- *Corresponding Author:
- J. Hu
Department of Hepatology, Shenzhen Bao’an Chinese Medicine Hospital, Guangzhou University of Chinese Medicine, Shenzhen, Guangdong 518133, China
E-mail: rzzyhjb@126.com
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “141-151” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Non-alcoholic steatohepatitis is an intermediate link in the transformation from non-alcoholic fatty liver disease to non-alcoholic cirrhosis. Jianpi Shenshi Jiangzhuo formula, an effective prescription for treating non-alcoholic fatty liver disease, is summarized by our research group on the basis of long-term clinical practice. Based on the network pharmacology and molecular docking, this study explores the mechanism of Jianpi Shenshi Jiangzhuo formula in non-alcoholic steatohepatitis treatment. The expression profiles of non-alcoholic steatohepatitis-associated genes were downloaded and the potential disease targets were obtained by differential gene analysis. 11 single drug ingredients of the Jianpi Shenshi Jiangzhuo formula were searched and the active ingredients were screened with oral bioavailability ≥30 % and drug-likeness ≥0.18 as the criteria. The corresponding targets were found and the Kyoto Encyclopedia of Genes and Genomes pathway and Gene Ontology functional pathway enrichment analyses were carried out to identify mitochondrial pathway genes. The drug target genes were then intersected with the differentially expressed genes of the disease to identify the overlapping genes, after which molecular docking between active ingredients and core targets was performed.
Keywords
Jianpi Shenshi Jiangzhuo formula, network pharmacology, molecular docking, nonalcoholic steatohepatitis
In case of obesity, the incidence of metabolic diseases has increased exponentially over the past few decades[1]. Insulin resistance, type 2 diabetes, hypertension, dyslipidemia, cardiovascular diseases, and fatty liver have been recognized as obesityrelated comorbidities[2]. Non-Alcoholic Fatty Liver Disease (NAFLD) is also a major independent risk factor for the development of cardiovascular diseases[3]. NAFLD is a series of medical conditions associated with fatty degeneration caused by the accumulation of >5 % of triglyceride vesicles in hepatocytes[4]. This process occurs due to the mismatch in the regulatory mechanisms involved in lipid metabolism, with some patients developing into more aggressive disease subtypes characterized by lobular inflammation and hepatic balloon degeneration, with or without fibrosis, called Non- Alcoholic Steatohepatitis (NASH)[5,6]. Approximately 22.5 % of NASH patients will develop hepatocellular carcinoma and 20 % will develop cirrhosis[7,8]. At present, there are no specific drugs to treat NASH.
The pathogenesis of NAFLD is mainly related to mitochondrial dysfunction, insulin resistance, oxidative stress and adipocytokine secretion disorder, etc., but the specific mechanism of action is not yet clear[9]. Therefore, improving the body's lipid metabolism is the key to NAFLD treatment. In addition, mitochondria are the most important organelles for fatty acid Beta (β) oxidation in the liver and play an important role in regulating fatty acid metabolism[10]. Mitochondria have been found to be critical in the pathogenesis of NAFLD[11]. Normal individuals maintain a relative balance between lipid uptake and synthesis through mitochondrial fatty acid β-oxidation and Very Low Density Lipoprotein (VLDL) secretion. Free fatty acids enter mitochondria for β-oxidation or esterification to triacylglycerols, and are eventually secreted in the form of VLDL. However, hindered or excessive oxidation of fatty acids and increased production of VLDL can lead to fatty liver[12,13]. Therefore, maintaining the normal function of hepatocyte mitochondria cannot be ignored in the treatment of NASH.
Studies have confirmed that Traditional Chinese Medicine (TCM) has a promising prospect in regulating blood lipids and improving lipid metabolism, and has become an important means of clinical treatment of NAFLD[14]. Jianpi Shenshi Jiangzhuo formula is an effective prescription for treating NAFLD, which is summarized by our research group on the basis of long-term clinical practice. Its main medicinal components include radix Pseudostellaria, roasted rhizoma Atractylodes macrocephala, rhizoma Atractylodes, Poria cocos, Magnolia officinalis, rhizoma Pinellia ternata, talcum powder, Tetrapanax papyrifer, radix Bupleuri, Fructus aurantii immaturus, radix Paeonia Rubra, radix Panax notoginseng, radix Rubia and Broussonetia. All the components are closely associated with the functions of stimulating spleen, promoting diuresis, soothing liver, regulating qi, eliminating dampness, resolving phlegm, promoting qi circulation and reducing turbidity. It has been reported that other decoctions containing the above TCM can play an important role in protecting liver cells[15].
In this study, the Gene Expression Omnibus (GEO) database and network pharmacology method were used to screen the effective components and targets of Jianpi Shenshi Jiangzhuo formula, and their intersections were mapped with the Differentially Expressed Genes (DEGs) of NASH. Moreover, were analyzed the possible pathways to screen the mitochondrial pathways, so as to predict the potential targets and mechanisms of action of Jianpi Shenshi Jiangzhuo formula in treating NASH, providing a theoretical basis for further research.
Materials and Methods
NASH-related target screening:
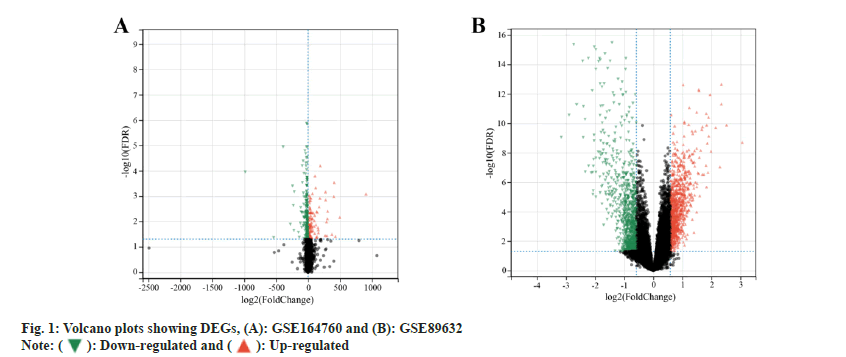
GEO (https://www.ncbi.nlm.nih.gov/geo/) microarray datasets GSE164760[16] and GSE89632[17] of NASH were selected to download the relevant Gene Set Enrichment (GSE) and GEO Platform (GPL) files, which were then sorted out, grouped and imported into R-software for differential analysis using the Limma package, and the DEGs were screened with an adjusted value of p<0.05 and Logarithm of Fold Change (logFC)>0.5 as the thresholds. The ggplot package was used to present the DEGs in a volcano plot.
