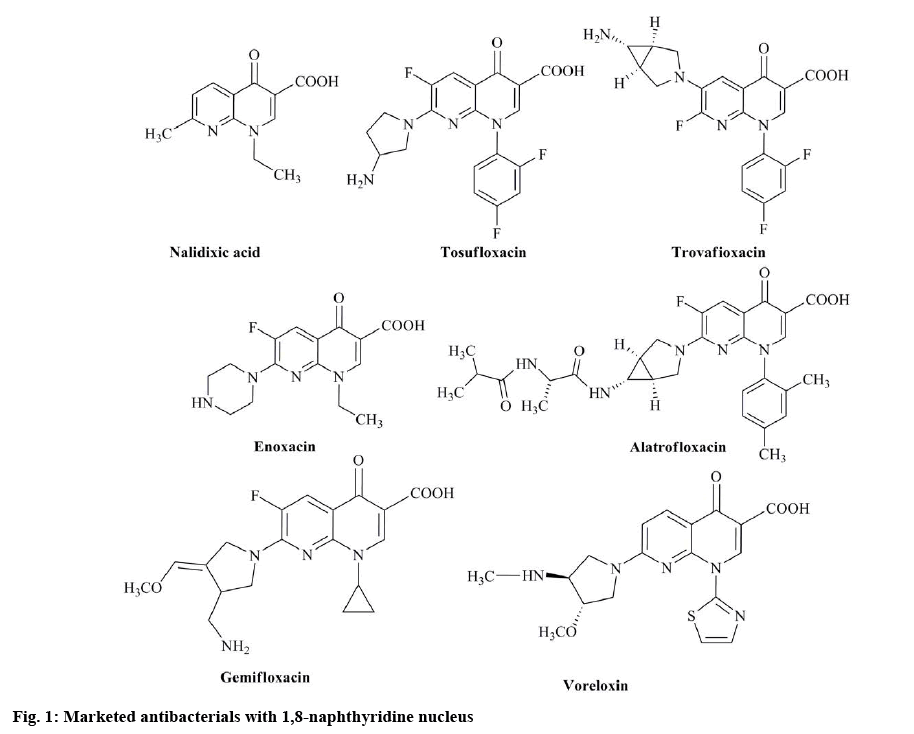
Despite their success against infectious diseases, fluoroquinolone type antibacterials have some problems. Multidrug resistance, in some cases, has been attributed to mutations leading to decreased drug permeability of fluoroquinolones[7]. In addition, the effects of quinolones such as plasmid curability and promotion of reverse and forward mutations and positive results in genotoxicity assays[8,9] suggest the possible bacterial mutagenicity of quinolones. Furthermore, the induction of DNA damage in both prokaryotic and eukaryotic cells and the induction of mutations in DNA has been reported as an essential risk for fluoroquinolones[10]. To overcome this challenge, extensive research being conducted on the synthesis of novel antibaterials. Research is also being carried out to introduce structural modifications in existing antibacterials. Based on these strategies, several drugs have been synthesized and improved[11,12]. In drug designing programs, an essential component of the search for new leads is the synthesis of molecules, which are novel yet resemble known biologically active molecules by virtue of the presence of critical structural features[13,14]. New quinolones that do not possess mutagenic potency might help eliminate some of those problematic issues. Based on the above information, a novel series of compounds in this class were synthesized with an objective to obtain effective yet non-mutagenic derivatives. The 1,8-naphthyridine nuclei are found in well-known antibacterial compounds[15] called quinolones (figure 1), which act as DNA gyrase inhibitors[16].
The 1,8-naphthyridine scaffolds have produced great interest in medicinal and organic chemistry fields. These derivatives are reported to possess a wide range of biological activities like A2A adenosine receptor ligands[17], antimycobacterial[18], A1 adenosine antagonists[19], antiinflammatory[20], cannabinoid receptor ligands[21] and antibacterial[22,23]. In the present work, efforts were made to synthesize 1,8-naphthyridine-3-carboxylic acid derivatives, which are modified at the terminal carboxylic acid end.
The synthesized compounds were evaluated for antimicrobial activity against a panel of reference strains of 21 microorganisms, including Gram-positive and Gram-negative bacteria and fungi belonging to yeasts. The microbial strains were procured from American Type Culture Collection (ATCC), routinely used for the evaluation of antimicrobials, and from clinical isolates. For comprehensive testing, molecular modeling and docking studies were performed on all compounds on the active sites often selected antimicrobial enzymes, in order to find possible targets. In the present study, synthesis, microbiological, and anticonvulsant evaluation of a group of 1,8-naphthyridine derivatives as potential hybrid compounds with antimicrobial and antifungal properties is being reported.
Materials and Methods
All experiments were carried out under air atmosphere unless stated otherwise. Reactants were generally the best quality commercial-grade products and were used without further purification. Melting points were measured with a laboratory melting point apparatus and were uncorrected. Proton nuclear magnetic resonance (1H NMR) spectra were recorded on a Bruker Avance II 400 MHz system (BrukerBioSpin AG, Fällanden, Switzerland). Chemical shifts were reported in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. The spin multiplicities were given as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet) or br (broad). MS and high-resolution mass spectra were obtained using electrospray ionization technique on a Bruker Mass spectrometry. The purity of the compounds was checked using thin layer chromatography analysis performed on silica gel GF254 purchased from Himediausing chloroform:methanol (4:1) as eluent.
