- *Corresponding Author:
- M. Tiwari
Department of Pharmacy, Laboratory of Medicinal and Pharmaceutical Chemistry, Shri G. S. Institute of Technology and Science, Indore, Madhya Pradesh 452003, India
E-mail: drmeenatiwari@gmail.com
| Date of Received | 31 July 2021 |
| Date of Revision | 10 June 2023 |
| Date of Acceptance | 15 September 2023 |
| Indian J Pharm Sci 2023;85(5):1329-1343 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To develop new and potent therapies against human immunodeficiency virus infections, a series of 3-acetyl- 2H-1-benzopyran-2-one derivatives (2a-n) were synthesized and evaluated for lens epithelium-derived growth factor/p75-dependent human immunodeficiency virus type-1 integrase inhibition activity. Knoevenagel reaction of o-hydroxy-benzaldehyde with a stable enolate initially formed a diester, which after transesterification formed 1 (3-acetyl-2H-1-benzopyran-2-one) as an intermediate. Claisen-Schmidt condensation of 1, with substituted bezaldehydes, lacking an alpha-hydrogen, using silica sulphuric acid as a catalyst formed 2an. In silico absorption, distribution, metabolism and excretion parameters of 2a-n were calculated, which were found well within their reference limits. In silico toxicity risk assessment showed that all synthesized compounds possess low toxicity risks for irritant effect, mutagenicity, reproductive effect and tumorigenicity. Amongst all synthesized compounds, 2a, 2g and 2h showed human immunodeficiency virus type-1 integrase inhibition with half-maximal inhibitory concentration value of 86.5, 90.0 and 98.5 μM respectively. Structure activity relationship showed that electron-withdrawing group at phenyl ring, attached to the benzopyran-2- one nucleus was important for human immunodeficiency virus type-1 integrase inhibition activity. Molecular docking study showed that the binding pattern of 2a-n in the binding pocket of human immunodeficiency virus type-1 integrase is similar to that of raltegravir. The findings of the present study may help in design and development of potent human immunodeficiency virus type-1 integrase inhibitors as novel antiretroviral agent.
Keywords
3-acetyl-2H-1-benzopyran-2-one synthesis, in silico absorption, distribution, metabolism and excretion, lens epithelium-derived growth factor-dependent human immunodeficiency virus type-1 integrase inhibition
Acquired Immunodeficiency Syndrome (AIDS) is the late-stage Human Immunodeficiency Virus type-1 (HIV-1) infection in which body's immune system severely damaged by HIV-1. Globally, 38.0 million people were living with HIV infection and 1.7 million people became newly infected with HIV by the end of year 2019[1]. Development of viral resistance against currently available antiretroviral drugs is the major cause of treatment failure. However, it provides an opportunity to scientific community to develop novel antiretroviral agents.
Human Immunodeficiency Virus type-1 Integrase (HIV-1 IN) is a key enzyme of HIV-1 life cycle and an important target for antiretroviral therapy[2]. Three United States Food and Drug Administration approved HIV-1 IN inhibitors namely, raltegravir, elvitegravir and dolutegravir are being used in the clinic while several others are in different stages of clinical trials[3]. HIV-1 IN inhibitors have high therapeutic index as IN enzyme has no counterpart in the host cell thus, do not interfere with normal cellular processes[4]. POL gene of HIV encodes IN, which belongs to the family of polynucleotidyl transferases[5]. It is a 32 kDa protein comprising of 288 amino acids and responsible for transfer of virally encoded deoxyribonucleic acid (DNA) into the host chromosome[6-8]. The integration process requires specific sequences at U3 and U5 termini of the viral DNA[9]. IN cleaves a dinucleotide from 3’ end of each terminus of the reverse-transcribed viral DNA and produces two sticky ends (i.e. reactive 3-hydroxyl ends), this process is known as “3’-processing”, while Strand Transfer (ST) is a transesterification reaction involving a nucleophilic attack of these reactive 3-hydroxyl groups of viral DNA on the phosphodiester backbone of host DNA[10]. Removal of unpaired dinucleotide from the 5'-ends of the viral DNA, repair of the singlestranded gaps created between the viral and target DNA and ligation of 3'-ends to 5'-ends of the host DNA completes the HIV integration process[9,10]. Divalent metals, Mg2+ or Mn2+, played crucial role in 3'-processing and ST steps of IN[11].
Lens Epithelium-Derived Growth Factor (LEDGF/ p75) is a transcriptional co-activator, identified as a cellular cofactor of HIV-1 IN[12]. LEDGF/ p75 directly interacts with IN at C-terminal region through the IN binding domain and promotes viral integration by attaching the pre-integration complex to host chromatin[13]. Now, LEDGF/p75- IN interaction has been validated as a target for antiretroviral therapy and resulted in development of inhibitors with dual mechanism of action i.e. inhibition of LEDGF/p75-IN protein-protein interaction and allosteric inhibition of catalytic function[14-16].
Since long time, coumarin class of natural products attracted the interest of medicinal chemists due to their abundance in plant kingdom, ease of synthesis and potential beneficial effects on human health[17]. Coumarins are flavonoid class of plant secondary metabolites, which exhibit variety of pharmacological properties such as anticoagulant, antioxidant, antibacterial, anti-HIV, antihyperlipidemic, etc.[17,18]. Various coumarin based antiretroviral agents have been reported to inhibit the retroviral enzymes i.e. reverse transcriptase, protease, and integrase[19-21]. The most widely used method for synthesis of coumarins is condensation of phenols with β-keto esters in the presence of acidic condensing agents[22-25]. Several acid catalysts like H2SO4, FeCl3, ZnCl2, POCl3, AlCl3, HCl, H3PO4, TFA, Silica Sulphuric Acid (SSA), oxalic acid, ClSO3H, SnCl2, HClO4, etc., are being used for this purpose[23,26]. It was found that yield of product in reactions catalyzed by SSA was higher as compared to other catalysts[22,26,27]. Due to various advantages associated with this non-hazardous and cheap reagent[28], SSA was used in the present study as a catalyst for the synthesis of 3-acetyl-2H-1-benzopyran-2-one derivatives.
Use of in silico models are consistently increasing these days to determine Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties of designed compounds at early stage of drug development[29]. The overall accuracy of these models and basic understanding is also increasing day by day[30,31]. Thus, a drug-likeness, in silico ADME profiling, and toxicity risk assessment (i.e. mutagenicity, tumorigenicity, irritant and reproductive effects) studies of 2a-n, were performed prior to the synthesis, to determine their pharmacokinetic and toxicity profile.
In previous studies, quantitative structure-activity relationship and docking studies were performed on coumarin derivatives as HIV-1 IN inhibitors and a series of 6-acetyl coumarin derivatives was synthesized as antiretroviral agents[32-34]. The present study explores the synthesis and in vitro biological evaluation i.e. LEDGF-dependent HIV- 1 IN inhibition activity, of 3-acetyl coumarin or 3-acetyl-2H-1-benzopyran-2-one derivatives. To understand the binding interaction of synthesized compounds (2a-n) in the binding pocket of HIV-1 IN, a molecular docking study is also performed. Based on the results and further exploration of present study, potent HIV-1 IN inhibitors can be developed as novel antiretroviral agents.
Materials and Methods
All reactions were performed in anhydrous conditions. Solvents and reagents were obtained from commercial suppliers and used as such without further purification. Thin Layer Chromatography (TLC) was used to monitor the reactions using mobile phase composition as hexane:ethyl acetate (3:7). Ultraviolet (UV) cabinet and saturated iodine chamber were used to detect the spots on TLC plates. Open capillary method was used to measure melting points (mp) of synthesized compounds and are uncorrected. Purification of synthesized compounds was done by Column chromatography. Fourier Transform Infrared (FTIR) spectra were recorded on Bruker Alpha spectrophotometer. Proton Nuclear Magnetic Resonance (1H-NMR) spectra were obtained at Bruker Avance II NMR spectrometer. Chemical shift values are given in δ in ppm using tetramethylsilane as an internal standard. Peak multiplicities are designated as singlet (s); doublet (d); doublet of doublet (dd) and multiplet (m). Mass spectra were recorded using Applied Biosystems/MDS SCIEX API 4000 mass spectrometer and elemental analysis was carried out using Vario EL III CHN elemental analyzer.