Screening of active ingredients and targets of the Jianpi Shenshi Jiangzhuo formula:
The active ingredients of radix Pseudostellaria, rhizoma Atractylodes, Poria cocos, Magnolia officinalis, Tetrapanax papyrifer, radix Bupleuri, Fructus aurantii immaturus, radix Paeonia Rubra, radix Panax notoginseng, radix Rubia and Broussonetia in Jianpi Shenshi Jiangzhuo formula were searched by using the Traditional Chinese Medicine Systems Pharmacology (TCMSP) platform (https://old.tcmsp-e.com/tcmsp.php). With Druglikeness (DL) ≥0.18 and Oral Bioavailability (OB) ≥30 % as screening parameters, the active ingredients with good drug properties were screened. Universal Protein Resource (UniProt) database (https://www.uniprot.org/) was used to correct the names of target genes of chemical components, so as to normalize the protein target information and remove duplicate genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) functional enrichment analysis:
The clusterProfiler package of R-software was used to conduct GO functional enrichment and KEGG pathway enrichment analysis of the non-repetitive target genes and DEGs in GEO microarray datasets. "Mitochondria" as keyword, mitochondria-related pathways were screened in the enrichment results in order to analyze the Molecular Function (MF), Cell Components (CC), Biological Processes (BP), and metabolic pathways of genes.
Identification of overlapping genes of drugs and diseases:
The previously obtained genes of the active ingredients and disease-related DEGs were mapped by Venn diagram, and the overlapping genes were obtained.
Molecular docking:
The active ingredients and core target genes of the Jianpi Shenshi Jiangzhuo formula were molecularly docked. Primarily, the structural formula of the active ingredient (MOL2 format) was downloaded from the TCMSP database. Next, the protein conformation of the target genes was searched in the Protein Data Bank (PDB) (https://www.rcsb.org/). Screening was carried out based on the following criteria; identification of protein structure by X-ray diffraction; crystal resolution of the protein <3 Å; being a well-defined protein. The proteins were then pretreated with Autotools to remove water molecules by hydrogenation. Energy lattice calculation and small molecule-protein docking were then performed using AutoGrid and AutoDock Vina software, respectively, and binding energy (affinity) scores for each docking was studied. The part with a binding energy <-7 was selected for plotting.
Results and Discussion
GEO differential gene analysis was carried out. The expression profiles of genes namely, GSE164760 and GSE89632 were retrieved from GEO database. Among 6 normal samples and 74 NASH samples which were included in the GSE164760 microarray dataset, 384 DEGs were identified, including 270 down-regulated and 114 up-regulated genes (fig. 1A). Similarly, in the GSE89632 dataset, 24 normal samples and 19 NASH samples were found, from which 2815 DEGs were identified, with 1386 down-regulated and 1429 up-regulated genes (fig. 1B).
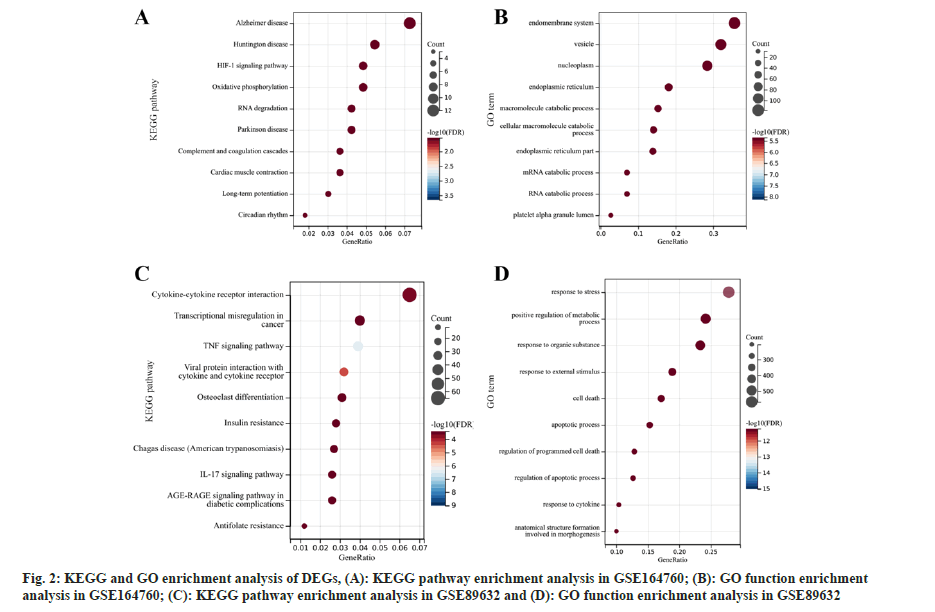
Pathway enrichment analysis of DEGs was studied. Subsequently, the GO function and KEGG pathway enrichment analyses of the DEGs found in the GSE164760 and GSE89632 microarray datasets were analyzed (fig. 2). According to KEGG pathway enrichment analysis, DEGs in GSE164760 were dominantly enriched in Alzheimer’s disease, Huntington disease, and Hypoxia-Inducible Factor 1 (HIF-1) signaling pathway (fig. 2A). GO function enrichment analysis showed that the DEGs in GSE164760 were mainly enriched in endomembrane system, vesicles, nucleoplasm and other functions (fig. 2B). However, the DEGs in GSE89632 were primarily enriched in cytokine-cytokine receptor interaction pathway (fig. 2C) and response to stress function (fig. 2D). Using the keyword “mitochondria” we searched and screened the mitochondria-related pathways. There was no mitochondrial pathway in GSE89632, while one associated pathway, “mitochondrial electron transport, cytochrome c to oxygen”, was found in GSE164760, which includes 5 DEGs namely, Cytochrome C Oxidase Subunit 8A (COX8A), COX6C, COX7B, COX7C and COX6A1.