Chemistry:
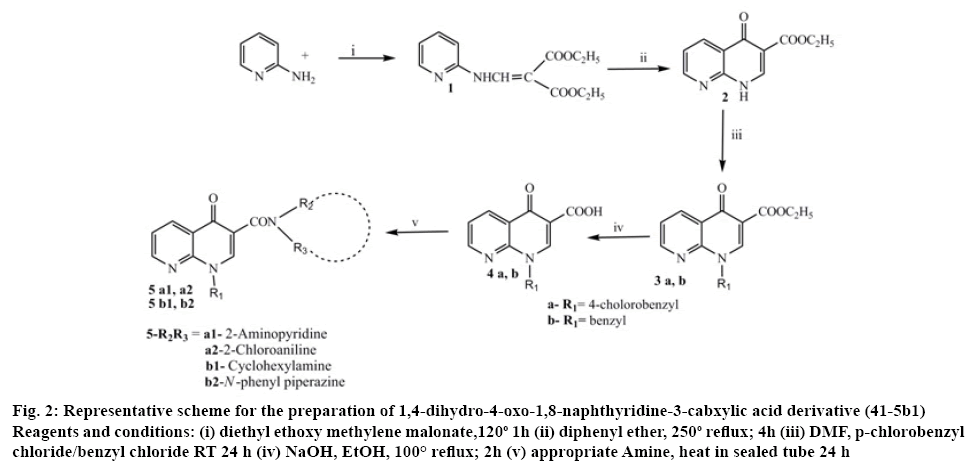
The target compounds were synthesized according to the reported method[17,19] (figure 2). 2-Aminopyridine (0.01 mol) and diethyl methoxy methylene malonate (0.01 mol) was heated at 120-130° for 2 h, the resulting ethanol was evaporated to obtain crude malonate (1), which was purified by recrystallization from light petroleum ether. Crude ester (1) 0.017 mol and diphenyl ether in access were heated at 240-250° for 1 h and the resultant solution was cooled to room temperature and washed with petroleum ether. The resulted white malonate (2) powder was collected and recrystallized from dimethylformamide. Sodium hydride, 1.81 mmol, 50 % in mineral oil and a solution of malonate (2) 1.5 mmol in 10 ml of dry dimethylformamide were stirred at room temperature. After 1 h, 4-chlorobenzyl chloride (for 3a) 0.01 mol and chlorobenzyl chloride (for 3b) 0.01 mol were added and the mixture was stirred for 24 h. The resultant solution was evaporated in vacuo and the addition of ethyl ether caused the carboxylate 3a and 3b to precipitate as a pure solid. The 1,8-naphthyridine- 3-carboxylic acid ethyl ester 3a and 3b 4.13 mmol and a mixture of 5 ml 10 % sodium hydroxide and 5 ml ethyl alcohol were refluxed for 2 h. After cooling, the solution was adjusted to pH 4 with 10 % aqueous hydrochloric acid. The resulting precipitate was filtered and washed with water. A mixture of 1,8-naphthyridine acid 4a and 4b 0.01 mol and 2-aminopyridine or 2-chloroaniline or cyclohexane amine or n-phenyl piperazine 0.1 mol heated in a sealed tube at 120° for 24 h. After cooling the reaction mixtures twas reated with ethyl ether to yield the pure titled compounds.
Figure 2: Representative scheme for the preparation of 1,4-dihydro-4-oxo-1,8-naphthyridine-3-cabxylic acid derivative (41-5b1)
Reagents and conditions: (i) diethyl ethoxy methylene malonate,120o 1H (ii) diphenyl ether, 250o reflux; 4h (iii) DMF, p-chlorobenzyl chloride/benzyl chloride RT 24 h (iv) NaOH, EtOH, 100° reflux; 2h (v) appropriate Amine, heat in sealed tube 24 h
Physicochemical characteristics of 1,8-naphthyridines (5a1, 5a2, 5b1 and 5b2):
1-(4-chlorobenzyl)-4-oxo-N-(pyridin-2-yl)-1,4- dihydro-1,8-naphthyridine-3-carboxamide (5a1); yield- 76.6 %; melting point (mp)- >300°; 1H NMR (DMSO-d6, δ, ppm): 5.32 (s, 2H CH2), 7.02 (d, 1H, H), 7.11 (d, 1H. H), 7.36 (d, 4H, Ph), 7.42 (d, 2H, H), 7.66 (d, 1H, H),7.76 (d, 2H H), 851 (1H. s), IR (νmax’ cm-1): 3112.7, 3086.0 (C-H aromatic) 1686.4 (C=O amide), 1651.1 (C=O ring), 734 (C-Cl); mass (m/z): 390.09 (M+), chemical formula: C21H15ClN4O2, molecular weight (MW) 390.82, anal: C, 64.54; H, 3.87; N, 14.34.
1-(4-chlorobenzyl)-N-(2-chlorophenyl)-4-oxo-1,4- dihydro-1,8-naphthyridine-3-carboxamide (5a2); yield- 76 %; mp- 193-195°; 1H NMR (DMSO-d6, δ, ppm): 9.80 (d,1H, NH), 9.15 (s, 1H, H2), 8.90 (dd,1H, H7), 8.67 (dd,1H, H5), 7.62 (m,1H, H6), 7.37 (m, 2H, Ph), 7.15 (m, 2H, Ph), 5.81 (s, 2H, CH2), 3.85 (m,1H, CH), ), 8.66 (d,1H, Ph), 8.60 (d, 2H, Ph), IR (νmax’ cm-1): 3112.7, 3086.0 (C-H aromatic) 1686.4 (C=O amide), 1651.1 (C=O ring), 737 (C-Cl); mass m/z 423 (M+), chemical formula: C22H15Cl2N3O2, MW 424.28, anal: C, 62.28; H, 3.56; N, 9.90.
1-benzyl-N-cyclohexyl-4-oxo-1,4-dihydro-1,8- naphthyridine-3-carboxamide (5b1); yield- 55 %; mp- 181-183°; 1H NMR (DMSO-d6, δ, ppm): 9.95 (d,1H, NH), 9.12 (s,1H, H2), 8.90 (d,1H, H7), 8.70 (d,1H, H5), 7.70 (m,1H, H6), 7.29 (m, 5H, Ar), 5.84 (s, 2H, CH2), 3.85 (m,1H, CH), 1.86-1.30 (m, 10H, cyclohexyl). IR (νmax’ cm-1): 3069.1, 2998.7 (C-H), 1714.0 (C=O keto), 1692.1 (C=O ring), 1081 (C-Cl); mass (m/z): 361 (M+), chemical formula: C22H23N3O2, MW: 361.44, anal: C, 73.11; H, 6.41; N, 11.63.