Preparation of SSA:
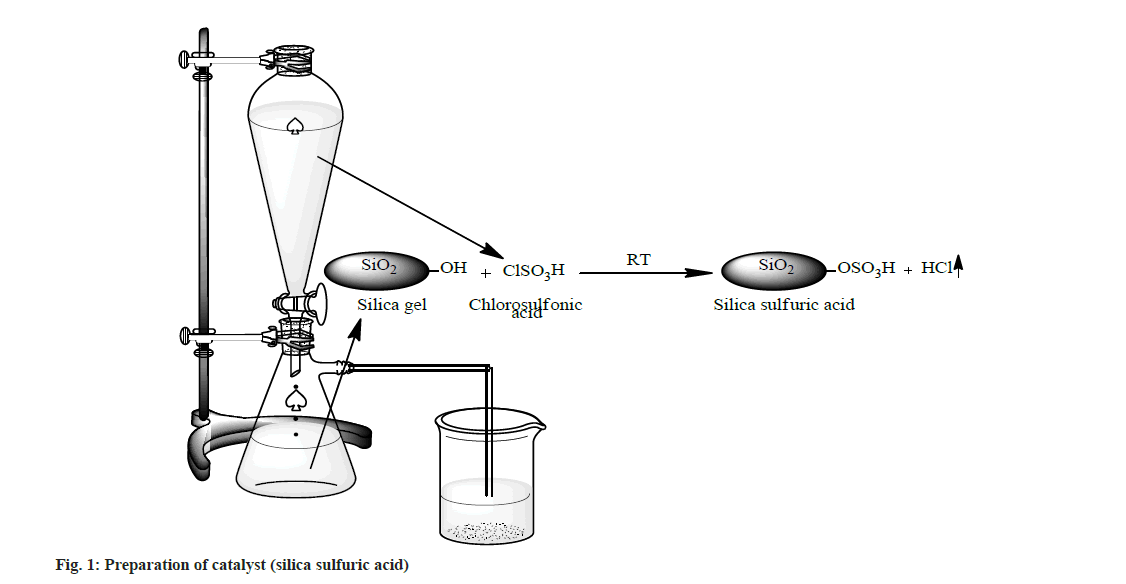
A suction flask of 500 ml capacity was fitted with a dropping funnel and a gas inlet tube. The flask was charged with approximately 50.0 g of silica gel (100-300 mesh size). Chlorosulphonic acid (23.3 g, 0.2 mol) was added drop wise for about 30 min at room temperature. Silanol group of silica gel reacted with chlorosulphonic acid and HCl gas immediately evolved from reaction vessel, which passed through the gas tube over an adsorbing solution (i.e. water). After complete addition of chlorosulphonic acid, the mixture was stirred with glass rod for 30 min. A white solid of silica sulfuric acid (66.1 g, ~100 %) was obtained (fig. 1).
Synthesis of 3-acetyl-2H-1-benzopyran-2-one (1):
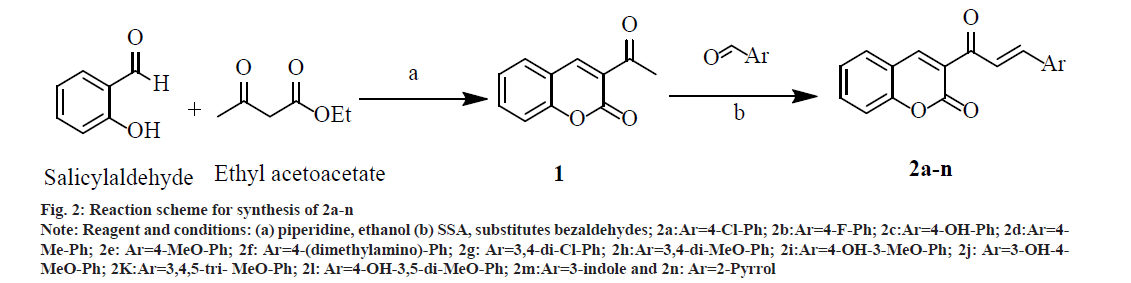
A mixture of 2.8 ml (20.0 mmol) salicylaldehyde, 3.3 ml (25.0 mmol) ethylacetoacetate and 5 ml absolute ethanol in a three necked Round Bottom Flask (RBF) was stirred on a magnetic stirrer. From the side arm of RBF 1.0 ml piperidine was added drop wise with continuous stirring. After some time, the mixture started thickening, to prevent this sufficient absolute ethanol was added and stirring was continued for 1 and half h. Reaction completion was confirmed by TLC. It was then allowed to crystallize into needle shaped crystals by cooling in freezer for 30 min. The crystals were washed with cold ethanol and recrystallized with water to get cream colored needle shaped crystals of 1 (fig. 2); % yield: 70; mp: 120-125°; Rf: 0.73; LogP: 1.12; IR (KBr, cm-1): 3029 (Ar-C-H), 1740 (O-C=O), 1678 (C=O), 1557, 1488, 1453 (C=C ring stretch), 1264, 1159, 1108 (Ar-C-H, bend, in plane), 737, 872 (Ar-C-H, bend, out of plane), and 424 (C=C ring bend, out of plane). 1H NMR (300 MHz, CDCl3) δ 8.48 (s, 1H, 4 –CH), 7.68 (s, 1H, 5 –CH), 7.53 (dd, 1H, 7 –CH), 7.30 (s, 1H, 8 –CH), 7.16 (s, 1H, 6 –CH), 2.36 (s, 3H, 3 – COCH3); Calculated mol. wt.: 188; found: electron ionization mass spectrometry (EIMS) m/z: 188 (M+ peak), 189 (M+1 peak); Molecular formula: C11H8O3; Elemental analysis, Calculated C, 70.21; H, 4.29; O, 25.51; Found C, 69.86; H, 3.99; O, 24.78.
Fig. 2: Reaction scheme for synthesis of 2a-n Note: Reagent and conditions: (a) piperidine, ethanol (b) SSA, substitutes bezaldehydes; 2a:Ar=4-Cl-Ph; 2b:Ar=4-F-Ph; 2c:Ar=4-OH-Ph; 2d:Ar=4- Me-Ph; 2e: Ar=4-MeO-Ph; 2f: Ar=4-(dimethylamino)-Ph; 2g: Ar=3,4-di-Cl-Ph; 2h:Ar=3,4-di-MeO-Ph; 2i:Ar=4-OH-3-MeO-Ph; 2j: Ar=3-OH-4- MeO-Ph; 2K:Ar=3,4,5-tri- MeO-Ph; 2l: Ar=4-OH-3,5-di-MeO-Ph; 2m:Ar=3-indole and 2n: Ar=2-Pyrrol
Synthesis of 3-acetyl-2H-1-benzopyran-2-one derivatives (2a-n):
Synthesis of 2a-n was done by mixing substituted bezaldehydes (10 mmol), intermediate 1 (7.5 mmol) and SSA (0.7 g) in a glass tube using a glassrod. The solid mixture was heated at 80° for 2 h with intermittent stirring every 15 min with a glass rod. The Solid mixture was cooled at room temperature and 50 ml of dichloromethane was added with gentle stirring. The dichloromethane extract was filtered and dried to give the solid mass. Recrystallization was done from ethanol, and further purification was done by column chromatography (fig. 2). The physicochemical properties and spectral data of 2a-n are as follows.
Synthesis of 3-(3-(4-chlorophenyl)acryloyl)-2Hchromen- 2-one (2a):
2a was synthesized by reaction of 1 with 4-chlorobenzaldehyde using SSA as a catalyst; yellowish white crystals; % yield: 78; mp: 171-175°; Rf: 0.43; LogP: 3.57; IR (KBr, cm-1): 3069 (Ar-C-H), 1719 (O-C=O), 1680 (C=O), 1610, 1488, 1453 (C=C ring stretch), 1090 (C-Cl), 1027, 1011 (Ar-C-H, bend, in plane), 735 (Ar-C-H, bend, out of plane) and 442 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.02 (d, 1H, 3' –CH), 7.67 – 7.59 (m, 1H, 5 –CH), 7.56- 7.36 (m, 5H, 7 –CH, 2'', 3'', 5'', 6'' –CH), 7.29-7.15 (m, 3H, 6, 8 –CH, 2’ –CH); Calculated mol. wt.: 310; found: EIMS m/z: 310 (M+ peak), 311 (M+1 peak); Molecular formula: C18H11ClO3; Elemental analysis, Calculated C, 69.58; H, 3.57; Cl, 11.41; O, 15.45; Found, C, 69.50; H, 3.49; O, 14.68.
Synthesis of 3-(3-(4-fluorophenyl)acryloyl)-2Hchromen- 2-one (2b)
2b was synthesized by reaction of 1 with 4-fluoro-benzaldehyde using SSA as a catalyst; off white solid; % yield: 79; mp: 164-169°; Rf: 0.35; UV (λmax): 297; IR (KBr, cm-1): 3042 (Ar- C-H), 1732 (O-C=O), 1657 (C=O), 1608, 1489, 1452 (C=C ring stretch), 1208 (C-F), 1121, 1279 (Ar-C-H, bend, in plane), 717, 872 (Ar-C-H, bend, out of plane), and 417 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.36 (s, 1H, 4 –CH), 8.02 (d, 1H, 3'–CH ), 7.58 (dd, 3H, 5 –CH, 2'', 6'' –CH), 7.45 (s, 1H), 7.26–7.08 (m, 5H, 6, 8 –CH, 2' –CH, 3'', 5'' –CH); Calculated Mol. Wt.: 294; Found: EIMS m/z: 294 (M+ peak), 295 (M+1 peak); Molecular formula: C18H11FO3; Elemental analysis, Calculated, C, 73.47; H, 3.77; F, 6.46; O, 16.31; Found, C, 73.78; H, 3.89; O, 16.01.
Synthesis of 3-(3-(4-hydroxyphenyl)acryloyl)-2Hchromen- 2-one (2c):
2c was synthesized by reaction of 1 with 4-hydroxy- benzaldehyde using SSA as a catalyst; yellowish white solid; % yield: 83; mp: 209-215°; Rf: 0.76; LogP: 2.78; IR (KBr, cm-1): 3424 (OH), 3039 (Ar-C-H), 1718 (O-C=O), 1666 (C=O), 1584, 1488, 1453 (C=C ring stretch), 1290, 1226, 1121 (Ar-C-H, bend, in plane), 828, 796, 717 (Ar-C-H, bend, out of plane) and 445 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.00 (d, 1H, 3'–CH), 7.63 (d, 1H, 5 –CH), 7.50–7.36 (m, 3H, 7, 2'', 6'' –CH), 7.26- 7.15 (m, 3H, 6, 8, 2'' –CH), 6.89 (d, 2H, 3'', 5'' –CH), 4.16 (s, 1H, 4'' –OH); Calculated Mol. Wt.: 292; Found: EIMS m/z: 292 (M+ peak), 293 (M+1 peak); Molecular formula: C18H12O4; Elemental analysis, Calculated C, 73.97; H, 4.14; O, 21.90; Found C, 73.68; H, 4.09; O, 21.46.
