- *Corresponding Author:
- L. Hui
College of Medical Laboratory, Dalian Medical University, Dalian 116044, China
E-mail: liuhui60@sina.com
| This article was originally published in a special issue, “Biomedical research applications in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2020:82(2)spl issue3;21-25 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the use of an experimental design based on pooled sample measurements to improve the efficiency of research sample analysis, a total of 40 serum samples were randomly selected from healthy human subjects and an additional 40 samples were collected from intensive care unit patients. The samples were mixed to obtain 20 samples containing 2 individual serum samples per vial and 10 samples containing 4 individual serum samples per vial for both the control group and the intensive care unit group. Ten analytes were measured using an autoanalyzer, which included proteins, enzymes and organic and inorganic molecules. The results obtained from the pooled samples were statistically similar to the results obtained from individually analyzed samples. A pooled-sample experimental design could be applied in the analysis of biological samples.
Keywords
Serum, biochemical analysis, experimental design, pooled samples
Life sciences and clinical research is often conducted through comparison of a control group with an experimental group that has received a form of intervention such as treatment with a therapeutic agent. Large samples are usually required for this type of research and the clinical and/or biochemical differences that appear between the 2 groups can be assessed by determining the sampling errors with statistical analysis. However, the use of a large sample means that large number of samples often have to be analysed, which may be laborious and require additional resources, such as expensive analytical kits. More importantly, it is often difficult to control the quality of the analysis between batches, particularly in the case of biological samples[1-4]. To address this issue, an experimental design was established wherein multiple samples were pooled in batches and co-analyzed, thus facilitating fast, high-quality analysis of fewer samples. However, it was unclear whether batch analysis altered the measurement of specific biochemical markers, such as the levels of proteins and enzymes, in a statistically significant way compared to the individually analyzed samples. Therefore, in this study, serum specimens from 40 healthy subjects were assessed with regards to some common clinical markers individually and in pooled batches of 2 and 4 samples. Interestingly, the clinical results obtained from samples assessed in pooled batches were similar to their individually analyzed counterparts.
Forty serum samples from randomly selected subjects who visited the hospital for a regular medical check-up were collected. Samples were then mixed in batches of 2 samples per vial to obtain one batch of 20 samples in total and 4 samples per vial to obtain another batch of 10 samples in total. In addition, 40 serum samples were randomly selected from patients in the Intensive Care Unit (ICU) and mixed in the same manner.
Serum analytes measured included total protein (TP), albumin (ALB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine (CRE), urea (URE), triglycerides (TRIG), cholesterol (CHOL), potassium (K) and sodium (Na). Serum samples were mixed as described, prior to subsequent analysis in the same run in an automatic biochemistry analyzer (ADVIA2400) in the clinical laboratory of the Dalian Medical University Hospital using standard commercial reagent kits.
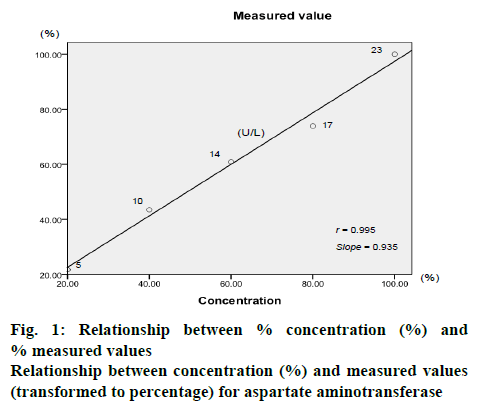
One serum sample was diluted with normal saline to concentrations of 100, 80, 60, 40 and 20 % and analytes were measured as described above. The results were transformed to percentage. The sample concentrations were plotted against measured values to form a scatter diagram as shown in figure 1. The outcome for an indicator was considered to be suited by a mixed sample when the r-value was close to 1 and the slope was close to 1.
Values of mean and standard deviation (SD) were calculated on the basis of the measured values in different groups. The expected mean value was the mean value of the single serum for the pooled serum group. The expected SD was the standard error (SE), which was calculated using the Eqn., SE-SD/√n, where SE is the standard error, SD is the SD of the single serum group and n represents the number of single serum samples in the pooled serum sample. The expected values of mean and SD were compared with the measured data. Serum samples from patients in the ICU and healthy population were used to compare effects between single and pooled sample measurements. The independent sample t-test was used to assess the difference between the means of 2 groups. The analyses were made using SPSS statistical analysis software (SPSS Inc., Chicago, IL). P-values <0.05 (two-tailed) were considered significant. P-values from each dataset were also used to evaluate application effect between single and pooled sample measurements; near values of P from single or pooled sample measurements implied a valid design of pooled sample measurements.