The active ingredients and targets of Jianpi Shenshi Jiangzhuo formula were screened. A total of 1067 related compounds were found from the TCMSP database for 11 single drug ingredients. With OB ≥30 % and DL ≥0.18 as the screening criteria, a total of 93 active ingredients were obtained. In the TCMSP database, the potential targets involved in each active ingredient were obtained, sorted out, and then deduplicated by UniProt database comparison and correction. A total of 224 non-repetitive potential targets were obtained.
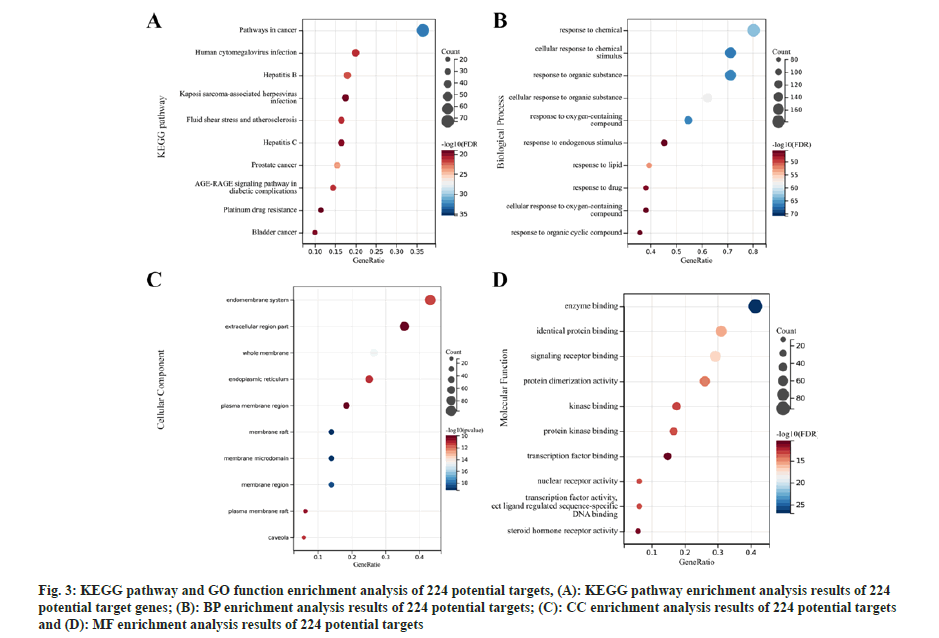
Further, functional enrichment analysis of potential targets of active ingredients was studied. A total of 2598 GO pathways (BP: 2332; CC: 70 and MF: 196) and 188 KEGG pathways were acquired through KEGG pathway and GO function enrichment analysis of the 224 potential targets. The top 10 functional pathways have been presented in fig. 3. From these enrichment results, mitochondria-related pathways were further screened by using the keyword “mitochondria”. 26 associated pathways were obtained, returning 189 mitochondria-related genes, 31 of which were found after removing duplicate genes (Table 1).
| Ontology | ID | Description | Gene ID | Count |
|---|---|---|---|---|
| CC | GO:0005741 | Mitochondrial outer membrane | BCL2L1/MCL1/CASP8/PGR/BAX/BCL2/MAOB/MAOA/GJA1/RAF1/HK2/ACSL4/HK1/ACSL1/MTOR | 15 |
| BP | GO:0006839 | Mitochondrial transport | TP53/BCL2L1/CASP8/BAX/BCL2/GSK3B/BBC3/HK2/ACACA/E2F1/NPEPPS/MAPK8/SREBF1/SREBF2 | 14 |
| BP | GO:0008637 | Apoptotic mitochondrial changes | AKT1/MMP9/TP53/BCL2L1/CASP8/BAX/BCL2/GSK3B/BBC3/HK2/E2F1/MAPK8 | 12 |
| BP | GO:0046902 | Regulation of mitochondrial membrane permeability | TP53/BCL2L1/CASP8/BAX/BCL2/GSK3B/BBC3/HK2/E2F1/MAPK8 | 10 |
| BP | GO:0007006 | Mitochondrial membrane organization | TP53/BCL2L1/CASP8/BAX/BCL2/GSK3B/BBC3/HK2/E2F1/MAPK8 | 10 |
| BP | GO:0051881 | Regulation of mitochondrial membrane potential | AKT1/BCL2L1/BAX/BCL2/KDR/OPRD1/PARP1/SOD1/CASP1 | 9 |
| BP | GO:1902108 | Regulation of mitochondrial membrane permeability involved in apoptotic process | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:0035794 | Positive regulation of mitochondrial membrane permeability | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:1902686 | Mitochondrial outer membrane permeability involved in programmed cell death | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:1902110 | Positive regulation of mitochondrial membrane permeability involved in apoptotic process | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:0097345 | Mitochondrial outer membrane permeability | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:1901028 | Regulation of mitochondrial outer membrane permeability involved in apoptotic signaling pathway | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:1901030 | Positive regulation of mitochondrial outer membrane permeability involved in apoptotic signaling pathway | TP53/CASP8/BAX/BCL2/GSK3B/BBC3/E2F1/MAPK8 | 8 |
| BP | GO:0001836 | Release of cytochrome C from mitochondria | AKT1/MMP9/TP53/BCL2L1/BAX/BCL2/BBC3 | 7 |
| BP | GO:0090151 | Establishment of protein localization to mitochondrial membrane | TP53/CASP8/BAX/BCL2/BBC3/E2F1/MAPK8 | 7 |
| BP | GO:0051204 | Protein insertion into mitochondrial membrane | TP53/CASP8/BAX/BCL2/BBC3/E2F1/MAPK8 | 7 |
| BP | GO:0001844 | Protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | TP53/CASP8/BAX/BCL2/BBC3/E2F1/MAPK8 | 7 |
