- *Corresponding Author:
- Jingyu Wang
Department of Pathology, The First Hospital of Jiaxing/Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang 314000, China
E-mail: 11018140@zju.edu.cn
| This article was originally published in a special issue, “Transformative Discoveries in Biomedical and Pharmaceutical Research” |
| Indian J Pharm Sci 2023:85(4) Spl Issue “277-284” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The abnormal expression of semaphorin 3D is associated with various tumors. Nevertheless, the function and mechanism in colorectal cancer have not been fully understood. This study aims at exploring the migration pattern and related mechanisms of semaphorin 3D in colorectal cancer cells in vitro. Human SW480 colorectal cancer cells lines were used in this study. Semaphorin 3D lentiviral overexpressed vector and control lentiviral vector were used to transfect SW480 cells. Quantification of semaphorin 3D, actin filament associated protein 1 like 1 messenger ribonucleic acid and protein levels were done by real-time quantitative polymerase chain reaction and western blotting. Ribonucleic acid sequencing is used for analysis of differential gene expression and transwell assay for determination of migration capacity. The research results showed that semaphorin 3D gene expression was dramatically overexpressed in SW480 cells that were transfected by semaphorin 3D lentiviral overexpressed vector. Compared to control group, cell migration capacity of semaphorin 3D overexpression SW480 cells were strikingly reduced. Ribonucleic acid sequencing, quantitative polymerase chain reaction and western blotting assays hinted actin filament associated protein 1 like 1 was significantly downregulated in semaphorin 3D overexpression cells. Based on these findings, overexpression of semaphorin 3D might prevent migration of colorectal cancer cells via down regulating actin filament associated protein 1 like 1. Therefore, semaphorin 3D may inhibit colorectal cancer cell migration and suppress colorectal cancer carcinogenesis and progression.
Keywords
Semaphorin 3D, colorectal cancer, actin filament associated protein 1 like 1, lentiviral vector
As a common gastrointestinal carcinoma, Colorectal Cancer (CRC) is associated with high malignancy, rapid progression and poor therapeutic efficacy[1]. In 2023, CA: A Cancer Journal for Clinicians reported that the number of expected new cases and deaths of CRC has ranked third in the United States for both men and women[2]. According to the latest study of Sung et al. the incidence of CRC in China has surged to the second place in cancer in 2020 and simultaneously its mortality rate has soared to the fifth[3]. CRC is less sensitive to chemotherapy[4] and metastasis is a vital cause of death in CRC patients. However, the molecular mechanism underlying CRC metastasis is yet to be elucidated[5]. Therefore, exploring the possible mechanisms of metastatic CRC and looking for new therapeutic targets would be the promising treatment approach for CRC.
Semaphorin 3D (SEMA3D) is a protein belonging to the semaphorins family. SEMA3D plays an important physiological role in heart[6], blood vessels[7], embryonic development[8], regeneration[9] and neural development[10]. However, abnormal expression of SEMA3D can cause corresponding diseases. Some studies have demonstrated that abnormal expression of SEMA3D can regulate the occurrence and development of Hirschsprung disease by affecting the development of enteric nervous system[11-13]. Nevertheless, whether SEMA3D is correlated with Ovine Johnes Disease (OJD) is still controversial[14,15]. With further study, the role played by SEMA3D in tumor carcinogenesis and progression is increasingly concerned. Recently, SEMA3D has been suggested to suppress the occurrence and progression of glioblastoma, breast and thyroid carcinomas[16-18]. On the contrary, Foley et al. suggested that SEMA3D can promote the metastasis of Pancreatic Ductal Adenocarcinoma (PDA)[19].
These studies indicated that SEMA3D may play a role in promoting or inhibiting cancer carcinogenesis and progression in different tumors. Yet, its mechanism of action in CRC has not been fully elucidated.
In our earlier research, SEMA3D expression was found to be markedly reduced in CRC tissues versus adjacent counterparts, with an inverse connection with lymph node metastasis, manifesting that SEMA3D is a favorable prognostic factor for CRC patients[20]. Therefore, SEMA3D may inhibit CRC development and metastases.
Herein, we further investigate whether SEMA3D suppresses CRC cells migration and preliminarily explores its inhibition mechanisms.
Materials and Methods
Source and cultivation of cells:
We Human CRC cell lines like RKO, SW620, HCT116, HT29, LOVO, CW2, LS513 and SW480 ordered from the American Type Culture Collection (ATCC, Manassas, Virginia, United States), were placed in an Roswell Park Memorial Institute (RPMI) 1640 medium, routinely supplemented with 10 % Fetal Bovine Serum (FBS) (HyClone, Tauranga, New Zealand) for cultivation at 37° with 95 % Oxygen (O2) and 5 % Carbon dioxide (CO2).