Dilution tests showed that the r- and slope-values were close to 1 for all analytes (Table 1). The measured results for single serum samples and pooled serum samples are listed in Table 2. The mean values from pooled samples were close to the mean values from the relevant single serum samples (Table 2). The SD values of pooled serum samples, such as mixtures of 2 or 4 individual samples, were close to the expected values (Table 3). The results of the individual and pooled-batch sample analysis from ICU patients and healthy subjects are shown in Table 4. The mean values from the pooled samples were found to be statistically similar at a significant level to the mean values obtained from individually analyzed samples. The P-values between the ICU group and the control group from the pooled serum samples (of 2 or 4 individual samples) were close to the P-values obtained from individually analyzed samples.
Table 1: Correlation Cooefficient and Slope Values of Analyses of Sample Dilutions
| Analyte | r | Slope | Analyte | r | Slope |
|---|---|---|---|---|---|
| AST | 0.995 | 0.935 | URE | 0.999 | 0.994 |
| ALT | 0.981 | 1.100 | K | 0.998 | 0.953 |
| TP | 1.000 | 0.995 | Na | 1.000 | 0.986 |
| ALB | 0.997 | 0.968 | CHOL | 0.999 | 0.996 |
| CER | 0.999 | 1.023 | TRIG | 0.997 | 1.010 |
TP is total protein, ALB is albumin, ALT is alanine aminotransferase, AST is aspartate aminotransferase, CRE is creatinine, URE is urea, TRIG is triglycerides, CHOL is cholesterol, K is potassium and Na is sodium
Table 2: Mean Values from Single and Pooled Serum Samples for All Analytes Tested
| Analyte | Single serum | Mixture of two sera | Mixture of four sera | ||
|---|---|---|---|---|---|
| Expected | Observed | Expected | Observed | ||
| AST (U/l) | 17 | 20 | 20 | 22.5 | 23 |
| 23 | |||||
| 32 | 25 | 25 | |||
| 18 | |||||
| ALT (U/l) | 14 | 17 | 17 | 16 | 16 |
| 20 | |||||
| 15 | 15 | 15 | |||
| 15 | |||||
| TP (g/l) | 84 | 84 | 84 | 79 | 79 |
| 84 | |||||
| 70 | 74 | 74 | |||
| 78 | |||||
| ALB (g/l) | 48 | 46.5 | 47 | 44.8 | 45 |
| 45 | |||||
| 41 | 43 | 43 | |||
| 45 | |||||
| CRE (mM) | 65 | 70 | 70 | 63.5 | 64 |
| 75 | |||||
| 62 | 57 | 57 | |||
| 52 | |||||
| UREA (mM) | 7.2 | 6.5 | 6.5 | 5.7 | 5.7 |
| 5.8 | |||||
| 4.6 | 4.9 | 4.8 | |||
| 5.1 | |||||
| (m) | 4.7 | 4.8 | 4.8 | 4.6 | 4.7 |
| 4.8 | |||||
| 4.6 | 4.4 | 4.5 | |||
| 4.2 | |||||
| Na (mM) | 134 | 140 | 142 | 142.8 | 146 |
| 145 | |||||
| 148 | 146 | 146 | |||
| 144 | |||||
| CHOL (mM) | 7.7 | 7 | 7 | 6.1 | 6.2 |
| 6.3 | |||||
| 4.8 | 5.2 | 5.2 | |||
| 5.6 | |||||
| TRIG (mM) | 2.2 | 2 | 2 | 2.1 | 2 |
| 1.8 | |||||
| 0.9 | 2.1 | 2.1 | |||
| 3.2 | |||||
Table 3: Standard Deviation of Single and Pooled Serum Samples for All Analytes Measured
| Analyte | Single serum (n=40) | Mixture of 2 sera (n=20) | Mixture of 4 sera (n=10) | ||