| BP | GO:0090199 | Regulation of release of cytochrome C from mitochondria | AKT1/MMP9/TP53/BCL2L1/BAX/BBC3 | 6 |
| BP | GO:1900739 | Regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | TP53/CASP8/BCL2/BBC3/E2F1/MAPK8 | 6 |
| BP | GO:1900740 | Positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | TP53/CASP8/BCL2/BBC3/E2F1/MAPK8 | 6 |
| BP | GO:0090200 | Positive regulation of release of cytochrome C from mitochondria | MMP9/TP53/BAX/BBC3 | 4 |
| BP | GO:0051882 | Mitochondrial depolarization | BCL2/KDR/PARP1/CASP1 | 4 |
| BP | GO:0051900 | Regulation of mitochondrial depolarization | BCL2/KDR/PARP1 | 3 |
| BP | GO:0000002 | Mitochondrial genome maintenance | TP53/PARP1 | 2 |
| BP | GO:0090201 | Negative regulation of release of cytochrome C from mitochondria | AKT1/BCL2L1 | 2 |
| BP | GO:0032042 | Mitochondrial Deoxyribonucleic Acid (DNA) metabolic process | TP53/PARP1 | 2 |
Table 1: 26 Pathways Related To Mitochondria
| TCM | Scientific name | Molecular ID | Molecule name | Symbol | OB | DL |
|---|---|---|---|---|---|---|
| Chishao | Radix Paeonia Rubra | MOL000842 | Sucrose | HK1 | 7.171 | 0.227 |
| Chishao | Radix Paeonia Rubra | MOL000842 | Sucrose | SREBF2 | 7.171 | 0.227 |
| Chishao | Radix Paeonia Rubra | MOL000842 | Sucrose | SREBF1 | 7.171 | 0.227 |
| Chaihu | Radix Bupleuri | MOL000098 | Quercetin | GJA1 | 46.433 | 0.275 |
| Chaihu | Radix Bupleuri | MOL000098 | Quercetin | HK2 | 46.433 | 0.275 |
| Chaihu | Radix Bupleuri | MOL000098 | Quercetin | MMP9 | 46.433 | 0.275 |
| Sanqi | Panax notoginseng (Burk.) F. H. Chen Ex C. Chow | MOL000098 | Quercetin | HK2 | 46.433 | 0.275 |
| Sanqi | Panax notoginseng (Burk.) F. H. Chen Ex C. Chow | MOL000098 | Quercetin | GJA1 | 46.433 | 0.275 |
| Sanqi | Panax notoginseng (Burk.) F. H. Chen Ex C. Chow | MOL000098 | Quercetin | MMP9 | 46.433 | 0.275 |
| Zhishi | Fructus aurantii immaturus | MOL005828 | Nobiletin | MMP9 | 61.669 | 0.517 |
| Zhishi | Fructus aurantii immaturus | MOL004328 | Naringenin | SREBF1 | 59.294 | 0.211 |
| Taizishen | Pseudostellaria radix | MOL000006 | Luteolin | MMP9 | 36.163 | 0.246 |
| Zhishi | Fructus aurantii immaturus | MOL000006 | Luteolin | MMP9 | 36.163 | 0.246 |
| Chushiziu | Broussonetia | MOL000006 | Luteolin | MMP9 | 36.163 | 0.246 |
| Sanqi | Panax notoginseng (Burk.) F. H. Chen Ex C. Chow | MOL005344 | Ginsenoside Rh2 | CASP1 | 36.32 | 0.559 |
| Chishao | Radix Paeonia Rubra | MOL001002 | Ellagic acid | MMP9 | 43.065 | 0.434 |
| Chushiziu | Broussonetia | MOL002773 | β-carotene | GJA1 | 37.184 | 0.584 |
| Chishao | Radix Paeonia Rubra | MOL002714 | Baicalein | MMP9 | 33.519 | 0.209 |
Table 2: 7 Corresponding Genes to the 9 Active Chemical Components of the Jianpi Shenshi Jiangzhuo Formula
Fig 3: KEGG pathway and GO function enrichment analysis of 224 potential targets, (A): KEGG pathway enrichment analysis results of 224 potential target genes; (B): BP enrichment analysis results of 224 potential targets; (C): CC enrichment analysis results of 224 potential targets and (D): MF enrichment analysis results of 224 potential targets
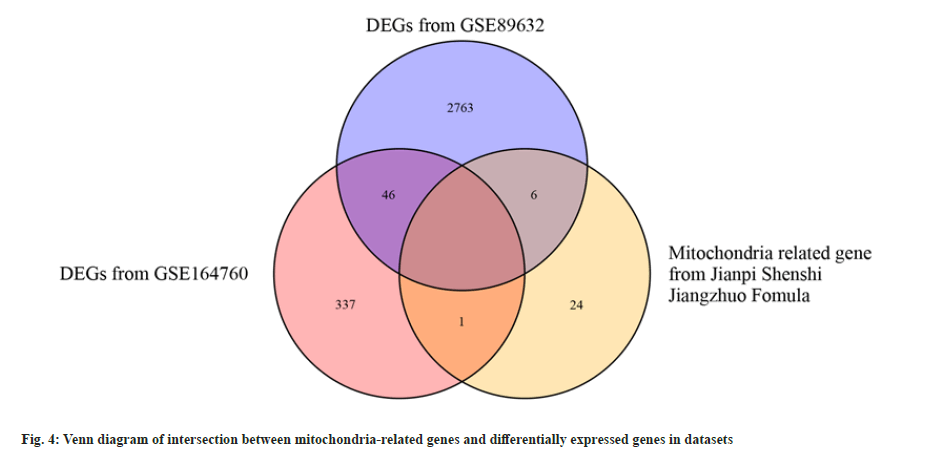
Molecular docking of active ingredients with core targets was studied. The target genes of the Jianpi Shenshi Jiangzhuo formula were intersected with the 5 NASH mitochondria-associated pathway genes, and no intersection was found. Therefore, the 31 mitochondria-related genes were used to intersect with DEGs of both the NASH datasets, i.e., GSE89632 and GSE164760. As shown in the fig. 4, 7 of the 31 mitochondria-related genes were found to be associated with NASH DEGs.