Real-time quantitative Polymerase Chain Reaction (qPCR):
We used an RNeasy Mini Kit (Qiagen, Catalog no. 74106) to isolate cellular total Ribonucleic Acid (RNA) and a PrimeScript Reverse Transcriptase (RT) reagent kit (Takara Biotechnology, Dalian, China, Catalog no. RR037A) to synthesize complementary Deoxyribonucleic Acid (cDNA). This was followed by qPCR using a SYBR Premix Ex (Taq Takara, Catalog no. RR420A) and the calculation of relative gene expression levels with 2-delta delta threshold cycle (2–∆∆Ct) (Internal control: Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH)). Primer sequences are GAPDH Forward (F): 5'-ACCACAGTCCATGCCATCAC-3'
and GAPDH Reverse (R): 5'-TCCACCACCCTGTTGCTGTA-3', SEMA3D F: 5'-TGGGACATCGAAGACAGCAT-3' and SEMA3D R: 5'-AAAGTGTGCTCCTGGGCTTT-3', Actin
Filament Associated Protein 1 Like 1 (AFAP1L1) F: 5'-GTGACCTGAGTGACCTTCGG-3' andAFAP1L1 R: 5'-AGAGTCATTATTGTGGGAGCTGA-3'.
Western Blotting (WB) analysis:
The WB program was performed as previously described[21] and the information of antibodies was followed. SEMA3D (Abcam, Catalog no. AB18074, 1:1000), AFAP1L1 (Santa Cruz Biotech, Inc., Catalog no. SC-514788, 1:1000), beta (β)-actin (Affinity, Catalog no. T0022, 1:5000) and β-actin was used as a control.
Lentiviral vector constructs and cell transfection:
Human SEMA3D lentiviral overexpressed vector and control lentiviral vector were constructed and packaged by Wuhan GeneCreate Biological Engineering Co., Ltd (Wuhan, China). Full-length SEMA3D cDNA was amplified and then inserted into lentiviral-protocadherin (pCDH)-Cytomegalovirus (CMV)-Multiple Cloning Sites (MCS)-human Elongation Factor 1 alpha (EF1α)-Green Fluorescent Protein (GFP)-P2A-Puro vector to obtain pCDH- SEMA3D overexpressed plasmid. Packaging plasmids (pMDLg-pRRE, pMD2.G and pRSV-Rev) were co-transfected into Human Embryonic Kidney 293 T antigen (HEK293T) to obtain pCDH-SEMA3D lentiviral particles. The SW480 cell strain was then infected with concentrated pCDH-SEMA3D lentiviral particles and screened by puromycin to obtain stable SEMA3D-overexpressed SW480 cell line. Lentiviral-pCDH-CMV-MCS-EF1α-GFP- P2A-Puro vector was used as negative control and quantification of SEMA3D messenger RNA (mRNA) was made by real-time qPCR.
RNA-Sequencing (RNA-Seq) and Differentially Expressed Gene (DEG) analyses:
We separated total RNA from SW480 SEMA3D overexpressed cells and negative control cells and then enriched mRNA with Oligo deoxythymidine (dT) beads and reverse transcribed it into cDNA. The cDNA fragments were then treated for purification and ligation with Illumina sequencing adapters. After the selection of ligated products of a proper size using Agarose gel electrophoresis, PCR amplification and sequencing using Illumina HiSeqTM 4000 were carried out.
DEGs were analyzed by edgeR package (http://www.r-project.org/) across overexpressed cells and negative controls. We identified genes with a False Discovery Rate (FDR)<0.05 and a fold change≥2 as significant DEGs and validated them by qPCR.
Cell migration assays:
Migratory capacities of cells with transfected or untransfected SEMA3D gene are evaluated by transwell chambers (8 μm pore, Corning, Catalog no. 3422). 600 μl 10 % FBS added RPMI 1640
medium was placed into the lower chambers as a chemoattractant and then 100 μl cells of 1×105 density with RPMI 1640 (serum-free) were suspended in upper chambers. After 24 h of incubation, cells located on the upper chamber surface were erased, while those on the lower surface were fixed with 4 % neutral formaldehyde and subsequently stained with 0.1 % crystal violet. After photographing, the lower surface cells were treated with 33 % acetic acid. Then the absorbance was determined at 570 nm in a 96-well plate by a microplate reader (Bio-TEK, ELx 800, Saxony).
Statistical analysis:
All experiments were run in triplicates and the obtained data was statistically described as mean±Standard Deviation (SD). GraphPad Prism software was used for statistical analysis and statistical significance was indicated by p<0.05.