Synthesis of 3-(3-p-tolylacryloyl)-2H-chromen-2- one (2d):
2d was synthesized by reaction of 1 with 4-methyl- benzaldehyde using SSA as a catalyst; yellowish brown crystals; % yield: 72; mp: 163-165°; Rf: 0.59; LogP: 3.65; IR (KBr, cm-1): 3168 (Ar-C-H), 2965 (C-H, CH3), 1735 (O-C=O), 1666 (C=O), 1598, 1516, 1453 (C=C ring stretch), 1286, 1217, 1160 (Ar-C-H, bend, in plane), 858, 788, 709 (Ar- C-H, bend, out of plane) and 459 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.03 (d, 1H, 3'–CH), 7.63 (d, 1H, 5 –CH), 7.57-7.40 (m, 3H, 7, 2'', 6'' –CH), 7.33- 7.15 (m, 5H, 6, 8, 2', 3'', 5'' –CH), 2.35 (s, 3H, 4'' –CH3); Calculated Mol. Wt.: 290; Found: EIMS m/z: 290 (M+ peak), 291 (M+1 peak); Molecular formula: C19H14O3; Elemental analysis, Calculated C, 78.61; H, 4.86; O, 16.53; Found C, 78.23; H, 4.54; O, 16.01.
Synthesis of 3-(3-(4-methoxyphenyl)acryloyl)-2Hchromen- 2-one (2e)
2e was synthesized by reaction of 1 with 4-methoxy- benzaldehyde using SSA as a catalyst; grayish white crystals; % yield: 85; mp: 160-170°; Rf: 0.65; Log P: 2.98; IR (KBr, cm-1): 3032 (Ar-C-H), 2929 (C-H, CH3), 1717 (O-C=O), 1679 (C=O), 1584, 1511, 1423 (C=C ring stretch), 1283, 1239, 1174 (Ar-C-H, bend, in plane), 1119 (C-O-C asymmetric stretch), 864, 787, 717 (Ar-C-H, bend, out of plane) and 447 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.00 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.54 (d, 2H, 2'', 6'' –CH), 7.45 (s, 1H, 7 –CH), 7.26- 7.15 (m, 3H, 6, 8, 2' –CH), 7.02 (d, 2H, 3'', 5'' –CH), 3.81 (s, 3H, 4'' –CH3); Calculated Mol. Wt.: 306; Found: EIMS m/z: 306 (M+ peak), 307 (M+1 peak); Molecular formula: C19H14O4; Elemental analysis, Calculated C, 74.50; H, 4.61; O, 20.89; Found C, 74.34; H, 4.54; O, 20.03.
Synthesis of 3-(3-(4-(dimethylamino)phenyl) acryloyl)-2H-chromen-2-one (2f):
2f was synthesized by reaction of 3 with p-dimethyl- amino-benzaldehyde using SSA as a catalyst; brownish white solid; % yield: 78; mp: 186-190°; Rf: 0.50; LogP: 3.23; IR (KBr, cm-1): 3008 (Ar- C-H), 2929, 2852 (C-H, CH3), 1740 (O-C=O), 1678 (C=O), 1487, 1453, 1410 (C=C ring stretch), 1295 (Ar-C-N), 1264, 1210, 1122 (Ar-C-H, bend, in plane), 1159 (C-N, Aliphatic), 872, 758, 737 (Ar-C-H, bend, out of plane) and 436 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.31 (s, 1H, 4 –CH), 7.67–7.55 (m, 2H, 5, 3' –CH), 7.45 (s, 1H, 7 –CH), 7.35 (d, 2H, 2'', 6'' –CH), 7.20 (dd, 2H, 6'', 8'' –CH), 6.98 (d, 1H, 2' –CH), 6.74 (d, 2H, 3'', 5'' –CH), 2.92 (s, 6H, 4'' –N(CH3)2); Calculated Mol. Wt.: 319; Found: EIMS m/z: 319 (M+ peak), 320 (M+1 peak); Molecular formula: C20H17NO3; Elemental analysis, Calculated C, 75.22; H, 5.37; N, 4.39; O, 15.03; Found C, 75.02; H, 5.43; N, 4.45; O, 14.66.
Synthesis of 3-(3-(3,4-dichlorophenyl)acryloyl)-2Hchromen- 2-one (2g):
2g was synthesized by reaction of 1 with 3, 4-dichloro- benzaldehyde using SSA as a catalyst; yellowish white crystals; % yield: 82; mp: 184-189°; Rf: 0.43; LogP: 4.28; IR (KBr, cm-1): 3089 (Ar- C-H), 1723 (O-C=O), 1668 (C=O), 1487, 1470, 1452 (C=C ring stretch), 1275, 1232, 1181 (Ar- C-H, bend, in plane), 1079 (C-Cl), 881, 882, 693 (Ar-C-H, bend, out of plane) and 443 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.02 (d, 1H, 3' –CH), 7.69- 7.58 (m, 2H, 5, 6'' –CH), 7.45 (s, 1H, 7 –CH), 7.37 (s, 2H, 2'', 3'' –CH), 7.29-7.15 (m, 3H, 6, 8, 2'' – CH); Calculated Mol. Wt.: 345; Found: EIMS m/z: 345 (M+ peak), 346 (M+1 peak); Molecular formula: C18H10Cl2O3; Elemental analysis, Calculated, C, 62.63; H, 2.92; Cl, 20.54; O, 13.91; Found, C, 62.93; H, 3.08; O, 13.23.
Synthesis of 3-(3-(3,4-dimethoxyphenyl)acryloyl)- 2H-chromen-2-one (2h):
2h was synthesized by reaction of 1 with 3, 4-dimethoxy- benzaldehyde using SSA as a catalyst; white solid; % yield: 78; mp: 162-167°; Rf: 0.53; LogP: 2.79; IR (KBr, cm-1): 3052 (Ar-C-H), 2931, 2836 (C-H, CH3), 1727 (O-C=O), 1656 (C=O), 1452, 1418 (C=C ring stretch), 1266, 1182, 1136 (Ar-C-H, bend, in plane), 1120 (C-O-C asymmetric stretch), 839, 798, 691 (Ar-C-H, bend, out of plane) and 442 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.00 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.45 (s, 1H, 7 –CH), 7.26-7.14 (m, 5H, 6, 8, 2', 2'', 6'' –CH), 6.98 (d, 1H, 5'' –CH), 3.82 (d, 6H, 3'', 4'' – CH); Calculated Mol. Wt.: 336; Found: EIMS m/z: 336 (M+ peak), 337 (M+1 peak); Molecular formula: C20H16O5; Elemental analysis, Calculated C, 71.42; H, 4.79; O, 23.78; Found C, 71.52; H, 4.89; O, 23.02.
Synthesis of 3-(3-(4-hydroxy-3-methoxyphenyl) acryloyl)-2H-chromen-2-one (2i):
2i was synthesized by reaction of 1 with 4-hydroxy- 3-methoxy-benzaldehyde using SSA as a catalyst; off white solid; % yield: 85; mp: 200- 204°; Rf: 0.72; LogP: 2.69; IR (KBr, cm-1): 3252 (O-H), 3005 (Ar-C-H), 2939 (C-H, -OCH3), 1722 (O-C=O), 1688 (C=O), 1453, 1421 (C=C ring stretch), 1284, 1208, 1178 (Ar-C-H, bend, in plane), 1120 (C-O-C asymmetric stretch), 843, 779, 714 (Ar-C-H, bend, out of plane) and 438 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.00 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.45 (s, 1H, 7 –CH), 7.25-7.14 (m, 3H, 6, 8, 2' –CH), 7.11-7.02 (m, 2H, 2'', 6'' –CH), 6.85 (d, 1H, 5''–CH), 3.94 (s, 1H, 4'' – OH), 3.83 (s, 3H, 3'' –OCH3); Calculated Mol. Wt.: 322; Found: EIMS m/z: 322 (M+ peak), 323 (M+1 peak); Molecular formula: C19H14O5; Elemental analysis, Calculated C, 70.80; H, 4.38; O, 24.82; Found C, 71.02; H, 4.89; O, 23.91.
Synthesis of 3-(3-(3-hydroxy-4-methoxyphenyl) acryloyl)-2H-chromen-2-one (2j):
2j was synthesized by reaction of 1 with 3-hydroxy- 4-methoxy-benzaldehyde using SSA as a catalyst; grayish white crystals; % yield: 79; mp: 200-208°; Rf: 0.59; LogP: 2.56; IR (KBr, cm-1): 3233 (O-H), 3003 (Ar-C-H), 2974 (C-H, -OCH3), 1731 (O-C=O), 1673 (C=O), 1578, 1445 (C=C ring stretch), 1277, 1219, 1167 (Ar-C-H, bend, in plane), 1120 (C-O-C asymmetric stretch), 865, 792, 681 (Ar-C-H, bend, out of plane) and 459 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.35 (s, 1H, 4 –CH), 8.00 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.45 (s, 1H, 7 –CH), 7.25-7.14 (m, 3H, 6, 8, 2' –CH), 7.14–7.05 (m, 2H, 2'', 6'' –CH), 6.85 (d, 1H, 5''–CH), 3.86 (s, 1H, 3'' – OH), 3.81 (s, 3H, 4'' –OCH3); Calculated Mol. Wt.: 322; Found: EIMS m/z: 322 (M+ peak), 323 (M+1 peak); Molecular formula: C19H14O5; Elemental analysis, Calculated C, 70.80; H, 4.38; O, 24.82; Found C, 70.54; H, 4.10; O, 24.13.