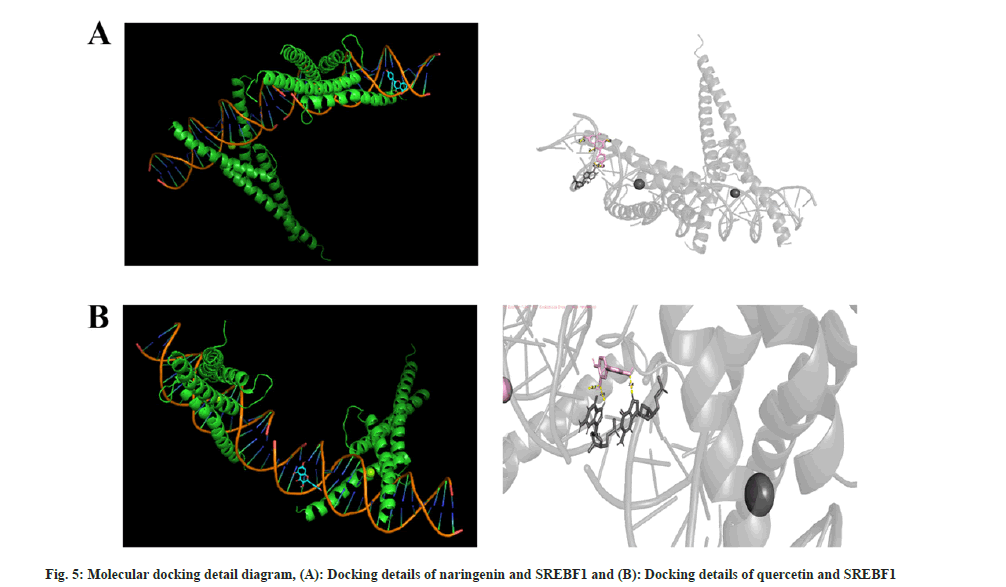
They are, Gap Junction protein Alpha 1 (GJA1), Sterol Regulatory Element-Binding Protein 1 (SREBF1), Caspase-1 (CASP1), SREBF2, Hexokinase 2 (HK2), Matrix Metalloproteinase-9 (MMP-9), and HK1, corresponding to the 9 active chemical components in Jianpi Shenshi Jiangzhuo formula (Table 2). Finally, the Vina tool was used for batch molecular docking, and the results showed that the genes that docked well with the active ingredients were SREBF1, CASP1, HK1, and HK2 (Table 3). The detailed simulation diagrams of molecular docking between naringenin and SREBF1 and quercetin and SREBF1 are shown in fig. 5. TCM treatment of NAFLD from a holistic perspective, integrates the patient's physique, the causes and laws of the disease to comprehensively regulate the qi-blood and yin-yang imbalance. Treatment based on syndrome differentiation, specific method and prescription for certain illness, experience of renowned doctor, integrated traditional Chinese and Western medicine therapy, and multi-channel and approach treatment of TCM have achieved unique therapeutic advantages in controlling etiology, lowering blood lipid and glucose, promoting the regression of liver fat deposits, reducing hepatocyte necrosis, inflammation and fibrosis[18]. Shenshi Jiangzhuo formula is the experiential effective prescription developed by Chen Qichang, a famous doctor in Henan Province in the late Qing Dynasty and the early Republic of China, which has the effects of inducing diuresis and excreting dampness, harmonizing and promoting blood circulation, removing blood stasis, promoting qi circulation and reducing turbidity.