Results and Discussion
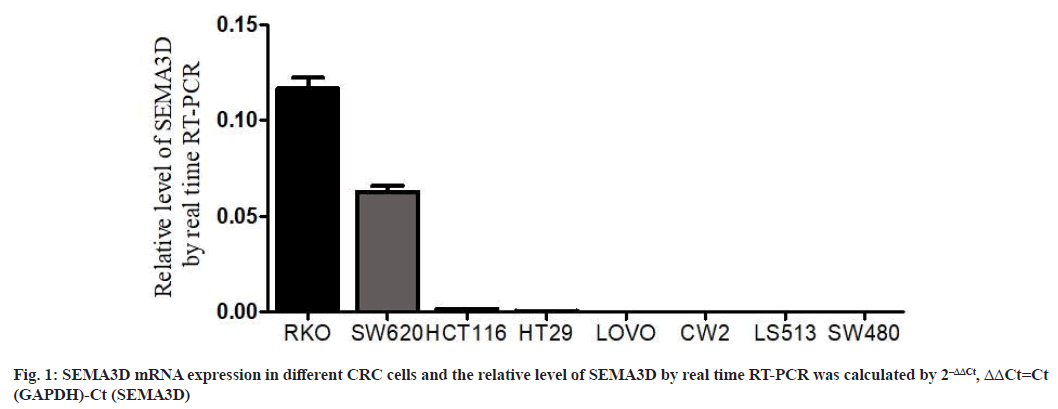
SEMA3D gene expression in CRC cell lines was shown in fig. 1. Real-time PCR quantified SEMA3D expression. SEMA3D mRNA levels were highly expressed in RKO and SW620 CRC cell lines, while lower in HCT116, HT29, LOVO, CW2, LS513 and SW480 CRC cell lines. Thus, SW480 cells with low SEMA3D expression were randomly selected as overexpression transfection cell lines for subsequent functional and mechanism studies.
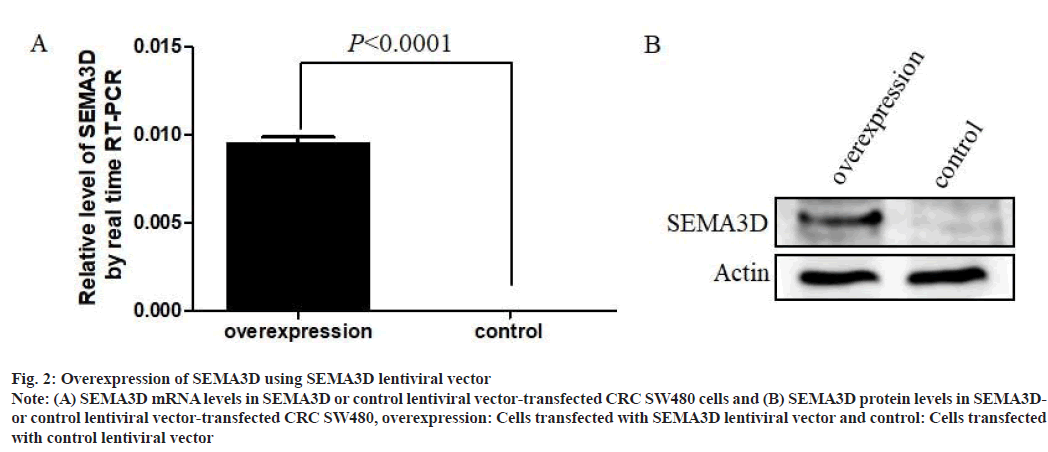
Overexpression of SEMA3D using lentiviral vector was shown in fig. 2. To demonstrate the role of SEMA3D in CRC cell line SW480, SEMA3D overexpressed lentiviral vector and control lentiviral vector were performed. Results showed that SEMA3D successfully overexpressed at mRNA levels in overexpressed SW480 cells compared to control vector (fig. 2A). Concurrently, WB analysis also demonstrated dramatical increase of SEMA3D protein in SEMA3D overexpressed SW480 cells, compared with control cells (fig. 2B). These results suggested that construction of stably transfected cell lines overexpressing SEMA3D is successful.
Fig. 2: Overexpression of SEMA3D using SEMA3D lentiviral vector
Note: (A) SEMA3D mRNA levels in SEMA3D or control lentiviral vector-transfected CRC SW480 cells and (B) SEMA3D protein levels in SEMA3Dor
control lentiviral vector-transfected CRC SW480, overexpression: Cells transfected with SEMA3D lentiviral vector and control: Cells transfected
with control lentiviral vector
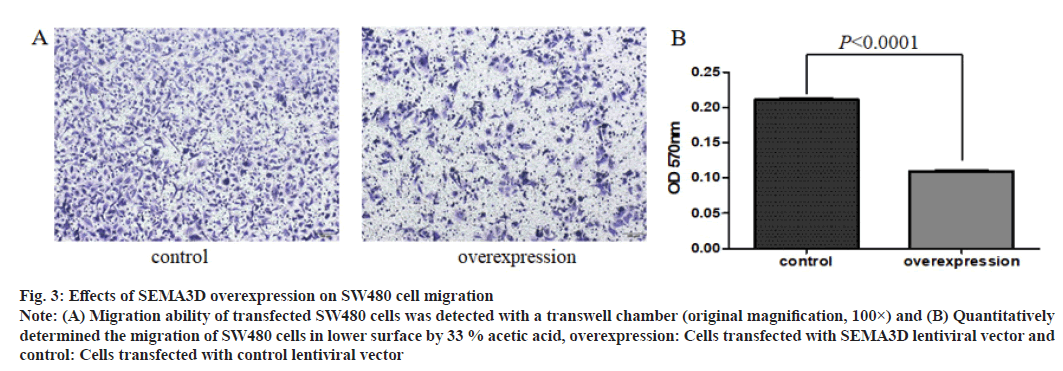
SEMA3D inhibits in vitro migration of CRC cells as shown in fig. 3. To manifest the migration function of SEMA3D in CRC, transwell assay was used. Compared to the control cells, the migration ability of SEMA3D overexpressed SW480 cells were notably decreased.