|---|---|---|---|---|---|
| Expected | Observed | Expected | Observed | ||
| AST | 6.2 | 4.4 | 4.2 | 3.1 | 3.0 |
| ALT | 10.8 | 7.6 | 7.6 | 5.4 | 4.6 |
| TP | 5.9 | 4.2 | 4.6 | 3.0 | 2.9 |
| ALB | 3.1 | 2.2 | 2.6 | 1.6 | 1.8 |
| CRE | 58.6 | 41.4 | 44.5 | 29.3 | 31.8 |
| URE | 3.2 | 2.3 | 2.2 | 1.6 | 1.6 |
| K | 0.5 | 0.3 | 0.4 | 0.2 | 0.3 |
| Na | 3.1 | 2.2 | 1.5 | 1.6 | 0.8 |
| CHOL | 1.1 | 0.8 | 0.8 | 0.6 | 0.6 |
| TRIG | 1.0 | 0.7 | 0.7 | 0.5 | 0.5 |
TP is total protein, ALB is albumin, ALT is alanine aminotransferase, AST is aspartate aminotransferase, CRE is creatinine, URE is urea, TRIG is triglycerides, CHOL is cholesterol, K is potassium and Na is sodium
Table 4: Results of Individually Analyzed Serum Samples and Batches of Pooled-Samples of ICU Patients and Healthy Subjects
| Analyte | Group | Single sample (n=40) | Mixture of 2 sera (n=20) | Mixture of 4 sera (n=10) | |||
|---|---|---|---|---|---|---|---|
| Mean±SD | P-value | Mean±SD | P-value | Mean±SD | P-value | ||
| AST | ICU | 45.3±43.5 | 0.001 | 45.2±31.2 | 0.003 | 46.1±20.8 | 0.002 |
| (U/l) | Control | 22.2±6.2 | 22.6±4.2 | 22.5±3.0 | |||
| ALT | ICU | 29.9±34.4 | 0.059 | 29.8±25.1 | 0.077 | 30.4±15.9 | 0.042 |
| (U/l) | Control | 19.0±10.8 | 19.1±7.6 | 19.0±4.6 | |||
| TP | ICU | 54.2±12.9 | 0 | 54.4±7.8 | 0 | 54.5±5.0 | 0 |
| (g/l) | Control | 76.9±5.9 | 76.9±4.6 | 77.6±2.9 | |||
| ALB | ICU | 28.1±8.8 | 0 | 27.9±5.6 | 0 | 27.9±4.3 | 0 |
| (g/l) | Control | 45.2±3.1 | 45.2±2.6 | 45.4±1.8 | |||
| CRE | ICU | 128.0±166.2 | 0.061 | 126.9±120.4 | 0.08 | 125.4±73.5 | 0.064 |
| (mM) | Control | 75.0±58.6 | 75.2±44.5 | 75.5±31.8 | |||
| URE | ICU | 9.67±7.0 | 0.01 | 9.6±4.7 | 0.01 | 9.5±3.3 | 0.017 |
| (mM) | Control | 6.4±3.2 | 6.4±2.2 | 6.5±1.6 | |||
| K | ICU | 3.9±0.8 | 0.001 | 3.9±0.5 | 0.001 | 3.9±0.4 | 0.002 |
| (mM) | Control | 4.4±0.5 | 4.4±0.4 | 4.5±0.3 | |||
| Na | ICU | 139.2±8.5 | 0.001 | 139.7±5.4 | 0 | 139.5±3.0 | 0 |
| (mM) | Control | 144.1±3.1 | 144.7±1.5 | 145.0±0.8 | |||
| CHOL | ICU | 3.7±1.4 | 0 | 3.8±1.0 | 0 | 3.8±0.6 | 0 |
| (mM) | Control | 5.6±1.1 | 5.6±0.8 | 5.6±0.6 | |||
| TRIG | ICU | 1.6±0.9 | 0.468 | 1.6±0.8 | 0.556 | 1.6±0.5 | 0.564 |
| (mM) | Control | 1.8±1.0 | 1.8±0.7 | 1.8±0.5 | |||
The scientific research usually addresses a group issue rather than individual issue, therefore, the design of pooled sample is accepted[5-8]. The pooled samples were similar to the diluted samples, when a serum sample was diluted to concentrations of 100, 80, 60, 40 and 20 %, the percentages of the various measured values should also be 100, 80, 60, 40 and 20 %; thus, the r- and slope-values between concentration and percentage of measured value should be close to 1. Results showed that the r- and slope-values were close to 1 for all indicators, suggesting that it was reasonable to measure pooled samples.