Synthesis of 3-(3-(3,4,5-trimethoxyphenyl)acryloyl)- 2H-chromen-2-one (2k):
2k was synthesized by reaction of 1 with 3,4,5-trimethoxy- benzaldehyde using SSA as a catalyst; yellow crystals; % yield: 82; mp: 166-172°; Rf: 0.69; LogP: 2.83; IR (KBr, cm-1): 2942 (Ar-C-H), 2841 (C-H, -OCH3), 1727 (O-C=O), 1685 (C=O), 1585, 1456, 1421 (C=C ring stretch), 1290, 1234, 1176 (Ar-C-H, bend, in plane), 1126 (C-O-C asymmetric stretch), 845, 794, 697 (Ar-C-H, bend, out of plane) and 442 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.34 (s, 1H, 4 –CH), 8.05 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.45 (s, 1H, 7 –CH), 7.30 – 7.15 (m, 3H, 6, 8, 2' –CH), 6.91 (s, 2H, 2'', 6'' –CH), 3.82 (s, 9H, 3'', 4'', 5'' –CH); Calculated Mol. Wt.: 366; Found: EIMS m/z: 366 (M+ peak), 367 (M+1 peak); Molecular formula: C21H18O6; Elemental analysis, Calculated C, 68.85; H, 4.95; O, 26.20; Found C, 68.75; H, 4.86; O, 25.64.
Synthesis of 3-(3-(4-hydroxy-3,5-dimethoxyphenyl) acryloyl)-2H-chromen-2-one (2l):
2l was synthesized by reaction of 1 with 4-hydroxy- 3,5-dimethoxy-benzaldehyde using SSA as a catalyst; off white solid; % yield: 79; mp: 197- 205°; Rf: 0.65; LogP: 2.42; IR (KBr, cm-1): 3281 (O-H), 3007 (Ar-C-H), 2967, 2841 (C-H, -OCH3), 1721 (O-C=O), 1670 (C=O), 1587, 1465, 1425 (C=C ring stretch), 1293, 1255, 1110 (Ar-C-H, bend, in plane), 1038 (C-O-C asymmetric stretch), 871, 829, 729 (Ar-C-H, bend, out of plane) and 454 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.36 (s, 1H, 4 –CH), 8.01 (d, 1H, 3' –CH), 7.63 (d, 1H, 5 –CH), 7.45 (s, 1H, 7 –CH), 7.25-7.15 (m, 3H, 6, 8, 2' –CH), 6.74 (s, 2H, 2'', 6'' –CH), 4.15 (s, 1H, 4'' –CH), 3.82 (s, 6H, 3'', 5'' –CH); Calculated Mol. Wt.: 352; Found: EIMS m/z: 352 (M+ peak), 353 (M+1 peak); Molecular formula: C20H16O6; Elemental analysis, Calculated C, 68.18; H, 4.58; O, 27.25; Found C, 67.99; H, 4.23; O, 26.95.
Synthesis of 3-(3-(1H-indol-3-yl)acryloyl)-2Hchromen- 2-one (2m):
2m was synthesized by reaction of 1 with 1H-indole- 3-carbaldehyde using SSA as a catalyst; brownish white solid; % yield: 87; mp: 210-216°; Rf: 0.43; Log P: 2.60; IR (KBr, cm-1): 3168 (Ar- C-H), 1740 (O-C=O), 1678 (C=O), 1488, 1453, 1410 (C=C ring stretch), 1295, 1235, 1122 (Ar- C-H, bend, in plane), 871, 787, 758 (Ar-C-H, bend, out of plane) and 438 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.33 (s, 1H, 4 –CH), 7.71–7.57 (m, 4H, 5, 3', 2'' –CH, 3'' –NH), 7.46 (s, 1H, 7 –CH), 7.39 – 6.96 (m, 7H, 6, 8, 2', 5'', 6'', 7'', 8'' –CH); Calculated Mol. Wt.: 315; Found: EIMS m/z: 315 (M+ peak), 316 (M+1 peak); Molecular formula: C20H13NO3; Elemental analysis, Calculated C, 76.18; H, 4.16; N, 4.44; O, 15.22; Found C, 76.40; H, 4.34; N, 4.67; O, 14.78.
Synthesis of 3-(3-(1H-pyrrol-2-yl)acryloyl)-2Hchromen- 2-one (2n):
2n was synthesized by reaction of 1 with 1H-pyrrole-2-carbaldehyde using SSA as a catalyst; brown crystals; % yield: 80; mp: 179-181°; Rf: 0.39; LogP: 1.67; IR (KBr, cm-1): 3067 (Ar- C-H), 1740 (O-C=O), 1678 (C=O), 1488, 1453, 1410 (C=C ring stretch), 1295, 1209, 1108 (Ar- C-H, bend, in plane), 872, 758 (Ar-C-H, bend, out of plane) and 452 (C=C, ring bend, out of plane); 1H NMR (300 MHz, CDCl3) δ 8.34 (s, 1H, 4 –CH), 8.12 (d, 1H, 3' –CH), 7.62 (dd, 1H, 5 –CH), 7.45 (td, 1H, 7 –CH), 7.30 (dd, 1H, 3'' –CH), 7.25 – 7.09 (m, 4H, 6, 8, 2' –CH, 2'' –NH), 6.45 (dd, 1H, 5'' –CH), 6.20 (s, 1H, 6'' –CH); Calculated Mol. Wt.: 265; Found: EIMS m/z: 265 (M+ peak), 266 (M+1 peak); Molecular formula: C16H11NO3; Elemental analysis, Calculated C, 72.45; H, 4.18; N, 5.28; O, 18.09; Found C, 72.15; H, 3.98; N, 5.44; O, 17.89.
Drug-likeness and in silico ADME/T study:
In silico drug-likeness, ADME properties and toxicity risks, were predicted for all synthesized compounds prior to their synthesis using Qikprop module of Schrödinger suite 2010[35] and DataWarrior[36]. Lipinski’s rule of five and Jorgensen’s rule of three were used to assess drug-likeness and lead-likeness of synthesized compounds, respectively[37-39]. As per Lipinski’s rule of five, a compound should have a MW (molecular weight) <500 Da, LogP ≤5, Hydrogen Bond Donors (HBD)≤5 and Hydrogen Bond Acceptors (HBA)≤10 while as per Jorgensen’s rule of three a compound should have LogP ≤3, MW <300 Da, HBD ≤3, HBA ≤3 and rotatable bonds ≤3.
The predicted ADME parameter included, LogS (aqueous solubility), #rot (number of rotatable bonds), #met (number of metabolic reactions), CNS activity, CaCo2 permeability (permeability through intestinal epithelium), LogBB (ability to cross blood brain barrier), LogKp (permeability through skin), LogKhsa (serum protein binding) and percent Human Oral Absorption (% HOA). The predicted toxicity risks included mutagenicity, tumorigenicity, irritant effect and reproductive effect.
Biological evaluation:
Purification of recombinant IN and LEDGF: Recombinant proteins were expressed and purified as explained[5,7,40,41]. In brief, the proteins were expressed in Escherichia coli (E. coli) strain BL21 (DE3). HIV-1 IN was purified in two step process. In first step, HIV-1 IN was eluted with an imidazole gradient from 20 mM to 500 mM concentration in a 50 mM (4-(2-Hydroxyethyl)- 1-Piperazineethanesulfonic acid (HEPES) (pH 7.5) buffer containing 1 M NaCl, 7.5 mM 3-[(3-Cholamidopropyl)dimethylammonium]- 1-Propanesulfonate (CHAPS), 2 mM β-mercaptoethanol using a Ni-Sepharose column. In second step HIV-1 IN was purified by a heparin column with a NaCl gradient from 0 to 1 M concentrations. Purification of His-LEDGF was done by loading precipitate of cell lysate onto a heparin column and was eluted with an increasing NaCl gradient (200 mM to 1 M) in a 50 mM HEPES (pH 7.5) buffer containing 2 mM β-mercaptoethanol and 7.5 mM CHAPS. Peak fractions were concentrated and loaded onto a Superdex 200 GL column and eluted in a buffer containing 50 mM HEPES (pH 7.5), 200 mM NaCl and 2 mM β-mercaptoethanol. Fractions containing HIV-1 IN and LEDGF were pooled and stored in 15 % glycerol at -80°.