1-benzyl-3-(4-phenylpiperazine-1-carbonyl)-1,8- naphthyridin-4(1H)-one (5b2); yield- 76.6 %; mp- 147- 149°; 1H NMR (DMSO-d6, δ, ppm): 9.87 (d,1H, NH), 8.94 (s,1H, H2), 8.53 (d,1H, H5), 7.46 (d,1H, H6), 7.18 (m, 2H, Ar), 6.90-6.71 (m, 3H, Ar), 4.70 (s, 2H, CH2), 3.03 (m, 4H, piperazinyl), 2.73 (m, 2H, CH2), 2.60 (m, 4H, piperazinyl), 1.85-1.08 (m, 10H, cyclohexyl). IR (νmax’ cm-1): 3069.1, 2998.7 (C-H), 1714.0 (C=O keto), 1692.1 (C=O ring), 1081(C-Cl); mass (m/z): 424 (M+), chemical formula: C26H24N4O2, MW: 424.49, anal: C, 71.01; H, 7.45; N, 14.79.
Antimicrobial activity:
The synthesized 1,8-naphthyridine derivatives (4a-5b2) were screened in vitro for antibacterial and antifungal activities using the broth microdilution method against reference strains of microorganisms including Gramnegative bacteria, Escherichia coli NCTC 5933, E. coli K88, E. coli NCTC 7360, E. coli LT37, E. coli 872, E. coli ROW 7/12, E. coli 3:37C, E. coli CD/99/1, Salmonella typhi Ty2, S. enterica TD 01, Shigella dysentery 8, S. soneii1, S. boydii D13629, S. flexneri Type 6, Vibrio cholerae NCTC 4693, V. cholerae NCTC5596, V. cholerae NCTC 10732 and V. cholerae NCTC 11501, Gram-positive bacteria, Staphylococcus aureus ML 267, Bacillus pumilus 82 and B. subtilis ATCC 6633 and fungi, Candida albicans ATCC 10231, Aspergillus niger ATCC 6275, Penicillium notatum ATCC 11625 and P. funiculosum NCTC 287. All microbial cultures were first subcultured on nutrient agar or Sabouraud agar for bacteria and fungi, respectively[24,25].
Reference preparations of derivatives were in dimethyl sulphoxide (DMSO) at 10X concentrations and covered a full range of 50-2000 μg/ml. This method was convenient and economical for pipette use. Bacto agar was used as a medium for bacterial screening. The compounds were added to liquefied agar medium at 45-50°, mixed and poured into Petri dishes and allowed to solidify. A series of Petri plates were prepared with increasing concentrations of the test sample. With the help of inoculating applicator, as many as 4 different strains were spot inoculated on each plate. After overnight incubation, minimum inhibitory concentration (MIC) endpoint was determined by placing a plate against a dark background and observing the lowest concentration of samples inhibiting visible growth[26]. The MIC of each derivative was recorded in μg/ml, wherever two or more colonies persisted beyond the endpoint or growth was present in higher concentration and not in lower concentration, the test was repeated. The inhibition of microbial growth was judged by comparison with a control culture prepared without any sample tested. Ciprofloxacin and griseofulvin were used as a reference antibacterial and antifungal compound, respectively.
The minimal bactericidal concentration (MBC) or minimal fungicidal concentrations (MFC) are defined as the lowest concentration of the compounds that are required to kill a particular bacterial or fungal species. MBC or MFC was determined by removing the culture using for MIC determinations from each well and spotting onto the appropriate agar medium. The plates were incubated. The lowest concentrations with no visible growth observed were assessed as a bactericidal or fungicidal concentration. All experiments were repeated thrice and representative data are presented[27].
In this study, no bioactivity was defined as a MIC> 1000 μg/ml, mild bioactivity as a MIC in the range 501-1000 μg/ml, moderate bioactivity with MIC from 126 to 500 μg/ml, good bioactivity as a MIC in the range 26-125 μg/ml, strong bioactivity with MIC between 10 and 25 μg/ml and very strong bioactivity as a MIC <10 μg/ml. The MBC/MIC or MFC/MIC ratios were calculated in order to determine bactericidal/fungicidal (MBC/MIC≤4, MFC/MIC≤4) or bacteriostatic/ fungistatic (MBC/MIC>4, MFC/MIC>4) effect of the tested compounds[28].
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) and molecular docking:
The toxicological properties of these compounds was predicted using the PreADMET Toxicity server. The compounds (4a-5b2) were predicted for Caco-2 cell permeability, MDCK cell and blood-brain barrier (BBB), human intestinal absorption, skin permeability and plasma protein binding[29,30]. Compounds containing the chlorine atom exhibited improved oral absorption, skin penetration, and membrane permeability[31].
To find a better explanation of the data of microbial inhibitory potency of the synthesized compounds at a molecular level and to shed light on the structural determinants for activity, docking studies were carried out against one of the prominent microbial targets, S. typhi OmpF complex with ciprofloxacin. Molecular docking is a vital tool applied for carrying out structurebased drug design by finding interactions of small ligands in the active site of target receptor/proteins[32]. The docked ligands can be ranked on the basis of their binding affinity and docking poses in the target site. The comparison between the docked ligands and the reference ligand on the basis of their docking score can be made easily. The ligands were drawn in Chem Draw Ultra 12.0 followed by MM2 minimization of ligands (using ChemBio3D Ultra 12.0) by keeping a check on the connection error in the bonds. Ligand preparation was done by adding Gasteiger charges; the different conformation of ligands was built by allowing rotation of all torsions during docking. Protein and Grid preparation was done using Autodock tools and Autodock Vina 1.1.2[33] was used to perform molecular docking. The active site was defined to include all atoms within 6.0 Å radius of the native ligand. The first ten top-ranked docking poses were saved for each docking run. To validate the molecular docking protocol, the respective reference/ligands were initially docked into the crystal structure of the enzymes.
Results and Discussion
The proposed 1,8-naphthyridine-3-carboxylic acid derivatives were prepared via multi-step reactions and the synthetic process was outlined in figure 2.
The intermediate ethyl 4-oxo-1,4-dihydro-1,8- naphthyridine-3-carboxylate (2) was prepared by refluxing malonate in diphenyl ether. The carboxylate, ethyl 1-(4-chlorobenzyl)-4-oxo-1,4-dihydro-1,8- naphthyridine-3-carboxylate and ethyl 1-benzyl-4-oxo- 1,4-dihydro-1,8-naphthyridine-3-carboxylate (3a and 3b) were prepared by refluxing 1,8-naphthyridine-3- carboxylate and p-chlorobenzyl chloride and benzyl chloride, respectively. Acid hydrolysis of 1,8-naphthyridine-3-carboxylate gave 1-substituted1,8- naphthyridine-3- carboxylic acids (4a and 4b). The title compounds were synthesized by heating the appropriate amine and N1-substituted1,8-naphthyridine-3- carboxylic acids in a sealed tube. The title compounds were obtained in fair to good yields.