Fig. 3: Effects of SEMA3D overexpression on SW480 cell migration
Note: (A) Migration ability of transfected SW480 cells was detected with a transwell chamber (original magnification, 100×) and (B) Quantitatively
determined the migration of SW480 cells in lower surface by 33 % acetic acid, overexpression: Cells transfected with SEMA3D lentiviral vector and
control: Cells transfected with control lentiviral vector
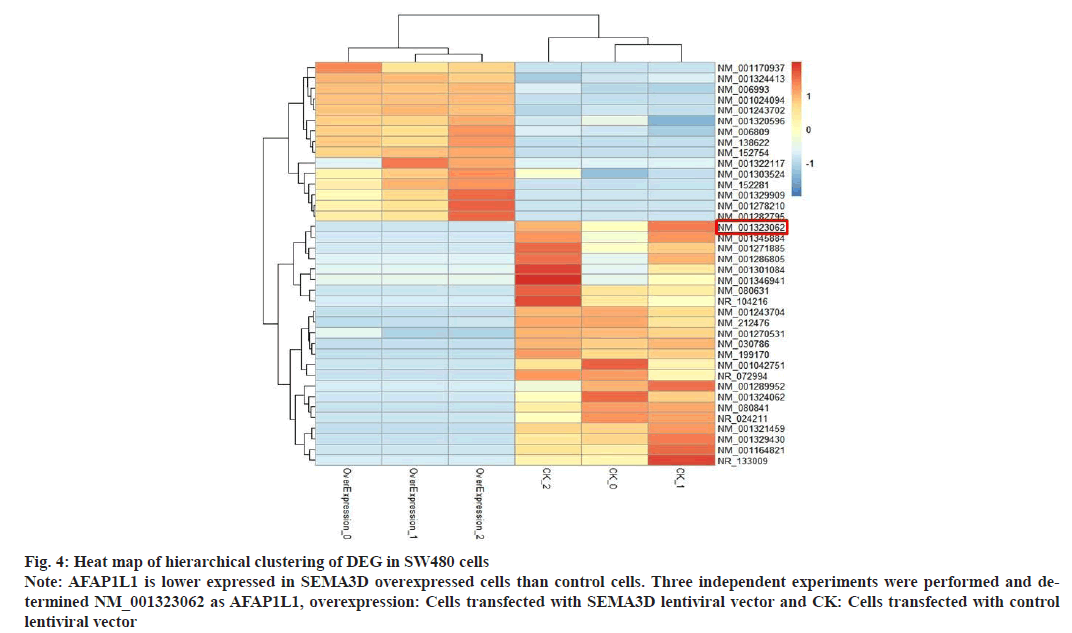
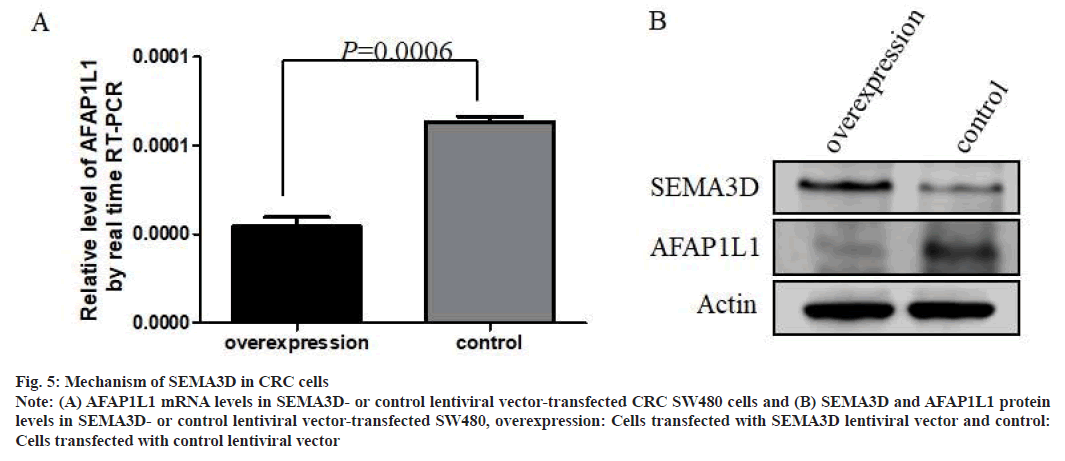
To probe into the regulatory mechanisms of SEMA3D in CRC cell migration, changes in downstream genes expression profiles in SW480 with overexpression of SEMA3D were analyzed. RNA-Seq analysis found that 38 genes were significantly changed after overexpression of SEMA3D (fig. 4). Consistently, qPCR further validated dramatically down-regulated AFAP1L1 mRNA in SEMA3D overexpressing SW480 cells (fig. 5A). According to WB analysis, AFAP1L1 protein was also remarkably down- regulated in SEMA3D overexpressing SW480 cells versus control group (fig. 5B).