HTRF HIV-1 IN LEDGF-dependent assay:
The assay was performed as reported[42-44]. Briefly, 50 nM IN was pre-incubated with increasing concentration of compounds for 1 h at room temperature in reaction buffer containing 1 % Glycerol, 1 mM Dithiothreitol, 20 mM HEPES pH 7.5, 20 mM MgCl2, 0.05 % Brij-35 and 0.1 mg/ml bovine serum albumin. After this time, 9 nM DNA donor substrate (5'-ACAGGCCTAGCACGCGTCG-Biot in-3' annealed with 5'-CGACGCGTGGTAGGCCTGTBiotin3'), 50 nM DNA acceptor substrate (5'-Cy5- ATGTGGAAAATCTCTAGCAGT-3' annealed with 5'-Cy5- TGAGCTCGAGATTTTCCACAT-3') and 50 nM LEDGF/p75 protein were added and incubated at 37° for 90 min. 4 nM of Europium- Streptavidin were added at the reaction mixture and the Homogenous Time-Resolve Fluorescence (HTRF) signal was recorded using a Perkin Elmer Victor 3 plate reader using an excitation wavelength of 314 nm and the wavelength of the acceptor and the donor substrates emission as 668 and 620 nm, respectively. Raltegravir (the strandtransfer inhibitor) was used as a positive control[45].
Docking study:
A co-crystallized structure of functional retroviral intasome containing full-length Prototype Foamy Virus (PFV) IN (complexed with raltegravir) and oligonucleotide mimics its preprocessed viral U5 DNA end (PDB ID: 3OYA), was retrieved form RCSB Protein data bank with a resolution of 2.5 Å[46]. To investigate the binding orientation of synthesized compounds 2a-n in binding pocket of HIV-1 IN, a molecular docking simulation was performed using GOLD software package[47]. The ligands and protein used in molecular docking were prepared using maestro v9.3 module of Schrodinger suite.
To prepare the ligand, a multistep minimization protocol, was performed using the OPLS_AA forcefield, which includes steepest descent, Polak- Ribiere Conjugate Gradient followed by Lig- Prep v2.5 module to obtain the low energy minimized structure.
The protein structure was prepared as follows. Hydrogens were added; bond orders were corrected; water molecules were removed; missing side chains and loops were added (using prime), H-bonding network and orientation of Asn, Gln and His residues were optimized, and entire complex was energy minimized (using Impref) until the Root Mean Square Deviation (RMSD) of the heavy atoms in the minimized structure relative to the X-ray structure exceeded 0.2 Å. This helps in maintaining the integrity of the prepared structures relative to the corresponding experimental structures, while eliminating severe bad contacts between heavy atoms.
The prepared protein and ligands were used for molecular docking study. In order to define the binding site, and validation of parameters used in GOLD, a re-docking approach was followed, in which the co-crystallized bound ligand (raltegravir) was extracted and docked back in the binding pocket of HIV-1 IN. The prediction of binding mode is considered successful if the RMSD between co-crystallized ligand and re-docked pose is below a certain value, usually 2.0 Å[48]. The exact superimposition was obtained with RMSD value 0.4505, which confirmed that docking parameters were validated. The synthesized compounds 2a-n were docked in the same binding pocket and interaction points of compounds 2a-n with HIV- 1 IN protein were compared with interaction points of raltegravir and their GoldScore fitness was reported. The GoldScore fitness is the scoring function of GOLD which is used for prediction of ligand binding positions and considers the factors like van der Waals energy, H-bonding energy, ligand torsion strain and metal interaction. The GoldScore represents goodness of docked pose. Higher GoldScore represents better docking results.
Results and Discussion
The prepared catalyst, SSA was characterized by titration, FTIR and Differential Scanning Colorimetry (DSC). Acid base titration determined the number of H+ site of SSA which was found 0.250±0.05 m equiv.g-1[49,50]. This value corresponds to the sulfur content (about 95 %) present in the sample. The FTIR spectra of silica gel showed bands at 3432, 1630, 1093, 803, 485 cm-1 while SSA showed bands at 3405, 1284, 1177, 1069, 850 cm-1. In SSA, sulfonic acid was covalently bonded on silica gel by Si-O-S bond, while sulfuric acid simply interacted with silica gel by a weak hydrogen bond[51]. FTIR absorption of O=S=O bands of sulfonic acid functional group, lies between 1180-1250 cm-1[26,52]. The bands at 1177 cm-1 and 1284 cm-1 were due to the symmetric and asymmetric stretching vibrations, respectively, of S=O functionality of the sulfonic acid, while the broad band in 3200-3400 cm-1 region was due to O-H stretching vibration of unreacted hydroxyl groups. DSC graph of silica gel showed no peak in the temperature range of 50-450° while SSA showed two peaks at 100-125° and 250-275°[53].
Condensation of salicylaldehyde with ethylacetoacetate using piperidine and alcohol as condensing agent led to the formation of 1, which was used as a starting material for synthesis of 2an. The chemical shift at δ 8.48, 7.68, 7.16, 7.53 and 7.30 was attributed to the single proton at 4, 5, 6, 7 and 8 positions of benzopyran-2-one ring in 1. The chemical shift at δ 2.36 was attributed to three protons of -COCH3 group attached to the coumarin ring of 1 at third position. Mass spectrum of 1 showed a characteristic peak at m/z of 188, as molecular ion peak. Fragmentation of molecular ion peak showed peaks at 173 and 145 which may be due to –CH3 (M-15) and –COCH3 (M-43) fragments.
Common FTIR bands in the frequency range of 3100-3000 (Ar-C-H stretch), 3500-3200 (OH), 1720-1650 (-C=O of coumarin), 1660-1600 (C=O of ketone), 1600-1575, 1500-1400 (C=C, ring stretch), 1300-1100 (Ar-C-H, bend, in plane), 1150-1085 (C-O-C, stretch), 900-675 (Ar-C-H, bend, out of plane), 600-420 cm-1 (C=C, ring bend, out of plane)[54], confirmed the presence of these groups in all synthesized compounds. NMR spectrum of compound 2a showed chemical shift values at δ 8.35, 7.67, 7.29 and 7.56 may be due to the single proton of the coumarin ring at 4, 5, 6 and 7 position. Protons at 2' and 3' position were characterized by their chemical shift values at 8.02 and 7.15. Protons of the phenyl ring at 2'', 3'', 5'' and 6'' showed a multiplet between 7.56-7.36. Mass Spectrum of 2a showed the molecular ion peak at 310, which characterized the molecular weight of compound. The fragment peak at 199 may be due to chlorobenzene fraction (M-111). The mass at 186 fractions may be due to breaking of the double bond at the linker and may be of 3-acetyl coumarin functionality. In mass spectrum of compound 2g, the molecular ion peak was found at 344 and fragment were found at 328, 309, 199, 186, 173 and 145. The fragment at m/z 199 may be due to dichlorobenzene fraction (M-145). Mass Spectrum of compound 2h showed molecular ion peak at 336 and daughter ion fragments were found at the m/z value 321, 305, 199, 173, 145 and 137.
To confirm the synthesis of 2a-n, a comparison of spectral data was made between intermediate 1 and final products 2a-n. Comparison of IR spectra of 1 and compound 2a showed sharp bands at 2800-3000 cm-1 in 1 due to presence of -CH3 group while few bands were found in 2a due to formation of C=C bond. The NMR spectrum of 1 showed chemical shift at δ 2-2.5 due to CH3 protons which was absent in the synthesized compounds 2a-n due to involvement of CH3 in formation of C=C.
In drug-likeness study none of the synthesized compounds i.e. 2a-n, violated the Lipinski’s rule of five and Jorgensen’s rule of three, which suggested that these compounds are more likely to be orally available (Table 1). The calculated ADME parameters of all compounds were well within their reference limits (Table 2). In silico toxicity study showed that all synthesized compounds have no toxicity risks for mutagenicity, tumorigenicity, irritant effects while having very low reproductive risk except compound 2f which have high mutagenic and tumorigenic risk, and compounds 2e which have high irritant effect (Table 3).