All new compounds were characterized using IR, 1H NMR and MS spectra. Their spectral analyses were consistent with the assigned structures and listed in the experimental section. The mass spectra for compounds gave a major fragment of [M+H]+ according to their molecular formula. As per the spectral data of compound, FTIR and 1H NMR showed the presence of characteristic peak at 1650-1692 and 3300 cm−1 (for CO and -NH stretching), 3100-3000 cm−1 (phenyl group), and 1380 cm−1 (for ether linkage) and δ around 9.87, 7.18, 4.70, 3.03, 2.60, and 1.85-1.08 ppm due to existence of aromatic. -NH, aromatic. -CH, piperazinyl, and cyclohexyl. In the case of compounds 3–5 as per the 1H NMR spectroscopy, spectral data at 8.70 and 8.90 ppm indicated the presence of p-chlorobenzyl and benzyl moiety. Further, the structures were established by mass spectral data in accordance with their molecular formula.
All synthesized compounds were screened for antibacterial and antifungal activities in an agar diffusion well method[34] and it was found that the 1,8-naphthyridines 4a-5b2, exhibited significant antibacterial and antifungal activity against both Grampositive and Gram-negative bacteria and fungal strains at various concentrations as shown in Tables 1 and 2. The agar diffusion assay method indicated that bacterial growth was inhibited by 1,8-naphthyridine derivatives to produce concentration-dependent inhibition zones[35].
Table 1: Comparison of zones of inhibition of compounds 4a-5b2
| Compounds | S. aureus ML 267 | B. pumilus 82 | B. subtilis ATCC 6633 | |
|---|---|---|---|---|
| 4a | 13.5, 13.5, 13.5 | 6.0, 6.0, 6.0 | 6.0, 6.0, 6.0 | |
| 4b | 14.0, 13.5, 13.0 | 6.0, 6.0, 6.5 | 6.0, 6.5, 6.0 | |
| 5a1 | 13.5, 14.5, 15.5 | 6.0, 6.0, 6.0 | 6.0, 6.0, 6.0 | |
| 5a2 | 14.5, 15.0, 14.0 | 7.0, 6.5, 7.0 | 7.0, 6.0., 6.5 | |
| 5b1 | 14.0, 14.5, 15.5 | 6.0, 6.0, 6.0 | 6.0, 6.0, 6.0 | |
| 5b2 | 13.5, 11.5, 12.0 | 6.0, 6.0, 6.0 | 6.0, 6.0, 6.0 | |
| Ciprofloxacin | 18.0, 18.0, 18.0 | 19.0, 18.0,19.5 | 18.0, 18.5, 17.5 | |
| Compounds | C. albicans ATCC 10231 | A. niger | ATCC 6275 | P. notatum ATCC 11625 |
| 4a | 14.5 | 12.5 | 12.5 | 13.5 |
| 4b | 14.0 | 12.0 | 12.5 | 13.0 |
| 5a1 | 11.5 | 11.0 | 10.0 | 9.5 |
| 5a2 | 10.0 | 9.0 | 8.0 | 8.0 |
| 5b1 | 13 | 12 | 12.5 | 13.5 |
| 5b2 | 12.5 | 11 | 12 | 12.5 |
| Griseofulvin | 16.0 | 15.0 | 13.5 | 14.0 |
Diameters of zones of inhibition in mm around discs of diameter 6 mm at a concentration of 200 µg/ml. Diameters of zones of inhibition in mm around discs of diameter 6 mm ata concentration of 2000 µg/ml of coded drug in saborauds dextrose agar media
Table 2: Comparison of zones of inhibition of compounds 4a-5b2 (gram –ve)
| Comp. | E. coli NCTC 5933 | E. coli K88 | E. coli NCTC 7360 | E. coli LT37 | E. coli 872 | E. coli ROW 7/12 | E. coli 3:37C | E. coli CD/99/1 | S. typhi Ty2 |
|---|---|---|---|---|---|---|---|---|---|
| 4a | 15, 15, 15 | 14.5,14.5,16.5 | 15.5,15.5,15.5 | 14.5,14.6,14.4 | 14.5,14.5,14.5 | 15,15,15 | 15,15,15 | 15,14,14.5 | 15,15,15 |
| 4b | 14.5,14.5,14.5 | 15.5,15.5,16 | 14.5,14.5,14.5 | 14.5,15.5,14 | 14.5,15.5,13.5 | 15,14.,16. | 15,15,15 | 15.0,14,16 | 14,14,14.5 |
| 5a1 | 15,15,15 | 15.5,15.5,15 | 15.5,15.5,15.5 | 14.5,13.5,15.5 | 14.5,14.5,14.5 | 15,15,15 | 15,14.5,14.5 | 15,15,15 | 15,14,16 |
| 5a2 | 15.5,15.5,15.5 | 16.5,15.5,14.5 | 16.5,15.5,15 | 15.5,15.5,15 | 15.15,15 | 15.5,15.5,15.5 | 15.0,14.0, 5.5 | 15,16,14.5 | 15,15,15 |
| 5b1 | 15,15,15 | 15.5,15,15 | 15.5,15.5,15.5 | 15.5,13.5,15.5 | 15.5,14.5,14.5 | 16,15,15 | 15.5,14.5,14.5 | 15.,15,15 | 12,12.5,13 |
| 5b2 | 15, 15, 15 | 14.5,14.5,16.5 | 15.5,15.5,15.5 | 14.5,14.6,14.4 | 14.5,14.5,14.5 | 15,15,15 | 15.0,15.0,15.0 | 15.0,14.0,14.5 | 12,12.5,12.5 |
| Ciprofloxacin | 16, 17, 15 | 17, 17, 17 | 16, 18, 17. | 16, 16, 16 | 16., 15., 16.5 | 16.5, 16.5, 16.5 | 16.5, 17.5, 15.5 | 17.0, 16., 17.5 | 16, 16, 16. |
| Comp. | S. enterica TD 01 | S. dysentery 8 | S. soneii 1 | S. boydii D13629 | S. flexneri Type 6 | V. cholerae NCTC 4693 | V. cholerae NCTC5596 | V. cholerae NCTC 10732 | V. cholerae NCTC 11501 |
| 4a | 16,17,15 | 13.5,13.5,13.5 | 16.5,16.5,16 | 15.5,15.5,15.5 | 15.5,16,15. | 14.5,14,15 | 15,15,16 | 15.5,14.5,16.5 | 15.5,14.5,16.5 |
| 4b | 13.5,14.5,12.5 | 13.5,14.5,12.5 | 14.5,15.5,13.5 | 15.0,15.5,16. | 15.5,15.5, 15.5 | 14.5,14.5,15.5 | 15,15,15 | 15.5,16.5,14.5 | 15.5,15.5,15.5 |
| 5a1 | 15,14,16 | 14,4,14 | 15.5,15.5,15.5 | 15.5,16.5,14.5 | 15.5,16.5,14.5 | 14.5,15.5,13.5 | 15,15,15 | 15.5,14.5,16.5 | 15.5,16.5,14.5 |
| 5a2 | 16.5,16.5,16 | ,14.5,14.5,14.5 | 15,15,14.5 | 15,15,15 | 15,14,14.5 | 13.5,14.5,12.5 | 14, 13.5, 14.5 | 14, 3.5, 14.5 | 14.5, 14.5, 14.5 |
| 5b1 | 15,14,16 | 14,14,14 | 16,16.5,16 | 15.5,16.5,14.5 | 15.5,16.5,14.5 | 14.5,15.5,13.5 | 15,15,15 | 15.5,14.5,16.5 | 15.5,16.5,14.5 |