Fig. 4: Heat map of hierarchical clustering of DEG in SW480 cells
Note: AFAP1L1 is lower expressed in SEMA3D overexpressed cells than control cells. Three independent experiments were performed and determined
NM_001323062 as AFAP1L1, overexpression: Cells transfected with SEMA3D lentiviral vector and CK: Cells transfected with control
lentiviral vector
Fig. 5: Mechanism of SEMA3D in CRC cells
Note: (A) AFAP1L1 mRNA levels in SEMA3D- or control lentiviral vector-transfected CRC SW480 cells and (B) SEMA3D and AFAP1L1 protein
levels in SEMA3D- or control lentiviral vector-transfected SW480, overexpression: Cells transfected with SEMA3D lentiviral vector and control:
Cells transfected with control lentiviral vector
The semaphorins protein family was originally thought to be an inducer of axonal growth cones that plays a vital part in nervous system development[22,23]. SEMA3D is a secreted protein of the semaphorins protein family that plays a crucial role in heart[6], blood vessels[7,24], embryonic development[8], neural regeneration[9] and development[10]. For example, Aghajanian et al. found that SEMA3D inhibits human umbilical vein epithelial cell migration by regulating the Phosphoinositide 3-Kinase/ Protein kinase B (PI3K/AKT) axis[24]. However, abnormal expression of SEMA3D can cause various diseases. In a comprehensive analysis of SEMA3D variation in Asian ancestry, Gunadi et al. found abnormal expression of SEMA3D in Indonesian Hirschsprung patients[25]. Furthermore, other research studies showed that SEMA3D is essential in carcinogenesis. The occurrence and development of glioma[16,26], breast[17], thyroid[18,27], pancreatic[19,28] and Non-Small Cell Lung Cancer (NSCLC)[29], as well as meningioma[30], cervical[31], bladder[32] and endometrial cancers[33] are all correlated with abnormal expression of SEMA3D. Sabag et al. showed that SEMA3D inhibited glioblastoma angiogenesis in the cerebral cortex of mice, thereby inhibiting tumor genesis[16]. In human meningioma, the mutation of Nuclear receptor Coactivator 3 (NCOA3) gene can up-regulate the expression of SEMA3D, which can be used as a biomarker for the occurrence and development of meningioma[30]. Sun et al. confirmed the inverse connection between SEMA3D expression and the prognosis of bladder cancer, which shows the potential role of SEMA3D in promoting bladder carcinogenesis[32]. However, another study shown that SEMA3D may play an inhibition role in clear-cell renal-cell carcinoma[34]. SEMA3D can also inhibit the onset and development of human breast carcinoma[17]. Gomez-Rueda et al. reported that SEMA3D was down-regulated in thyroid cancer, which may be a new diagnostic marker for thyroid cancer with cytological uncertainty cases[18]. Hai et al. reached similar conclusions by finding that SEMA3D prevented papillary thyroid carcinoma from proliferating and migrating through the Mitogen-Activated Protein Kinase/Extracellular signal-Regulated Kinase (MAPK/ERK) axis[27]. These studies suggested that SEMA3D may have a tumor suppressive effect.
Foley et al. indicated that SEMA3D was upregulated by Annexin A2 (AnxA2) in PDA and the patients with high expression of SEMA3D were more prone to metastasis than those with low expression[19]. Another research manifested that knockdown of SEMA3D could reduce invasion and metastasis in mice. Increased expression of SEMA3D could promote perineural invasion in human PDA and SEMA3D might be a positive factor in PDA[28]. Wang et al. demonstrated the important role of SEMA3D in the malignant transformation of Peptic Ulcer Disease (PUD) and Gastrointestinal Cancer (GC)[35]. According to the study of Li et al. SEMA3D may inhibit liver carcinogenesis and its progression by blocking PI3K/AKT axis via Filamin A (FLNA) [36]. Through whole-genome profiles assay of Malay CRC patients with intact Mismatch Repair (MMR) proteins, Juhari et al. illustrated that the mutation of SEMA3D can result in the development of CRC[37]. In addition, SEMA3D has been shown to be associated with multidrug resistance in leukemia and NSCLC[38,39].
All these studies suggested that SEMA3D may play promotion or inhibition roles in different types of tumors and may also be essential in drug resistance in tumors. Our previous work has shown markedly reduced SEMA3D expression in CRC tissues compared to paired paracancerous tissues and an inverse association of SEMA3D with lymph node metastasis, suggesting that SEMA3D may be an inhibitor of CRC metastasis and a relatively independent prognostic indicator[20]. Therefore, SEMA3D is a promising prognostic factor for CRC patients.