| PDB ID | Gene | PubChem ID | Molecule name | Binding energy (kcal/mol) |
|---|---|---|---|---|
| Lam9 | SREBF1 | 932 | Naringenin | -7.4 |
| Lam9 | SREBF1 | 5988 | Sucrose | -5.5 |
| Lam9 | SREBF1 | 5280343 | Quercetin | -7.2 |
| 5HEX | HK2 | 5280343 | Quercetin | -6.8 |
| 5HEX | HK2 | 932 | Naringenin | -7.4 |
| 5HEX | HK2 | 5988 | Sucrose | -5.2 |
| Lam9 | SREBF1 | 72344 | Nobiletin | -7.8 |
| 5HEX | HK2 | 72344 | Nobiletin | -6.8 |
| Lam9 | SREBF1 | 119307 | Ginsenoside Rh2 | -8 |
| 5HEX | HK2 | 119307 | Ginsenoside Rh2 | -6.8 |
| Lam9 | SREBF1 | 5280445 | Luteolin | -7.4 |
| 5HEX | HK2 | 5280445 | Luteolin | -7.2 |
| Lam9 | SREBF1 | 5280489 | β-carotene | -8.2 |
| Lcza | HK1 | 5280489 | β-carotene | -5.7 |
| 2hbq | CASP1 | 5280489 | β-carotene | -4.2 |
| 5HEX | HK2 | 5280489 | β-carotene | -7.8 |
| Lam9 | SREBF1 | 5281605 | Baicalein | -7.5 |
| 5HEX | HK2 | 5281605 | Baicalein | -7.2 |
Table 3: Molecular Docking of Components and Core Targets
Network pharmacology is a new data analysis technology emerging in recent years. This study applies this technology and data mining to validate the scientificity and feasibility of Jianpi Shenshi Jiangzhuo formula in the treatment of NASH. Among the TCM components of Jianpi Shenshi Jiangzhuo formula, rhizoma Atractylodes and Magnolia officinalis can eliminate dampness to resolve phlegm, improve digestion, relieve chest and diaphragm discomfort caused by gas upwelling[19]; radix Bupleuri and Fructus aurantii immaturus have the functions of soothing the liver, regulating qi, resolving phlegm, dissipating distention and fullness, promoting blood circulation, removing blood stasis, and eliminating alcohol and facilitating digestion[20,21]. Poria cocos, also known as Yuling and Wanlinggui, is sweet and light in taste, with the functions of stimulating spleen, relieving edema, calming the mind, and relieving diarrhea[22]; rhizoma Pinellia ternata is capable of drying dampness to resolve phlegm and removing blood stasis[23]; radix Panax notoginseng, radix Paeonia Rubra and radix Rubia can promote blood circulation, remove blood stasis, nourish blood, promote blood circulation, and dredge menstruation[24-26]; talcum powder and Tetrapanax papyrifer have the effects of clearing damp, promoting diuresis, and relieving difficult urination, edema, fullness, and phlegm and retained fluid-induced dizziness[27,28]; Broussonetia has the function of nourishing kidney, clearing liver-heat and improving eyesight[29]. The combination of the above drugs can effectively treat NAFLD and give full play to their respective medicinal effects, complementing each other.
Mitochondrial function is closely related to the metabolic function of the three major nutrients in the liver, so it is also important to the pathology of NASH[30]. Lipid deposition in NASH cells is bound to physically destroy the formation of mitochondria in the cells and negatively affect mitochondrial function, suggesting a close pathological relationship between them. In this study, 93 active ingredients and 224 non-repetitive potential targets were obtained by screening. Subsequently, KEGG pathway and GO function analyses of the 224 potential targets were performed, and mitochondrial pathways were screened, returning 31 target genes. Seven overlapped mitochondrial genes, GJA1, SREBF1, CASP1, SREBF2, HK2, MMP9, and HK1, corresponding to 9 active chemical components (sucrose, quercetin, nobiletin, naringenin, luteolin, ginsenoside Rh2, ellagic acid, β-carotene, and baicalein) in Jianpi Shenshi Jiangzhuo formula, were obtained by combining the DEGs in the NASH-associated datasets. Among them, quercetin is a flavonoid found in the human diet that has been shown to have a wide range of activities in preventing common diseases[31]. In addition, studies in mice showed that quercetin treatment ameliorated inflammation and fibrosis in mice with NASH[32]. Nobiletin improves the efficacy of hypercholesterolemia and NAFLD in mice on a high-cholesterol diet[33]. It may also be involved in the prevention of NAFLD and its associated risk factors by increasing the expression of Peroxisome Proliferator-Activated Receptor (PPAR) target genes such as PPAR-Alpha (α), PPAR-Gamma (γ) and regulating energy homeostasis[34]. The role of luteolin is linked to changes in the gut microbiota that help prevent progression from simple steatosis to NASH[35].
While ginsenoside Rh2 which is a natural compound, reduces the number and size of lipid droplets in 3T3-L1 adipocytes, which can be used as a natural pharmaceutical approach for NAFLD[36]. Similarly, the use of ellagic acid can activate the liver Adenosine- Monophosphate Activated-Protein Kinase (AMPK) pathway to treat liver diseases and NAFLD and prevent hepatic steatosis[37]. Baicalein, on the other hand, affects the intestinal microbial community structure and liver transcriptome expression through its own metabolism, thus affecting the metabolism of NAFLD by liver fatty acids[38].
Finally, we used the Vina tool to dock molecules in batches. The results showed that SREBF1, CASP1, HK1, and HK2 were the genes that docked well with the active ingredients. SREBF1 belongs to the Sterol Regulatory Element Binding Proteins (SREBPs) family, can stimulate the expression of lipogenic genes such as acetyl-CoA carboxylase and Fatty Acid Synthase (FAS)[39]. There is increasing evidence that hepatic steatosis is associated with increased expression of SREBF1-mediated pathway, and inhibition of SREBF1 can reduce cellular fat accumulation in vivo and in vitro[40]. In a word, the loss of CASP1 ameliorates the damaging effects of obesity induced by a high-fat diet. CASP1 plays a central regulatory role in high-fat diet-induced NASH, and mice with CASP1 deficiency are protected from high-fat induced hepatic steatosis, inflammation, and early fibrosis[41]. Studies have shown higher expression of HK1 in liver samples from patients with NASH compared to samples from patients with NAFLD[42]. All the selected target genes mentioned above belong to mitochondria-related genes. It also means that these active ingredients of Jianpi Shenshi Jiangzhuo formula may have therapeutic effects on NASH by regulating these genes and pathways.
However, our study has some limitations. First, the results need to be validated by further experiments. Second, a more comprehensive database of target genes of TCM is needed to make the results of network pharmacology analysis more reliable. Third, even combining the results of network pharmacology and molecular docking, the exact therapeutic mechanism of Jianpi Shenshi Jiangzhuo formula and NASH it is still not completely comprehensible, which relies on multidisciplinary co-development.
In summary, this study focused on the active ingredients of Jianpi Shenshi Jiangzhuo formula and explored the mechanism of action of this formula in the treatment of NASH through combining differential analyses of NASH datasets in GEO and network pharmacology to reveal the potential therapeutic role of the mitochondrial pathway in NASH, providing a new reference for the application of Jianpi Shenshi Jiangzhuo formula in the treatment of NASH.