| 5b2 | 16,17,15. | 13,11.5,12.5 | 16.5,16.5,16 | 15.5,15.5,15.5 | 15.5,16,15 | 14.5,14,15 | 15,15,16 | 15.5,14.5,16.5 | 15.5,14.5,16.5 |
| Ciprofloxacin | 19.0, 19.5, 18.5 | 20, 20, 20 | 19.5, 19.5, 19.5 | 20, 20, 20 | 20.5, 20.5, 20.5 | 17.5, 17.5, 17.5 | 18.5,18, 19.5 | 18.5, 19, 19.5 | 18.5,18, 19 |
Diameters of zones of inhibition in mm around discs of diameter 6 mm at a concentration of 200 µg/ml
The bacterial inhibition zones produced by the synthesized 1,8-naphthyridine derivatives ranged between 14-20 mm. The degree of the antibacterial activities of the synthesized compounds were also assayed by serial double dilution method to determine the MIC. Minimum concentrations, which inhibited the growth of these microorganisms ranged from 5-8 μg/ml for both Gram-positive and Gram-negative bacteria (Tables 3 and 4).
Table 3: Determination of MIC of the samples 4a-5b2
| Comp. | Concentrations of compounds (µg/ml) | S. aureus ML 267 | B. pumilus 82 | B. subtilis ATCC 6633 | Comp. | Growth in nutrient agar with different concentrations of compounds (µg/ml) | S. aureus ML 267 | B. pumilus 82 | B. subtilis ATCC 6633 |
|---|---|---|---|---|---|---|---|---|---|
| 4a | 0* | + | + | + | 5a2 | 0* | + | + | + |
| 5 | + | + | + | 5 | + | + | + | ||
| 10 | + | + | + | 10 | + | + | + | ||
| 25 | + | + | + | 25 | + | + | + | ||
| 50 | + | + | + | 50 | + | + | + | ||
| 100 | - | + | + | 100 | - | + | + | ||
| 200 | - | + | + | 200 | - | + | + | ||
| 400 | - | + | + | 400 | - | + | + | ||
| 800 | - | + | + | 800 | - | - | - | ||
| 4b | 0* | + | + | + | 5b1 | 0* | + | + | + |
| 5 | + | + | + | 5 | + | + | + | ||
| 10 | + | + | + | 10 | + | + | + | ||
| 25 | + | + | + | 25 | + | + | + | ||
| 50 | + | + | + | 50 | + | + | + | ||
| 100 | - | + | + | 100 | + | + | + | ||
| 200 | - | + | + | 200 | + | + | + | ||
| 400 | - | + | + | 400 | - | + | + | ||
| 800 | - | + | + | 800 | - | + | + | ||
| 5a1 | 0* | + | + | + | 5b2 | 0* | + | + | + |
| 5 | + | + | + | 5 | + | + | + | ||
| 10 | + | + | + | 10 | + | + | + | ||
| 25 | + | + | + | 25 | + | + | + | ||
| 50 | - | + | + | 50 | + | + | + | ||
| 100 | - | + | + | 100 | + | + | + | ||
| 200 | - | + | + | 200 | + | + | + | ||
| 400 | - | + | + | 400 | - | + | + | ||
| 800 | - | + | + | 800 | - | + | + |
‘0*’ Control (sterile DMSO); ‘+’growth; ‘-’ no growth; ‘±’ inhibited growth
Table 4: Determination of mic of the samples 4a-5b2
| Comp. | Concentrations of compounds (µg/ml) | E. coli NCTC 5933 | E. coli | E. coli NCTC 7360 | E. coli | E. coli | E. coli ROW 7/12 | E. coli | E. coli | S. typhi Ty2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| K88 | LT37 | 872 | 3:37C | CD/99/1 | |||||||
| 4a | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | - | - | - | - | - | - | - | - | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 4b | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | - | + | - | - | - | - | - | + | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5a1 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | + | + | - | - | - | - | - | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5a2 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | + | + | + | + | + | + | + | + | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5b1 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | + | + | + | + | + | + | + | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5b2 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | - | - | - | - | - | - | - | - | + | ||
| 25 | - | - | - | - | - | - | - | - | + | ||
| 50 | - | - | - | - | - | - | - | - | + | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| Comp | Concentrations of compounds (µg/ml) | S. enterica | S. dysentery 8 | S. soneii 1 | S. boydii D13629 | S. flexneri Type 6 | V. cholerae NCTC 4693 | V. cholerae NCTC | V. cholerae NCTC 10732 | V. cholerae NCTC 11501 | |
| TD 01 | 5596 | ||||||||||
| 4a | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | - | + | + | + | + | - | - | - | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 4b | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | + | + | + | + | - | - | - | - | ||
| 50 | + | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5a1 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | - | + | + | + | - | - | - | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5a2 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | + | + | + | + | + | + | + | + | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5b1 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | + | - | - | + | + | - | - | - | - | ||
| 50 | - | - | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
| 5b2 | 0* | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | + | + | ||