As SD the most widely accepted and the best measure of dispersion, change in even one value affects the value of SD. The SD of the values was considered for the pooled serum samples should decrease as the number of samples within the batch increases and should converge towards the SE of individual samples. As expected, our results show that the SDs of the pooled serum samples were approximately equal to the SE. SE=SD/√n, where n represents the number of single serum samples within a pooled-batch sample. This finding suggested that the extent of the decrease conformed to the statistical relationship SE=SD/√n for SDs of pooled serum samples.
Patients in the ICUs of hospitals are mostly those in the end stages of a terminal disease, which is usually accompanied by multiple organ failure. Thus, serum samples from these patients were used in this study to assess multi-sample batch analysis compared to samples from control subjects. Serum samples were assessed for the levels of proteins, enzymes and the presence of organic and inorganic molecules. The results showed that the serum chemistry of ICU patients was mostly abnormal and there were wide variations in the sample-to-sample measurements obtained; the P-values obtained from the individually analyzed and batch-analyzed samples were similar at a significant level. The SE for the pooled serum samples was also found to decrease as the number of samples in the batch increased, conforming to SE=SD/√n. This further indicated that the measurements obtained from the pooled samples are equivalent to individual sample measurements.
In general, it was found that the mean biochemical values obtained from pooled samples were similar, but not significantly different to the mean values obtained from individually analysed samples, which indicated that the majority of the biochemical indicators assessed in this study are independent and not affected by the other factors present in serum. This finding suggested that a pooled-batch protocol is suitable for the analysis of biological samples. Limitation of the design of pooled sample measurements was that skewed distributions of variables or unexpected outliers could not be found from pooled sample measurements, the results could be significantly biased; therefore, the results from pooled sample measurements should be considered as a preliminary study. Another limitation was that pooled sample measurements cannot be used for exploring other links between variables; whereby, pooled sample measurements was not suggested for exploring relationship between variables.
In summary, assessing clinical samples as part of pooled batches could simplify experimental processes and improve the efficiency of analysis, especially for assessments that are typically accompanied by large errors due to variation between different batches, such as enzyme-linked immunosorbent assays. Indeed, this study has shown that a pooled-sample study design could be of great significance to enable multiple samples to be analyzed in the same experiment and reduce errors. Although a pooled-sample experimental set-up could be used cautiously, the result of statistical analysis could be accepted when P-value is very small. For experiments with a limited quantity of sample, such as those with mouse serum, using a pooled-sample experimental design could enable additional analyses to be carried out simultaneously or reduce the rate of sample collection. Thus, the experimental design described in this study could be of value to the field of life sciences research.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Levine DM, Maine GT, Armbruster DA, Mussell C, Buchholz C, O’Connor G, et al. The need for standardization of tacrolimus assays. Clin Chem 2011;57:1739-47.
- de los Santos MJ, Ruiz A. Protocols for tracking and witnessing samples and patients in assisted reproductive technology. Fertil Steril 2013;100:1499-502.
- World Health Organization. WHO Expert Committee on Biological Standardization. World Health Organ Tech Rep Ser 2013;1-231.
- Kinns H, Pitkin S, Housley D, Freedman DB. Internal quality control: best practice. J Clin Pathol 2013;66:1027-32.
- Mori A, Oda H, Onozawa E, Shono S, Takahashi T, Yamashita S, et al. Evaluation of portable blood glucose meters using canine and feline pooled blood samples. Pol J Vet Sci 2016;19:707-13.
- Kinyua J, Negreira N, Miserez B, Causanilles A, Emke E, Gremeaux L, et al. Qualitative screening of new psychoactive substances in pooled urine samples from Belgium and United Kingdom. Sci Total Environ 2016;573:1527-35.
- Caudill SP. Confidence interval estimation for pooled-sample bio monitoring from a complex survey design. Environ Int 2015;85:40-5.
- Wang J, Skoog T, Einarsdottir E, Kaartokallio T, Laivuori H, Grauers A, et al. Investigation of rare and low-frequency variants using high-throughput sequencing with pooled DNA samples. Sci Rep 2016;6:33256.