In the current work, overexpressing SEMA3D in SW480 cells evidently reduced cancer cell migration ability compared with control cells, suggesting that SEMA3D may inhibit CRC cell migration. To probe into the mechanism underlying SEMA3D inhibition of CRC migration, RNA-Seq and DEG assays were used in overexpressed SEMA3D cells and control cells. Thirty-eight DEGs were identified. Subsequently, the validated results by qPCR showed visibly down-regulated AFAP1L1 mRNA in SEMA3D overexpressed SW480 cells compared to control cells, which is consistent with the sequencing results. AFAP1L1 protein was also remarkably down- regulated in SEMA3D overexpressed CRC cells, as indicated by the WB analysis.
AFAP1L1 is an adaptor protein belonging to Actin Filament Associated Protein (AFAP) family. AFAP1L1 can interact with cortactin by binding to the Src Homology 3 (SH3) domain and promoting formation of invadosome[40]. Furu et al. have shown that AFAP1L1 is a marker for predicting metastasis in spindle cell sarcomas. Knocking out AFAP1L1 gene in sarcomas cells can inhibit cell invasion, while in overexpressed AFAP1L1 sarcoma cell lines, the growth rate and invasion ability were significantly enhanced[41]. Further studies by the same team suggested that the transcription factor Specificity proteins 3 (Sp3) directly regulates the expression of AFAP1L1, thereby promoting the metastasis of sarcoma cells[42].
Another recent research came to similar conclusion that AFAP1L1 can control migration and invasion in sarcoma cells via a novel pathway that AFAP1L1 is phosphorylated by Vav guanine nucleotide exchange factor 2 (Vav2) and Non-catalytic region of tyrosine kinase adaptor protein 2 (Nck2), and then activate Lyn/Src family tyrosine kinase signaling pathway[43]. In the research of NSCLC, Wang et al. found that AFAP1L1 can accelerate the cell cycle process, promote cell proliferation and inhibit apoptosis[44]. Zimmermann et al. identified that AFAP1L1 can fuse with Platelet-Derived Growth Factor Receptor beta (PDGFRβ) gene in myeloid neoplasm through next-generation sequencing which can provide opportunities for the development of therapeutic drugs[45].
In the study of CRC, Takahashi et al. discovered that AFAP1L1 expression was distinctly increased in tumor tissues compared to adjacent normal counterparts. AFAP1L1 can interact with vinculin protein to regulate cell morphology, movement and promote the development of CRC, indicating that AFAP1L1 plays a crucial part in CRC metastases via modulating cell morphology and promoting cell movement[46]. Nevertheless, in future studies, we should discuss the multiple biological functions of SEMA3D in CRC cells such as epithelial- mesenchymal transition, proliferation and apoptosis. In addition, the role of SEMA3D in CRC in vivo still needs further investigation.
Taken together, combined with our research results and literature review, we speculated that SEMA3D might inhibit the migration of CRC cells and ultimately inhibit CRC progression by inhibiting AFAP1L1 expression.
Acknowledgements:
This research was supported by Jiaxing Municipal Science and Technology Project (2019AY32026, 2019AD32056), Jiaxing Key Discipiline of Medicine- Pathology (2023-ZC-017) and Pioneer innovation team of Jiaxing Institute of Arteriosclerotic Disease (XFCX-DMYH).
Conflict of interests:
The authors declared no conflict of interest.
References
- Mármol I, Sánchez-de-Diego C, Dieste AP, Cerrada E, Yoldi MJR. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci 2017;18(1):1-39.
[Crossref] [Google Scholar] [PubMed]
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73(1):17-48.
[Crossref] [Google Scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Lin R, Ling H, Ke Y, Zeng Y, Xiong Y, et al. Dual inhibition of CDK4 and FYN leads to selective cell death in KRAS-mutant colorectal cancer. Signal Transduct Target Ther 2019;4(1):1-3.
[Crossref] [Google Scholar] [PubMed]
- Yao P, Li Y, Shen W, Xu X, Zhu W, Yang X, et al. ANKHD1 silencing suppresses the proliferation, migration and invasion of CRC cells by inhibiting YAP1-induced activation of EMT. Am J Cancer Res 2018;8(11):2311-24.
[Google Scholar] [PubMed]
- Lupu IE, Redpath AN, Smart N. Spatiotemporal analysis reveals overlap of key proepicardial markers in the developing murine heart. Stem Cell Reports 2020;14(5):770-87.
[Crossref] [Google Scholar] [PubMed]
- Hamm MJ, Kirchmaier BC, Herzog W. Sema3D controls collective endothelial cell migration by distinct mechanisms via Nrp1 and PlxnD1. J Cell Biol 2016;215(3):415-30.
[Crossref] [Google Scholar] [PubMed]
- Scott MK, Yue J, Biesemeier DJ, Lee JW, Fekete DM. Expression of class III semaphorins and their receptors in the developing chicken (Gallus gallus) inner ear. J Comp Neurol 2019;527(7):1196-209.
[Crossref] [Google Scholar] [PubMed]
- Ahi EP, Sefc KM. Towards a gene regulatory network shaping the fins of the Princess cichlid. Sci Rep 2018;8(1):1-13.