| 10 | + | + | + | + | + | + | + | + | + | ||
| 25 | - | + | + | + | + | - | - | - | - | ||
| 50 | - | + | - | - | - | - | - | - | - | ||
| 100 | - | - | - | - | - | - | - | - | - | ||
| 200 | - | - | - | - | - | - | - | - | - | ||
| 400 | - | - | - | - | - | - | - | - | - | ||
| 800 | - | - | - | - | - | - | - | - | - | ||
‘0*’ Control (sterile DMSO); ‘+’growth; ‘-’ no growth; ‘±’ inhibited growth
Compound 4a with para-chlorobenzyl at C1 was most active against E. coli, Vibrio, Salmonella, moderately active against. S. aureus, Shigella sp. and resistant against Bacillus sp. 4b was most active against E. coli, Vibrio, moderately active against S. aureus, Shigella sp, Salmonella and resistant against Bacillus sp. 5a1 was most active against E. coli, Vibrio, moderately active against S. aureus, Shigella sp., Salmonella and resistant against Bacillus sp. 5a2 with chlorine substituted aniline was most active against Salmonella, Shigella sp., Vibrio sp., E. coli and moderately sensitive to, Bacillus sp. 5b1 was most active against E. coli, Vibrio, moderately active against S. aureus, Shigella sp., Salmonella and resistant against Bacillus sp. 5b2 was most active against E. coli, Vibrio, Salmonella, moderately active against S. aureus, Shigella sp. and resistant against Bacillus sp. The all tested compounds had no activity towards B. pumilus 82 and B. subtilis ATCC 6633. On the basis of MIC values, it was shown also that tested compounds have a wide spectrum of anticandidal activity (Table 5). The fungicidal effects of compounds were very strong or strong against Candida sp. with MIC of 50-2000 μg/ml.
Table 5: Determination of MICS of the samples 4a-5b
| Comp. | Different concentrations of compounds (µg/ml) | C. albicans ATCC 10231 | A. niger | ATCC 6275 | P. notatum ATCC 11625 | Comp. | Different concentrations of compouns (µg/ml) | C. albicans ATCC 10231 | A. niger | ATCC 6275 | P. notatum ATCC 11625 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4a | 0* | + | + | + | + | 5a2 | 0* | + | + | + | + |
| 50 | + | + | + | + | 50 | + | + | + | + | ||
| 100 | + | + | + | + | 100 | + | + | + | + | ||
| 200 | + | + | + | + | 200 | + | + | + | + | ||
| 400 | - | + | - | - | 400 | + | + | + | + | ||
| 800 | - | + | - | - | 800 | - | + | + | + | ||
| 1000 | - | - | - | - | 1000 | - | + | - | - | ||
| 1500 | - | - | - | - | 1500 | - | - | - | - | ||
| 2000 | - | - | - | - | 2000 | - | - | - | - | ||
| 4b | 0* | + | + | + | + | 5b1 | 0* | + | + | + | + |
| 50 | + | + | + | + | 50 | + | + | + | + | ||
| 100 | + | + | + | + | 100 | + | + | + | + | ||
| 200 | + | + | + | + | 200 | + | + | + | + | ||
| 400 | - | + | + | + | 400 | + | + | + | + | ||
| 800 | - | + | - | - | 800 | - | - | + | + | ||
| 1000 | - | - | - | - | 1000 | - | - | - | - | ||
| 1500 | - | - | - | - | 1500 | - | - | - | - | ||
| 2000 | - | - | - | - | 2000 | - | - | - | - | ||
| 5a1 | 0* | + | + | + | + | 5b2 | 0* | + | + | + | + |
| 50 | + | + | + | + | 50 | + | + | + | + | ||
| 100 | + | + | + | + | 100 | + | + | + | + | ||
| 200 | + | + | + | + | 200 | + | + | + | + | ||
| 400 | - | + | + | + | 400 | + | + | + | + | ||
| 800 | - | + | - | - | 800 | + | + | + | + | ||
| 1000 | - | - | - | - | 1000 | - | - | - | - | ||
| 1500 | - | - | - | - | 1500 | - | - | - | - | ||
| 2000 | - | - | - | - | 2000 | - | - | - | - |
‘0*’ Control (sterile DMSO); ‘+’growth; ‘-’ no growth; ‘±’ inhibited growth
The toxicity was predicted using the PreADMET Toxicity server and the result shown in Table 6. The compounds were found non-mutagenic. The compounds were predicted to give negative Ames test and these parameters indicated that the synthesized compounds were safe. Also, the hERG inhibition prediction indicated medium cardiotoxic potential. According to the results of lipophilicity-activity relationship analysis, log P is not the main factor that influences cholinesterase inhibitory activity of this set of compounds. However, the lack of significant correlation does not diminish the importance of determined ClogP values, as log P is an essential physicochemical parameter related to drug intestinal absorption and BBB permeation. The lipophilicity of these compounds is within the range defined by the rule-of-five, thus predicting good intestinal permeability. The lipophilicity indicated that ClogP values ranged from 0.73 to 4.09. The chlorine atom at position-2 of the 1-(4-chlorobenzyl)-N-(2- chlorophenyl)-4-oxo-1,4-dihydro-1,8-naphthyridine- 3-carboxamide (5a2) core was enhanced the overall hydrophobic nature and this might have enabled these molecules to penetrate the complex mycobacterial cell wall. As the target molecules’ values are less than 5, it indicated a reasonable probability that these compounds would be well absorbed.