Funding:
This study was supported by the Shenzhen Excellent Science and Technology Innovation Talent Training Project (number: RCBS20210706092217036) and Discipline construction project of Guangdong Medical University (grant number: 4SG21229GDGFY01)
Conflict of interests:
The authors declared no conflict of interests.
References
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64(1):73-84.
[Crossref] [Google Scholar] [PubMed]
- Kumar R, Priyadarshi RN, Anand U. Non-alcoholic fatty liver disease: Growing burden, adverse outcomes and associations. J Clin Transl Hepatol 2020;8(1):76-86.
[Crossref] [Google Scholar] [PubMed]
- Kasper P, Martin A, Lang S, Kuetting F, Goeser T, Demir M, et al. NAFLD and cardiovascular diseases: A clinical review. Clin Res Cardiol 2021;110(7):921-37.
[Crossref] [Google Scholar] [PubMed]
- Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American association for the study of liver diseases. Hepatology 2018;67(1):328-57.
[Crossref] [Google Scholar] [PubMed]
- Ajmera V, Kim BK, Yang K, Majzoub AM, Nayfeh T, Tamaki N, et al. Liver stiffness on magnetic resonance elastography and the MEFIB index and liver-related outcomes in nonalcoholic fatty liver disease: A systematic review and meta-analysis of individual participants. Gastroenterology 2022;163(4):1079-89.
[Crossref] [Google Scholar] [PubMed]
- Cen J, Han Y, Liu Y, Hu H. Evaluated glomerular filtration rate is associated with non-alcoholic fatty liver disease: A 5-year longitudinal cohort study in Chinese non-obese people. Front Nutr 2022;9:1-16.
[Crossref] [Google Scholar] [PubMed]
- Witkowski M, Moreno SI, Fernandes J, Johansen P, Augusto M, Nair S. The economic burden of non-alcoholic steatohepatitis: A systematic review. Pharmacoeconomics 2022;40(8):751-76.
[Crossref] [Google Scholar] [PubMed]
- Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. Nonalcoholic steatohepatitis: A review. JAMA 2020;323(12):1175-83.
[Crossref] [Google Scholar] [PubMed]
- Karkucinska‐Wieckowska A, Simoes IC, Kalinowski P, Lebiedzinska‐Arciszewska M, Zieniewicz K, Milkiewicz P, et al. Mitochondria, oxidative stress and nonalcoholic fatty liver disease: A complex relationship. Eur J Clin Invest 2022;52(3):1-19.
[Crossref] [Google Scholar] [PubMed]
- Adeva-Andany MM, Carneiro-Freire N, Seco-Filgueira M, Fernández-Fernández C, Mouriño-Bayolo D. Mitochondrial β-oxidation of saturated fatty acids in humans. Mitochondrion 2019;46:73-90.
[Crossref] [Google Scholar] [PubMed]
- di Ciaula A, Passarella S, Shanmugam H, Noviello M, Bonfrate L, Wang DQ, et al. Nonalcoholic Fatty Liver Disease (NAFLD). Mitochondria as players and targets of therapies? Int J Mol Sci 2021;22(10):1-45.
[Crossref] [Google Scholar] [PubMed]
- Panov A, Mayorov VI, Dikalov S. Metabolic syndrome and β-oxidation of long-chain fatty acids in the brain, heart, and kidney mitochondria. Int J Mol Sci 2022;23(7):1-19.
[Crossref] [Google Scholar] [PubMed]
- Barbier-Torres L, Fortner KA, Iruzubieta P, Delgado TC, Giddings E, Chen Y, et al. Silencing hepatic MCJ attenuates Non-alcoholic Fatty Liver Disease (NAFLD) by increasing mitochondrial fatty acid oxidation. Nat Commun 2020;11(1):1-15.
[Crossref] [Google Scholar] [PubMed]
- Xu Y, Guo W, Zhang C, Chen F, Tan HY, Li S, et al. Herbal medicine in the treatment of non-alcoholic fatty liver diseases-efficacy, action mechanism, and clinical application. Front Pharmacol 2020;11:1-19.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Liu H. Efficacy of Gegenqinlian Xiezhuo decoction in the treatment of non-alcoholic fatty liver of spleen deficiency and blood stasis syndrome. J Contemp Med 2022;4(2):1-5.
- Pinyol R, Torrecilla S, Wang H, Montironi C, Piqué-Gili M, Torres-Martin M, et al. Molecular characterisation of hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J Hepatol 2021;75(4):865-78.
[Crossref] [Google Scholar] [PubMed]
- Arendt BM, Comelli EM, Ma DW, Lou W, Teterina A, Kim T, et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology 2015;61(5):1565-78.
[Crossref] [Google Scholar] [PubMed]
- Shi T, Wu L, Ma W, Ju L, Bai M, Chen X, et al. Nonalcoholic fatty liver disease: Pathogenesis and treatment in traditional Chinese medicine and Western medicine. Evid Based Complement Alternat Med 2020:1-16.
[Crossref] [Google Scholar] [PubMed]
- Jun X, Fu P, Lei Y, Cheng P. Pharmacological effects of medicinal components of Atractylodes lancea (Thunb.) DC. Chin Med 2018;13:1-10.
[Crossref] [Google Scholar] [PubMed]
- Yang F, Dong X, Yin X, Wang W, You L, Ni J. Radix Bupleuri: A review of traditional uses, botany, phytochemistry, pharmacology, and toxicology. Biomed Res Int 2017;2017:7597596.
[Crossref] [Google Scholar] [PubMed]
- Wu J, Huang G, Li Y, Li X. Flavonoids from aurantii Fructus immaturus and aurantii Fructus: Promising phytomedicines for the treatment of liver diseases. Chin Med 2020;15(1):1-18.