[Crossref] [Google Scholar] [PubMed]
- Goulart CO, Mendonça HR, Oliveira JT, Savoldi LM, dos Santos Heringer L, dos Santos Rodrigues A, et al. Repulsive environment attenuation during adult mouse optic nerve regeneration. Neural Plast 2018;2018:1-11.
[Crossref] [Google Scholar] [PubMed]
- Jiang Q, Arnold S, Heanue T, Kilambi KP, Doan B, Kapoor A, et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with Ret are critical to Hirschsprung disease liability. Am J Hum Genet 2015;96(4):581-96.
[Crossref] [Google Scholar] [PubMed]
- Luzon-Toro B, Fernandez RM, Torroglosa A, de Agustín JC, Mendez-Vidal C, Segura DI, et al. Mutational spectrum of semaphorin 3A and semaphorin 3D genes in Spanish Hirschsprung patients. PLoS One 2013;8(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Jiang Q, Turner T, Sosa MX, Rakha A, Arnold S, Chakravarti A. Rapid and efficient human mutation detection using a bench‐top next‐generation DNA sequencer. Hum Mutat 2012;33(1):281-9.
[Crossref] [Google Scholar] [PubMed]
- Yaman Y, Aymaz R, Keleş M, Bay V, Hatipoğlu E, Kaptan C, et al. Evaluation of CD109, PCP4 and SEMA3D genes for their association with Ovine Johne’s disease in Turkish sheep. Anim Biotechnol 2021;32(4):519-25.
[Crossref] [Google Scholar] [PubMed]
- Moioli B, D’Andrea S, de Grossi L, Sezzi E, de Sanctis B, Catillo G, et al. Genomic scan for identifying candidate genes for paratuberculosis resistance in sheep. Anim Prod Sci 2015;56(7):1046-55.
- Sabag AD, Bode J, Fink D, Kigel B, Kugler W, Neufeld G. Semaphorin-3D and semaphorin-3E inhibit the development of tumors from glioblastoma cells implanted in the cortex of the brain. PLoS One 2012;7(8):e42912.
[Crossref] [Google Scholar] [PubMed]
- Kigel B, Varshavsky A, Kessler O, Neufeld G. Successful inhibition of tumor development by specific class-3 semaphorins is associated with expression of appropriate semaphorin receptors by tumor cells. PLoS One 2008;3(9):1-10.
[Crossref] [Google Scholar] [PubMed]
- Gomez-Rueda H, Palacios-Corona R, Gutiérrez-Hermosillo H, Trevino V. A robust biomarker of differential correlations improves the diagnosis of cytologically indeterminate thyroid cancers. Int J Mol Med 2016;37(5):1355-62.
[Crossref] [Google Scholar] [PubMed]
- Foley K, Rucki AA, Xiao Q, Zhou D, Leubner A, Mo G, et al. Semaphorin 3D autocrine signaling mediates the metastatic role of annexin A2 in pancreatic cancer. Sci Signal 2015;8(388):1-13.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Ding M, Qian N, Song B, Yu J, Tang J, et al. Decreased expression of semaphorin 3D is associated with genesis and development in colorectal cancer. World J Surg Oncol 2017;15(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Wang C, Weng M, Xia S, Zhang M, Chen C, Tang J, et al. Distinct roles of programmed death ligand 1 alternative splicing isoforms in colorectal cancer. Cancer Sci 2021;112(1):178-93.
[Crossref] [Google Scholar] [PubMed]
- Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 1993;75(7):1389-99.
[Crossref] [Google Scholar] [PubMed]
- Luo Y, Raible D, Raper JA. Collapsin: A protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 1993;75(2):217-27.
[Crossref] [Google Scholar] [PubMed]
- Aghajanian H, Choi C, Ho VC, Gupta M, Singh MK, Epstein JA. Semaphorin 3D and semaphorin 3E direct endothelial motility through distinct molecular signaling pathways. J Biol Chem 2014;289(26):17971-9.
[Crossref] [Google Scholar] [PubMed]
- Gunadi, Kalim AS, Budi NY, Hafiq HM, Maharani A, Febrianti M, et al. Aberrant expressions and variant screening of SEMA3D in Indonesian Hirschsprung patients. Front Pediatr 2020;8:1-7.
[Crossref] [Google Scholar] [PubMed]
- Khan FH, Pandian V, Ramraj S, Aravindan S, Herman TS, Aravindan N. Reorganization of metastamiRs in the evolution of metastatic aggressive neuroblastoma cells. BMC Genomics 2015;16(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Hai R, You Q, Wu F, Qiu G, Yang Q, Shu L, et al. Semaphorin 3D inhibits proliferation and migration of papillary thyroid carcinoma by regulating MAPK/ERK signaling pathway. Mol Biol Rep 2022;49(5):3793-802.
[Crossref] [Google Scholar] [PubMed]
- Jurcak NR, Rucki AA, Muth S, Thompson E, Sharma R, Ding D, et al. Axon guidance molecules promote perineural invasion and metastasis of orthotopic pancreatic tumors in mice. Gastroenterology 2019;157(3):838-50.