Table 6: Prediction of ADMET properties of compounds (4a-5b2)
| SN ADME/T properties | Compounds | |||||
|---|---|---|---|---|---|---|
| 4a | 4b | 5a1 | 5a2 | 5b1 | 5b2 | |
| In vivo blood-brain barrier penetration (C. brain/C. blood | 0.34 | 0.74 | 0.05 | 0.08 | 0.35 | 0.05 |
| Water solubility in buffer system (SK atomic types, mg/l) | 2.9 | 0.16 | 0.38 | 0.65 | 0.42 | 21.56 |
| in vitro Caco-2 cell permeability (nm/sec) | 23.13 | 24.63 | 49.40 | 26.32 | 31.014 | 20.21 |
| Human intestinal absorption (HIA, %) | 95.94 | 94.54 | 97.52 | 96.34 | 97.13 | 93.83 |
| in vitro MDCK cell permeability (nm/sec) | 34.26 | 12.65 | 0.39 | 5.96 | 0.38 | 61.28 |
| in vitro plasma protein binding (%) | 87.45 | 86.98 | 95.12 | 99.324 | 92.82 | 77.54 |
| in vitro skin permeability (logKp, cm/h) | -3.56 | -3.29 | -2.94 | -3.94 | -2.89 | -4.16 |
| Ames test | mutagen | mutagen | mutagen | mutagen | mutagen | mutagen |
| in vitro hERG inhibition | medium risk | medium risk | medium risk | medium risk | medium risk | medium risk |
ADMET prediction carried out by using preADMET server
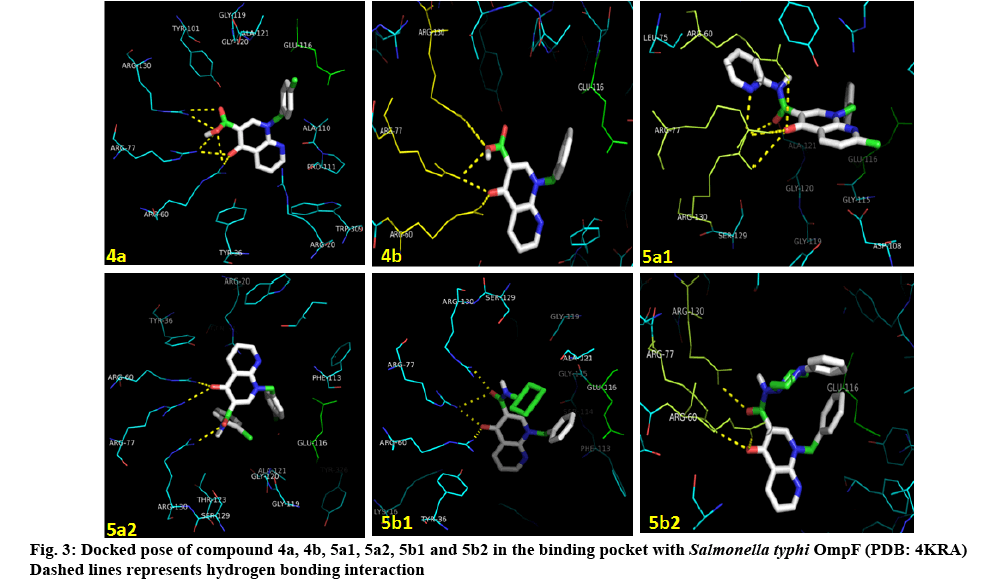
Molecular docking studies were carried out to understand the binding profile of synthesized 1,8-naphthyridine derivatives (4a-5b2) and to support their in vitro antimicrobial activity. One of the established targets for antimicrobial therapy is DNA gyrase. The fluoroquinolone class of antimicrobial agents act through inhibiting DNA gyrase. DNA gyrase binding pocket for fluoroquinolones is available from recently reported data (protein crystal structure complexes) originating from different microorganisms (S. pneumoniae topo IV-DNALFX (levofloxacin) [pdb code: 3K9F], A. baumaniitopo IV-DNAMOX (moxifloxacin) [pdb code: 2XKK] and S. typhi OmpF complex with ciprofloxacin [pdb code: 4KRA]). Among these structures, S. typhi OmpF complex with ciprofloxacin[36] was used in our docking studies. Automated docking was used to determine the orientation of inhibitors bound in the active site. The docking of series of ligands indicated that all the synthesized compounds have the potential of binding with one or the other amino acids in the active pocket as evident from the docking scores provided in (Table 7). The docked ligands were found to have similar binding poses to the co-crystallized ligands. Theoretically, the entire synthesized compounds showed moderate to good binding scores ranging from –7.1 to –8.4 kcal/ mol. Most of the ligands are showing better docking scores in comparison to nalidixic acid, which is used as a reference for biological interpretations. Among these compounds, 5b1 show highest docking score of -8.4 kcal/mol (figure 3).
Table 7: clogP values and docking score
| Compounds ClogP Docking score kcal/mol | ||
|---|---|---|
| 4a | 3.51 | -7.1 |
| 4b | 3.74 | -7.5 |
| 5a1 | 1.59 | -7.4 |
| 5a2 | 3.63 | -7.7 |
| 5b1 | 4.09 | -8.4 |
| 5b2 | 0.99 | -7.2 |
| Ciprofloxacin | -0.73 | -10.5 |
According to the obtained data from docking studies, it could be concluded that 5b1 might act as an effective antimicrobial agent similar to ciprofloxacin. But, further experimental investigation procedures are needed to confirm this prediction. Compound 4a, 4b,and 5a1 were active against E. coli, Vibrio, Salmonella. Compound 5a2 was active against Salmonella, Shigella, Vibrio sps., and E. coli. The fungicidal effects of compounds were either very strong or strong against Candida sp. Compounds showed moderate to good binding scores. Most of the ligands have shown better docking scores in comparison to nalidixic acid.
Acknowledgments:
The authors acknowledge the SAIF, Punjab University, Chandigarh for performing NMR and Mass spectra of the synthesized compounds.
Conflicts of interest:
There are no conflicts of interests in this manuscript.
Financial support and sponsorship:
Nil.
References
- Black JG. Microbiology: Principles and Applications. New Jersey: Prentice-Hall, Englewood Cliffs; 1993. p. 360.
- Pelczar MJ, Chan ECS, Krieg NR. Microbiology. New Delhi: Tata Mcgraw–Hill Publishing Company Ltd.; 2005. p. 3.
- Singh S, Wanjari PJ, Bhatia S, Sonwane VC, Chakraborti AK, Bharatam PV. Design synthesis, biological evaluation and toxicity studies of N, N–disubstituted biguanides as quorum sensing inhibitors. Med Chem Res 2015;24:1974-87.