| Compound code | MW | HBD | HBA | LogP | PSA | ROT | ROF |
|---|---|---|---|---|---|---|---|
| 2a | 310.735 | 0 | 4.5 | 3.382 | 62.453 | 0 | 0 |
| 2b | 294.281 | 0 | 4.5 | 3.113 | 62.451 | 0 | 0 |
| 2c | 292.289 | 1 | 5.25 | 2.474 | 85.393 | 0 | 0 |
| 2d | 290.318 | 0 | 4.5 | 3.204 | 62.476 | 0 | 0 |
| 2e | 306.317 | 0 | 5.25 | 2.861 | 70.705 | 0 | 0 |
| 2f | 319.359 | 0 | 5.5 | 3.263 | 65.877 | 0 | 0 |
| 2g | 345.181 | 0 | 4.5 | 3.829 | 62.446 | 0 | 0 |
| 2h | 336.343 | 0 | 6 | 3.087 | 74.931 | 0 | 0 |
| 2i | 322.315 | 1 | 6 | 2.498 | 92.486 | 0 | 0 |
| 2j | 322.315 | 1 | 6 | 2.489 | 92.65 | 0 | 0 |
| 2k | 366.37 | 0 | 6.75 | 3.109 | 85.041 | 0 | 0 |
| 2l | 352.343 | 1 | 6.75 | 2.713 | 97.037 | 0 | 0 |
| 2m | 315.328 | 1 | 4.5 | 3.303 | 76.291 | 0 | 0 |
| 2n | 265.268 | 1 | 4.5 | 2.402 | 75.777 | 0 | 0 |
| Raltegravir | 444.442 | 2 | 11 | 1.673 | 166.711 | 0 | 1 |
| RR* | <500 | ≤5 | ≤10 | ≤5 | < 140 | 0 | 1 |
Note: RR: Reference Range for 95 % of known drugs
Table 1: Drug Likeness Study of Synthesized Compounds
| Compound code | #rot | #met | LogS | CNS | Caco-2 | LogBB | LogKp | LogKhsa | % HOA |
|---|---|---|---|---|---|---|---|---|---|
| 2a | 4 | 0 | -4.135 | 0 | 1278.389 | -0.398 | -1.513 | 0.006 | 100 |
| 2b | 4 | 0 | -3.738 | 0 | 1278.498 | -0.447 | -1.478 | -0.076 | 100 |
| 2c | 5 | 1 | -3.702 | -2 | 413.855 | -1.109 | -2.373 | -0.065 | 88.266 |
| 2d | 4 | 1 | -3.98 | 0 | 1276.463 | -0.581 | -1.545 | 0.053 | 100 |
| 2e | 5 | 1 | -3.624 | 0 | 1276.02 | -0.639 | -1.447 | -0.146 | 100 |
| 2f | 5 | 2 | -4.201 | 0 | 1232.011 | -0.694 | -1.536 | 0.012 | 100 |
| 2g | 4 | 0 | -4.786 | 0 | 1277.313 | -0.269 | -1.648 | 0.125 | 100 |
| 2h | 6 | 2 | -3.83 | -1 | 1276.124 | -0.737 | -1.483 | -0.141 | 100 |
| 2i | 6 | 2 | -3.943 | -2 | 418.027 | -1.233 | -2.442 | -0.057 | 88.486 |
| 2j | 6 | 2 | -3.895 | -2 | 416.201 | -1.222 | -2.455 | -0.057 | 88.402 |
| 2k | 7 | 3 | -3.666 | -1 | 1344.679 | -0.76 | -1.528 | -0.192 | 100 |
| 2l | 7 | 3 | -3.908 | -2 | 705.496 | -1.048 | -2.027 | -0.076 | 93.811 |
| 2m | 4 | 0 | -4.452 | -1 | 776.494 | -0.789 | -1.717 | 0.265 | 100 |
| 2n | 4 | 0 | -3.379 | -1 | 745.303 | -0.764 | -1.99 | -0.073 | 92.419 |
| Raltegravir | 7 | 4 | -4.629 | -2 | 84.453 | -2.04 | -4.262 | -0.278 | 58.268 |
| RR* | 0to15 | <7 | > -5.7 | -2 to 2 | <25poor >500good | -3 to 1.2 | -8to-1 | ±1.5 | <25poor |
Note: RR: Reference Range for 95 % of known drugs
Table 2: In Silico Adme Profile Of Synthesized Compounds
| Compound code | Mutagenic risk | Tumorigenic risk | Reproductive risk | Irritant risk |
|---|---|---|---|---|
| 2a | None | None | Low | None |
| 2b | None | None | Low | None |
| 2c | None | None | Low | None |
| 2d | None | None | Low | None |
| 2e | None | None | Low | High |
| 2f | High | High | Low | None |
| 2g | None | None | Low | None |
| 2h | None | None | Low | None |
| 2i | None | None | Low | None |
| 2j | None | None | Low | None |
| 2k | None | None | Low | None |
| 2l | None | None | Low | None |
| 2m | None | None | Low | None |
| 2n | None | None | Low | None |
| Raltegravir | None | None | None | None |
Table 3: In Silico Toxicity Risk Assessment Of Synthesized Compounds
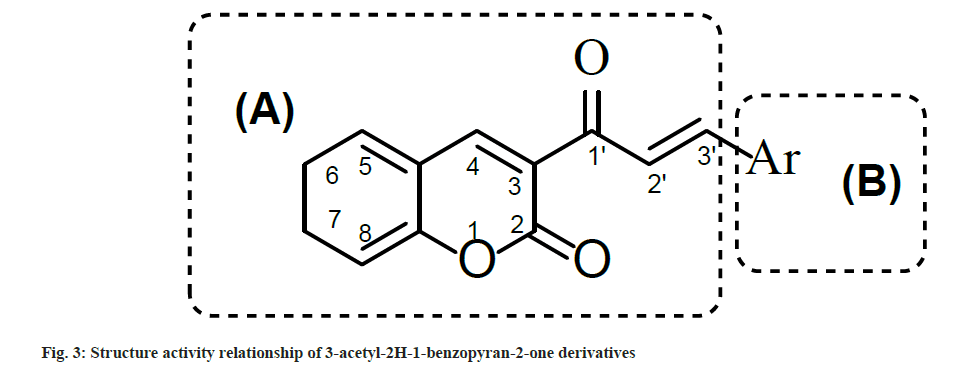
Synthesized compounds were tested for their ability to inhibit the HIV-1 IN activity in the presence of LEDGF/p75 in biochemical assay (Table 4). Results showed that compounds with electron withdrawing groups i.e. 2a, 2g and 2h inhibited LEDGF-dependent HIV-1 IN with IC50 values to 86.5, 90.0 and 98.5 μM, respectively. To understand the structure activity relationship of 2a-n, the main structure was divided in two parts i.e. A and B. Part A contain the 3-acetyl-2H-1-benzopyran-2- one functionality which provide a backbone for lead structure of 3-acetyl-2H-1-benzopyran-2- one derivatives and was kept unsubstituted While part B contain the Aryl functionality which was substituted with a variety of substituents (fig. 2) and was crucial for LEDGF-dependent HIV-1 IN inhibition activity (fig. 3).
| Compound code | aIC50 (µM) |
|---|---|
| 2a | 86.5±8.5 |
| 2b | >100 (68 %)b |
| 2c | >100 (81 %) |
| 2d | >100 (60 %) |
| 2e | >100 (89 %) |
| 2f | >100 (80 %) |
| 2g | 90±2 |
| 2h | 98.5±0.5 |
| 2i | >100 (83 %) |
| 2j | >100 (82 %) |
| 2k | >100 (62 %) |
| 2l | >100 (85 %) |
| 2m | NT |
| 2n | NT |
| Raltegravir | 0.055±0.002 |
Note: a: Compound concentration required to inhibit the HIV-1 IN catalytic activities in the presence of LEDGF by 50%; b: Percentage of control measured in the presence of 100 µM concentration and NT: Not Tested
Table 4: Ledgf/P75-Dependent Hiv-1 Integrase Inhibition of Synthesized Compounds
Only Compound having para-chloro (Cl) substitution (compound 2a), 3,4-dichloro substitution (compound 2g) and 3,4-dimethoxy substitution (compound 2h) on aryl ring of part B showed LEDGF/p75-dependent HIV-1 integrase inhibition activity. Rest all the compounds having substituents like flouro (F) group, hydroxyl (OH), methyl (CH3), methoxy (OCH3) and dimethylamino (N-(CH3)2) groups were less active as compared to Compounds 2a, 2g and 2h. Possibly, the reason behind such type of activity is the electronegativity of the substituents which was favorable for activity and the compounds which have less electronegative substituents on the aryl nucleus were less active against LEDGF/p75-dependent HIV-1 IN.