[Crossref] [Google Scholar] [PubMed]
- Zhang W, Chen L, Li P, Zhao J, Duan J. Antidepressant and immunosuppressive activities of two polysaccharides from Poria cocos (Schw.) wolf. Int J Biol Macromol 2018;120:1696-704.
[Crossref] [Google Scholar] [PubMed]
- Liu XQ, Wu H, Yu HL, Zhao TF, Pan YZ, Shi RJ. Purification of a lectin from Arisaema erubescens (Wall.) schott and its pro-inflammatory effects. Molecules 2011;16(11):9480-94.
[Crossref] [Google Scholar] [PubMed]
- Ng TB. Pharmacological activity of sanchi ginseng (Panax notoginseng). J Pharm Pharmacol 2006;58(8):1007-19.
[Crossref] [Google Scholar] [PubMed]
- Tan YQ, Chen HW, Li J, Wu QJ. Efficacy, chemical constituents, and pharmacological actions of Radix Paeoniae Rubra and Radix Paeoniae alba. Front Pharmacol 2020;11:1-11.
[Crossref] [Google Scholar] [PubMed]
- Priya DM, Siril EA. Traditional and modern use of Indian madder (Rubia cordifolia L.): An overview. Int J Pharm Sci Rev Res 2014;25(1):154-64.
- Merritt MA, Green AC, Nagle CM, Webb PM, Australian Cancer Study (Ovarian Cancer) and Australian Ovarian Cancer Study Group. Talcum powder, chronic pelvic inflammation and NSAIDs in relation to risk of epithelial ovarian cancer. Int J Cancer 2008;122(1):170-6.
[Crossref] [Google Scholar] [PubMed]
- Kwok CT, Chow FW, Cheung KY, Zhang XY, Mok DK, Kwan YW, et al. Medulla Tetrapanacis water extract alleviates inflammation and infection by regulating macrophage polarization through MAPK signaling pathway. Inflammopharmacology 2023:1-12.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Huang L, Qin L, Huang B. Antioxidative and anti-inflammatory properties of chushizi oil from Fructus Broussonetiae. J Med Plant Res 2011;5(28):6407-12.
- Kemper C, Sack MN. Linking nutrient sensing, mitochondrial function, and PRR immune cell signaling in liver disease. Trends Immunol 2022;43(11):886-900.
[Crossref] [Google Scholar] [PubMed]
- di Petrillo A, Orrù G, Fais A, Fantini MC. Quercetin and its derivates as antiviral potentials: A comprehensive review. Phytother Res 2022;36(1):266-78.
[Crossref] [Google Scholar] [PubMed]
- Marcolin E, San-Miguel B, Vallejo D, Tieppo J, Marroni N, González-Gallego J, et al. Quercetin treatment ameliorates inflammation and fibrosis in mice with nonalcoholic steatohepatitis. J Nutr 2012;142(10):1821-8.
[Crossref] [Google Scholar] [PubMed]
- Wang SW, Lan T, Sheng H, Zheng F, Lei MK, Wang LX, et al. Nobiletin alleviates non-alcoholic steatohepatitis in MCD-induced mice by regulating macrophage polarization. Front Physiol 2021;12:1-12.
[Crossref] [Google Scholar] [PubMed]
- Naeini F, Namkhah Z, Ostadrahimi A, Tutunchi H, Hosseinzadeh-Attar MJ. A comprehensive systematic review of the effects of naringenin, a citrus-derived flavonoid, on risk factors for nonalcoholic fatty liver disease. Adv Nutr 2021;12(2):413-28.
[Crossref] [Google Scholar] [PubMed]
- Sun WL, Yang JW, Dou HY, Li GQ, Li XY, Shen L, et al. Anti-inflammatory effect of luteolin is related to the changes in the gut microbiota and contributes to preventing the progression from simple steatosis to nonalcoholic steatohepatitis. Bioorg Chem 2021;112:1-9.
[Crossref] [Google Scholar] [PubMed]
- Kim MS, Kung S, Grewal T, Roufogalis BD. Methodologies for investigating natural medicines for the treatment of Nonalcoholic Fatty Liver Disease (NAFLD). Curr Pharm Biotechnol 2012;13(2):278-91.
[Crossref] [Google Scholar] [PubMed]
- Al-Tamimi JZ, Alshammari GM, Al-Faris NA, Alagal RI, Aljabryn DH, Albekairi NA, et al. Ellagic acid protects against non-alcoholic fatty liver disease in streptozotocin-diabetic rats by activating AMPK. Pharm Biol 2022;60(1):25-37.
[Crossref] [Google Scholar] [PubMed]
- Li P, Hu J, Zhao H, Feng J, Chai B. Multi-omics reveals inhibitory effect of baicalein on non-alcoholic fatty liver disease in mice. Front Pharmacol 2022;13:1-16.
[Crossref] [Google Scholar] [PubMed]
- Ferre P, Foufelle F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes Metab 2010;12:83-92.
[Crossref] [Google Scholar] [PubMed]
- Frederico MJ, Vitto MF, Cesconetto PA, Engelmann J, de Souza DR, Luz G, et al. Short-term inhibition of SREBP-1c expression reverses diet-induced non-alcoholic fatty liver disease in mice. Scand J Gastroenterol 2011;46(11):1381-8.
[Crossref] [Google Scholar] [PubMed]
- Dixon LJ, Flask CA, Papouchado BG, Feldstein AE, Nagy LE. Caspase-1 as a central regulator of high fat diet-induced non-alcoholic steatohepatitis. PLoS One 2013;8(2):1-10.
[Crossref] [Google Scholar] [PubMed]
- Bragoszewski P, Habior A, Walewska-Zielecka B, Ostrowski J. Expression of genes encoding mitochondrial proteins can distinguish nonalcoholic steatosis from steatohepatitis. Acta Biochim Pol 2007;54(2):341-8.
[Crossref] [Google Scholar] [PubMed]