- Zha GF, Leng J, Darshini N, Shubhavathi T, Vivek HK, Asiri AM, et al. Synthesis, SAR and molecular docking studies of benzo[d]thiazole-hydrazones as potential antibacterial and antifungal agents. Bioorg Med Chem Lett 2017;27:3148-55.
- Ball P. Quinolone generations: natural history or natural selection? J Antimicrob Chemother 2000;46:17-24.
- Oliphant CM, Green GM. Quinolones: a comprehensive review. Am Fam Physician 2002;65:455-64.
- Bisacchi GS. Origins of the quinolone class of antibacterials: An expanded "Discovery Story". J Med Chem 2015;58:4874-82.
- Chugunova E, Akylbekov N, Bulatova A, Gavrilov N, Voloshina A, Kulik N, et al. Synthesis and biological evaluation of novel structural hybrids of benzofuran derivatives and fluoroquinolones. Eur J Med Chem 2016;116:165-72.
- Thompson LA, Ellman JA. Synthesis and applications of small molecule libraries. Chem Rev 1996;96:555-600.
- Andriote VT. The 1998 Garrod lecture. Current and future antifungal therapy: new targets for antifungal agents. J Antimicrob Chemother 1999;44:151-62.
- Mamber SW, Kolek B, Brookshire KW, Bonner DP, Fung-Tomc J. Activity of quinolones in the Ames Salmonella TA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob Agents Chemother 1993;37:213-19.
- Albertini S, Chételat AA, Miller B, Muster W, Pujadas E, Strobel R, Gocke E. Genotoxicity of 17 gyrase- and four mammalian topoisomerase II-poisons in prokaryotic and eukaryotic test systems. Mutagenesis 1995;10:343-51.
- Yim G, McClure J, Surette MG, Davies JE. Modulation of Salmonella gene expression by subinhibitory concentrations of quinolones. J Antibiot 2011;64:73-80.
- Fort FL. Mutagenicity of quinolone antibacterials. Drug Saf 1992;7(3):214-22.
- Saleh MY, Ayoub AI. Synthesis of new derivatives of 2?chloro?3?formyl? 1,8?naphthyridine. Eur J Chem 2014;5:475-80.
- Smith J. The mode of action of 4-quinolones and possible mechanisms of resistance. J Antimicrob Chemother 1986;18(Suppl D):21-9.
- Gurjar VK, Pal D. Design, synthesis, biological evaluation, and in silico ADMET studies of 1,8-naphthyridine derivatives as an H1-receptor inhibitor. Int J Pharm Biol Arch 2018;9:265-73.
- Manera C, Betti L, Cavallini T, Giannaccini G, Martinelli A, Ortore G, et al. 1,8-Naphthyridine-4-one derivatives as new ligands of A2A adenosine receptors. Bioorg Med Chem Lett 2005;15:4604-10.
- Ferrarini PL, Manera C, Mori C, Badawneh M, Saccomanni G. Synthesis and evaluation of antimycobacterial activity of 4-phenyl-1,8-naphthyridine derivatives. Il Farmaco 1998;53:741-6.
- Dianzani C, Collino M, Gallicchio M, Braccio MD, Roma G, Fantozzi R. Effects of anti-inflammatory [1, 2, 4]triazolo[4, 3-a] [1,8]naphthyridine derivatives on human stimulated PMN and endothelial cells: an in vitro study. J Inflam 2006;3:4.
- Ferrarini PL, Calderone V, Cavallini T, Manera C, Saccomanni G, Pani L, et al. Synthesis and biological evaluation of 1,8-naphthyridine-4(1H)-on-3-carboxamide derivatives as new ligands of cannabinoid receptors. Bioorg Med Chem 2004;12:1921-33.
- Lv K, Liu ML, Feng LS, Sun LY, Sun YX, Wei ZQ, et al. Synthesis and antibacterial activity of naphthyridine derivatives containing mono/difluoro-methyloxime pyrrolidine scaffolds. Eur J Med Chem 2012;47:619-25.
- Debnath A, Mogha NK, Masram DT. Metal complex of the first-generation quinolone antimicrobial drug nalidixic acid: structure and it's biological evaluation. Appl Biochem Biotechnol 2015;175:2659-67.
- Isenberg HD. Clinical Microbiology Procedures Handbook. Vol. 1. Washington, DC:American Society for Microbiology; 1992.
- Schwalve R, Moore LS, Goodwin AC. Antimicrobial Susceptibility Testing Protocols. Boca Raton, Florida, United States: CRC Press; 2007.
- Grover G, Kini SG. Synthesis and evaluation of new quinazolone derivatives of nalidixic acid as potential antibacterial and antifungal agents. Eur J Med Chem 2006;41:256-62.
- Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protocol 2008;3:163-75.
- O’Donnell F, Smyth TJ, Ramachandran VT, Smyth WF. A study of the antimicrobial activity of selected synthetic and naturally occurring quinolones. Int J Antimicrob Agents 2010;35:30-38.
- Clark DE. In silico prediction of blood-brain barrier permeation. Drug Discov Today 2003;8:927-33.
- Didziapetris R, Japertas P, Avdeef A, Petrauskas A. Classification analysis of P-glycoprotein substrate specificity. J Drug Target 2003;11:391-406.
- Gentry CL, Egleton RD, Gillespie T, Abbruscato TJ, Bechowski HB, Hruby VJ, et al. The effect of halogenation on blood-brain barrier permeability of a novel peptide drug. Peptides 1999;20:1229-38.
- Shoichet BK, McGovern, SL, Wei B, Irwin JJ. Lead discovery using molecular docking. Curr Opin Chem Biol 2002;6:439-46.
- Trott O, Olson AJ. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31:455-61.
- Borman AM, Fraser M, Palmer MD, Szekely A, Houldsworth M, Patterson Z, et al. MIC distributions and evaluation of fungicidal activity for amphotericin B, itraconazole, voriconazole, posaconazole and caspofungin and 20 species of pathogenic filamentous fungi determined using the CLSI broth microdilution method. J Fungi 2017;3:27.
- Cappuccino JG, Sherman N. Microbiology: a laboratory manual. California: Benjamin-Cummings Pub Co.; 1999.
- Madhuranayaki T, Balasubramaniam D, Krishnaswamy S. Salmonella typhi OmpF complex with Ciprofloxacin. 2014. Available from: https://www.wwpdb.org/pdb?id=pdb_00004kra.