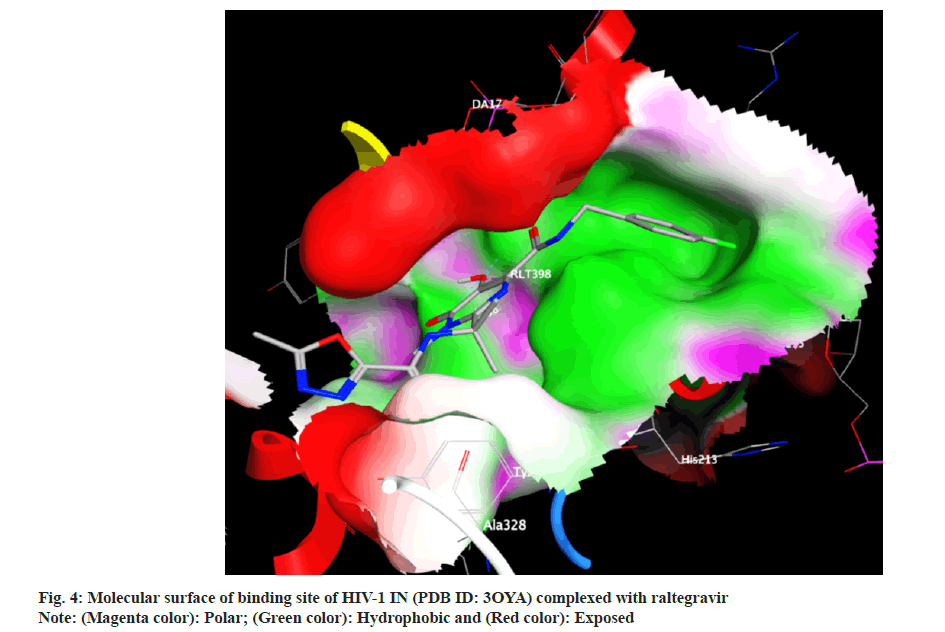
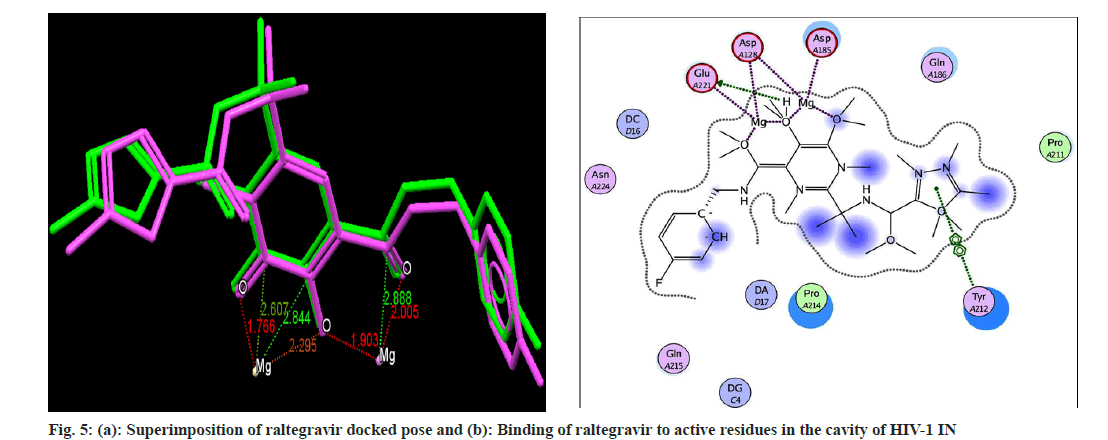
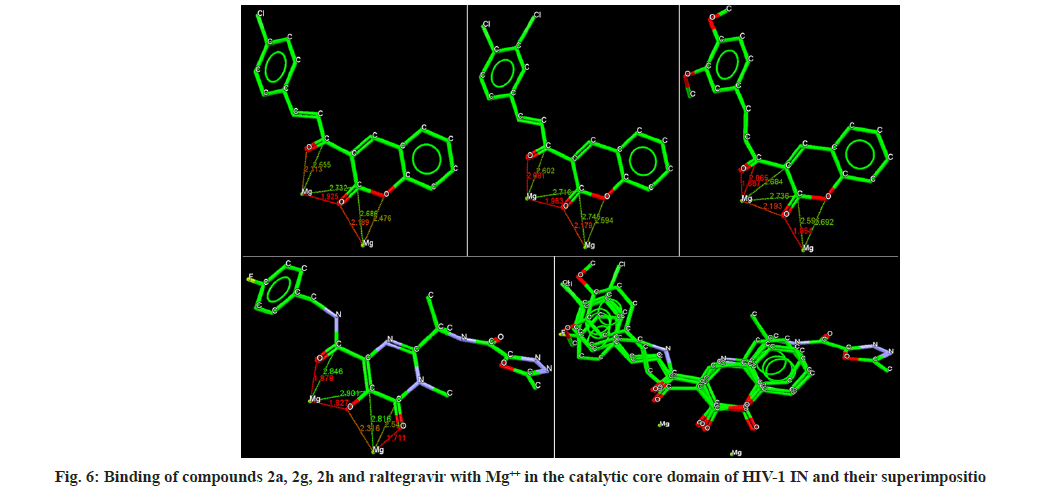
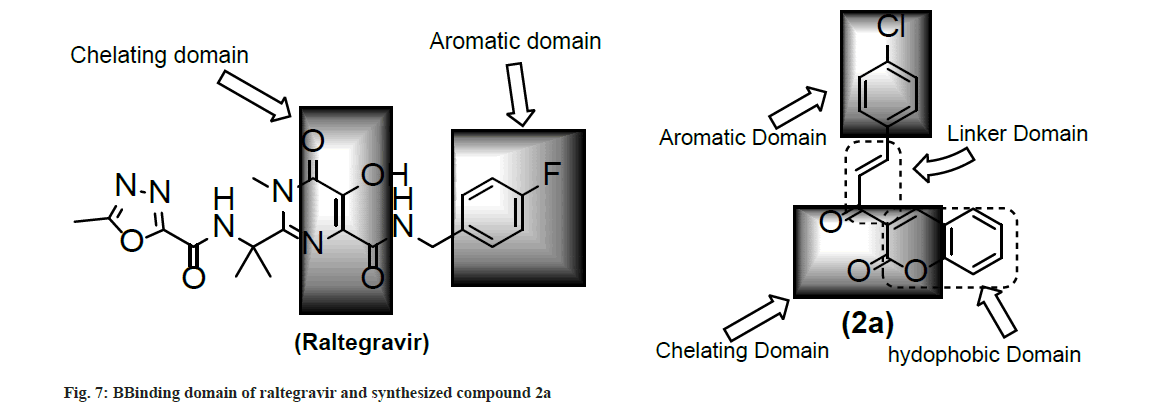
Docking analysis of raltegravir showed its binding to the catalytic metal cations (Mg+2) which is responsible for inactivation of HIV-1 IN by blocking the active site via dislocating the terminal nucleotide of viral DNA. The specifically positioned oxygen atoms in raltegravir form the basis of HIV-1 IN inhibition (fig. 4 and fig. 5). All the synthesized compounds were docked into the same binding pocket of HIV-1 IN using same protocol to predict their binding orientation. Docking of synthesized compounds revealed similar binding pattern as of raltegravir and the position of the oxygen atoms was also same as the docked pose of raltegravir. Oxygen atom at 1, 2 and 1' positions of benzopyran-2-one unit of synthesized compounds 2a-n were important for binding with metal ions (i.e. Mg+2) (fig. 6). Like raltegravir, synthesized compounds also have similar binding groups like aromatic domain, and chelating domain (fig. 7). The GoldScore fitness of synthesized compounds is given in Table 5. It was seen that electron withdrawing groups at phenyl ring were favorable for activity.
| Compound code | GoldScore fitness |
|---|---|
| 2a | 89.760 |
| 2b | 77.302 |
| 2c | 79.234 |
| 2d | 76.993 |
| 2e | 79.538 |
| 2f | 77.315 |
| 2g | 86.652 |
| 2h | 79.446 |
| 2i | 80.494 |
| 2j | 81.336 |
| 2k | 77.065 |
| 2l | 79.260 |
| 2m | 82.602 |
| 2n | 73.333 |
| Raltegravir | 104.861 |
Table 5: Goldscore Fitness Value of Synthesized Compounds
In this study, novel 3-acetyl-2H-1-benzopyran- 2-one derivatives 2a, 2g and 2h were identified as HIV-1 IN inhibitors. Compounds 2a, 2g and 2h inhibited the IN-LEDGF/p75 binding and, thus impaired the HIV-1 IN strand transfer catalytic reaction in the presence of LEDGF/p75 protein. It was found that specifically positioned heteroatoms (preferably oxygen) that coordinated two metal cofactors and electron withdrawing groups at phenyl ring is important for HIV-1 IN inhibition. However, it is still necessary to better understand the exact structural requirement and mechanism of compounds for HIV-1 IN inhibition, and subsequent modification in synthesized compounds to improve activity, which is under progress in our laboratory. In conclution, the 3-acetyl-2H-1-benzopyran- 2-one scaffold can be considered a novel and attractive structure for the development of novel HIV-1 IN inhibitors.
Acknowledgements:
Authors are thankful to Director SGSITS for providing state-of-art facilities and Prof. Enzo for technique supports, which helped in successful completion of this research work.
Financial support and sponsorship:
This work was partially supported by University Grant Commission, New Delhi, India as Major Research Project to M. Tiwari.
Conflict of interests:
The authors declared that there are no conflicts of interest.
References
- Fact sheet 2023. Global HIV statistics.2021.
- Puras Lutzke R, Plasterk R. HIV integrase: A target for drug discovery. Genes Funct 1997;1(5):289-307.
[Crossref] [Google Scholar] [PubMed]
- Smith SJ, Zhao XZ, Burke Jr TR, Hughes SH. HIV-1 integrase inhibitors that are broadly effective against drug-resistant mutants. Antimicrob Agents Chemother 2018;62(9):10-128.
[Crossref] [Google Scholar] [PubMed]
- Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov 2005;4(3):236-48.
[Crossref] [Google Scholar] [PubMed]
- Carcelli M, Rogolino D, Gatti A, Pala N, Corona A, Caredda A, et al. Chelation motifs affecting metal-dependent viral enzymes: N′-acylhydrazone ligands as dual target inhibitors of HIV-1 integrase and reverse transcriptase ribonuclease h domain. Front Microbiol 2017;8:440.
[Crossref] [Google Scholar] [PubMed]
- Sala M, Spensiero A, Esposito F, Scala MC, Vernieri E, Bertamino A, et al. Development and identification of a novel anti-HIV-1 peptide derived by modification of the N-terminal domain of HIV-1 integrase. Front Microbiol 2016;7:845.
[Crossref] [Google Scholar] [PubMed]
- Esposito F, Sechi M, Pala N, Sanna A, Koneru PC, Kvaratskhelia M, et al. Discovery of dihydroxyindole-2-carboxylic acid derivatives as dual allosteric HIV-1 integrase and reverse transcriptase associated ribonuclease H inhibitors. Antiviral Res 2020;174:104671.
[Crossref] [Google Scholar] [PubMed]
- Cocohoba J, Dong BJ. Raltegravir: The first HIV integrase inhibitor. Clin Ther 2008;30(10):1747-65.
[Crossref] [Google Scholar] [PubMed]
- Mouscadet JF, Delelis O, Marcelin AG, Tchertanov L. Resistance to HIV-1 integrase inhibitors: A structural perspective. Drug Resist Updat 2010;13(4-5):139-50.
[Crossref] [Google Scholar] [PubMed]
- de Luca L, de Grazia S, Ferro S, Gitto R, Christ F, Debyser Z, et al. HIV-1 integrase strand-transfer inhibitors: design, synthesis and molecular modeling investigation. Eur J Med Chem 2011;46(2):756-64.
[Crossref] [Google Scholar] [PubMed]
- Fan X, Zhang FH, Al-Safi RI, Zeng LF, Shabaik Y, Debnath B, et al. Design of HIV-1 integrase inhibitors targeting the catalytic domain as well as its interaction with LEDGF/p75: a scaffold hopping approach using salicylate and catechol groups. Bioorg Med Chem 2011;19(16):4935-52.
[Crossref] [Google Scholar] [PubMed]
- Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem 2004;279(47):48883-92.
[Crossref] [Google Scholar] [PubMed]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, de Clercq E, Debyser Z, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 2003;278(35):33528-39.
[Crossref] [Google Scholar] [PubMed]
- Al-Mawsawi LQ, Christ F, Dayam R, Debyser Z, Neamati N. Inhibitory profile of a LEDGF/p75 peptide against HIV-1 integrase: Insight into integrase–DNA complex formation and catalysis. FEBS Lett 2008;582(10):1425-30.
[Crossref] [Google Scholar] [PubMed]
- Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 2010;6(6):442-8.
[Crossref] [Google Scholar] [PubMed]
- Serrao E, Debnath B, Otake H, Kuang Y, Christ F, Debyser Z, et al. Fragment-based discovery of 8-hydroxyquinoline inhibitors of the hiv-1 integrase–lens epithelium-derived growth factor/p75 (in–ledgf/p75) interaction. J Med Chem 2013;56(6):2311-22.
[Crossref] [Google Scholar] [PubMed]
- Srivastav VK, Egbuna C, Tiwari M. Plant secondary metabolites as lead compounds for the production of potent drugs. In: Phytochemicals as lead compounds for new drug discovery 2020: pp. 3-14.
- Matos MJ, Santana L, Uriarte E, Abreu OA, Molina E, Yordi EG. Coumarins: An important class of phytochemicals. Phytochemicals-isolation, characterisation and role in human health 2015;25:533-8.
- Kostova I. Coumarins as inhibitors of HIV reverse transcriptase. Curr HIV Res 2006;4(3):347-63.
[Crossref] [Google Scholar] [PubMed]
- Structure-activity relationships of synthetic coumarins as HIV-1 inhibitors. Bioinorg Chem Appl 2006;2006:68274-74.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Neamati N, Hong H, Mazumder A, Wang S, Sunder S, et al. Coumarin-based inhibitors of HIV integrase. J Med Chem 1997;40(2):242-9.
[Crossref] [Google Scholar] [PubMed]
- Parhami A, Khalafi-Nezhad A, Haghighi SM, Bargebid R, Zare A, Moosavi-Zare AR, et al. Silica supported boric tri-sulfuric anhydride as a novel and efficient catalyst for solvent-free synthesis of coumarins via Pechmann condensation. Arkivoc 2012;9:111-21.
- Smitha G, Sanjeeva Reddy C. ZrCl4?catalyzed Pechmann reaction: Synthesis of coumarins under solvent?free conditions. Synthetic Commun 2004;34(21):3997-4003.
- Dabiri M, Baghbanzadeh M, Kiani S, Vakilzadeh Y. Alum (KAl(SO4)2·12H2O) catalyzed one-pot synthesis of coumarins under solvent-free conditions. Monats Chem 2007;138:997-9.
- Reddy BM, Reddy VR, Giridhar D. Synthesis of coumarins catalyzed by eco-friendly W/ZrO2 solid acid catalyst. Synthetic Commun 2001;31(23):3603-7.
- Pathak S, Debnath K, Pramanik A. Silica sulfuric acid: A reusable solid catalyst for one pot synthesis of densely substituted pyrrole-fused isocoumarins under solvent-free conditions. Beilstein J Org Chem 2013;9(1):2344-53.
- Nazeruddin GM, Pandharpatte MS, Mulani KB. PEG-SO3H: A mild and efficient recyclable catalyst for the synthesis of coumarin derivatives. C R Chim 2012;15(1):91-5.
- Dabiri M, Salehi P, Baghbanzadeh M, Zolfigol MA, Agheb M, Heydari S. Silica sulfuric acid: An efficient reusable heterogeneous catalyst for the synthesis of 2, 3-dihydroquinazolin-4 (1H)-ones in water and under solvent-free conditions. Catalysis Commun 2008;9(5):785-8.
- Paul Gleeson M, Hersey A, Hannongbua S. In silico ADME models: A general assessment of their utility in drug discovery applications. Curr Top Med Chem 2011;11(4):358-81.
[Crossref] [Google Scholar] [PubMed]
- Moroy G, Martiny VY, Vayer P, Villoutreix BO, Miteva MA. Toward in silico structure-based ADMET prediction in drug discovery. Drug Discov Today 2012;17(1-2):44-55.
[Crossref] [Google Scholar] [PubMed]
- Dearden JC. In silico prediction of ADMET properties: How far have we come? Expert Opin Drug Metab Toxicol 2007;3(5):635-9.
[Crossref] [Google Scholar] [PubMed]
- Srivastav VK, Tiwari M. k-nearest neighbor molecular field analysis based 3D-QSAR and in silico ADME/T studies of cinnamoyl derivatives as HIV-1 integrase inhibitors. Med Chem Res 2015;24:684-700.
- Srivastav VK, Tiwari M. QSAR and docking studies of coumarin derivatives as potent HIV-1 integrase inhibitors. Arab J Chem 2017;10:S1081-94.
- Srivastav VK, Tiwari M, Zhang X, Yao XJ. Synthesis and antiretroviral activity of 6-acetylcoumarin derivatives against HIV-1 infection. Indian J Pharm Sci 2018;80(1):108-17.
- QikProp 3.3, Schrodinger-Suite LLC, New York. 2010.
- Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 2015;55(2):460-73.
[Crossref] [Google Scholar] [PubMed]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2012;64:4-17.
[Crossref] [Google Scholar] [PubMed]
- Congreve M, Carr R, Murray C, Jhoti H. A'rule of three'for fragment-based lead discovery? Drug Discov Today 2003;8(19):876-7.
[Crossref] [Google Scholar] [PubMed]
- Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Adv Drug Deliv Rev 2002;54(3):355-66.
[Crossref] [Google Scholar] [PubMed]
- Esposito V, Esposito F, Pepe A, Gomez Monterrey I, Tramontano E, Mayol L, et al. Probing the importance of the G-quadruplex grooves for the activity of the anti-HIV-integrase aptamer T30923. Int J Mol Sci 2020;21(16):5637.
[Crossref] [Google Scholar] [PubMed]
- Pala N, Esposito F, Tramontano E, Singh PK, Sanna V, Carcelli M, et al. Development of a Raltegravir-based photoaffinity-labeled probe for human immunodeficiency virus-1 integrase capture. ACS Med Chem Lett 2020;11(10):1986-92.
[Crossref] [Google Scholar] [PubMed]
- Esposito F, Ambrosio FA, Maleddu R, Costa G, Rocca R, Maccioni E, et al. Chromenone derivatives as a versatile scaffold with dual mode of inhibition of HIV-1 reverse transcriptase-associated Ribonuclease H function and integrase activity. Eur J Med Chem 2019;182:111617.
[Crossref] [Google Scholar] [PubMed]
- Sanna C, Scognamiglio M, Fiorentino A, Corona A, Graziani V, Caredda A, et al. Prenylated phloroglucinols from Hypericum scruglii, an endemic species of Sardinia (Italy), as new dual HIV-1 inhibitors effective on HIV-1 replication. PLoS One 2018;13(3):e0195168.
[Crossref] [Google Scholar] [PubMed]
- Tintori C, Esposito F, Morreale F, Martini R, Tramontano E, Botta M. Investigation on the sucrose binding pocket of HIV-1 Integrase by molecular dynamics and synergy experiments. Bioorg Med Chem Lett 2015;25(15):3013-6.
[Crossref] [Google Scholar] [PubMed]
- Corona A, di Leva FS, Rigogliuso G, Pescatori L, Madia VN, Subra F, et al. New insights into the interaction between pyrrolyl diketoacids and HIV-1 integrase active site and comparison with RNase H. Antiviral Res 2016;134:236-43.
[Crossref] [Google Scholar] [PubMed]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci 2010;107(46):20057-62.
[Crossref] [Google Scholar] [PubMed]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol 1997;267(3):727-48.
[Crossref] [Google Scholar] [PubMed]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins 2003;52(4):609-23.
[Crossref] [Google Scholar] [PubMed]
- Wilson K, Lee AF, Macquarrie DJ, Clark JH. Structure and reactivity of sol–gel sulphonic acid silicas. Appl Catal A Gen 2002;228(1-2):127-33.
- Shimizu KI, Hayashi E, Hatamachi T, Kodama T, Kitayama Y. SO3H-functionalized silica for acetalization of carbonyl compounds with methanol and tetrahydropyranylation of alcohols. Tetrahedron Lett 2004;45(26):5135-8.
- Guo R, Zhu C, Sheng Z, Li Y, Yin W, Chu C. Silica sulfuric acid mediated acylation of amines with 1, 3-diketones via CC bond cleavage under solvent-free conditions. Tetrahedron Lett 2015;56(45):6223-6.
- Manna J, Roy B, Sharma P. Efficient hydrogen generation from sodium borohydride hydrolysis using silica sulfuric acid catalyst. J Power Source 2015;275:727-33.
- Siddiqui ZN, Khan K, Ahmed N. Nano fibrous silica sulphuric acid as an efficient catalyst for the synthesis of β-enaminone. Catal Lett 2014;144:623-32.
- Pavia DL, Lampman GM, Kriz GS, Vyvyan JA. Introduction to spectroscopy: A Guide for students of organic chemistry. Cengage learning; 2019:29-82.