[Crossref] [Google Scholar] [PubMed]
- Meert AP, Ameye L, Leclercq N, Paesmans M, Remmelink M, Sculier JP, et al. Difficulties and limitations in conducting translational research in thoracic oncology. A practical example. Rev Mal Respir 2016;33(7):594-9.
[Crossref] [Google Scholar] [PubMed]
- Hussein D, Dallol A, Quintas R, Schulten HJ, Alomari M, Baeesa S, et al. Overlapping variants in the blood, tissues and cell lines for patients with intracranial meningiomas are predominant in stem cell-related genes. Heliyon 2020;6(11):1-14.
[Crossref] [Google Scholar] [PubMed]
- Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 2015;47(2):158-63.
[Crossref] [Google Scholar] [PubMed]
- Sun Y, Zhu D, Xing H, Hou Y, Liu Y. Screening of characteristic biomolecules related to bladder cancer based on construction of ceRNA regulation network. World J Urol 2020;38(11):2835-47.
[Crossref] [Google Scholar] [PubMed]
- Yoshida E, Terao Y, Hayashi N, Mogushi K, Arakawa A, Tanaka Y, et al. Promoter-level transcriptome in primary lesions of endometrial cancer identified biomarkers associated with lymph node metastasis. Sci Rep 2017;7(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Xie R, Wu J, Shang B, Cao C, Bi X, Shi H, et al. Transmembrane transporter sema3D serves as a tumor suppressor in localized clear cell renal cell carcinoma. J Oncol 2022;2022:1-11.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Wei Y, An L, Wang K, Hong D, Shi Y, et al. SEMA3D plays a critical role in peptic ulcer disease-related carcinogenesis induced by H. pylori infection. Int J Gen Med 2022;15:1239-60.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Xu C, Sun B, Zhong F, Cao M, Yang L. Sema3D restrained hepatocellular carcinoma progression through inactivating PI3K/AKT signaling via interaction with FLNA. Front Oncol 2022;12:1-15.
[Crossref] [Google Scholar] [PubMed]
- Juhari WK, Noordin KBAA, Zakaria AD, Rahman WF, Mokhter WM, Hassan MR, et al. Whole-genome profiles of Malay colorectal cancer patients with intact MMR proteins. Genes 2021;12(9):1-13.
[Crossref] [Google Scholar] [PubMed]
- Lehne G, Grasmo-Wendler UH, Berner JM, Meza-Zepeda LA, Adamsen BL, Flack A, et al. Upregulation of stem cell genes in multidrug resistant K562 leukemia cells. Leuk Res 2009;33(10):1379-85.
[Crossref] [Google Scholar] [PubMed]
- Berghmans T, Ameye L, Lafitte JJ, Colinet B, Cortot A, CsToth I, et al. Prospective validation obtained in a similar group of patients and with similar high throughput biological tests failed to confirm signatures for prediction of response to chemotherapy and survival in advanced NSCLC: A prospective study from the european lung cancer working party. Front Oncol 2015;4:1-16.
[Crossref] [Google Scholar] [PubMed]
- Snyder BN, Cho Y, Qian Y, Coad JE, Flynn DC, Cunnick JM. AFAP1L1 is a novel adaptor protein of the AFAP family that interacts with cortactin and localizes to invadosomes. Eur J Cell Biol 2011;90(5):376-89.
[Crossref] [Google Scholar] [PubMed]
- Furu M, Kajita Y, Nagayama S, Ishibe T, Shima Y, Nishijo K, et al. Identification of AFAP1L1 as a prognostic marker for spindle cell sarcomas. Oncogene 2011;30(38):4015-25.
[Crossref] [Google Scholar] [PubMed]
- Kajita Y, Kato Jr T, Tamaki S, Furu M, Takahashi R, Nagayama S, et al. The transcription factor Sp3 regulates the expression of a metastasis-related marker of sarcoma, actin filament-associated protein 1-like 1 (AFAP1L1). PLoS One 2013;8(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Tie SR, McCarthy DJ, Kendrick TS, Louw A, Le C, Satiaputra J, et al. Regulation of sarcoma cell migration, invasion and invadopodia formation by AFAP1L1 through a phosphotyrosine-dependent pathway. Oncogene 2016;35(16):2098-111.
[Crossref] [Google Scholar] [PubMed]
- Wang M, Han X, Sun W, Li X, Jing G, Zhang X. Actin filament-associated protein 1-like 1 mediates proliferation and survival in non-small cell lung cancer cells. Med Sci Monit 2018;24:215-24.
[Crossref] [Google Scholar] [PubMed]
- Zimmermann N, Nassiri M, Zhou J, Miller AM, Zhang S. Myeloid neoplasm with a novel cryptic PDGFRB rearrangement detected by next-generation sequencing. Cancer Genet 2020;244:55-9.
[Crossref] [Google Scholar] [PubMed]
- Takahashi R, Nagayama S, Furu M, Kajita Y, Jin Y, Kato T, et al. AFAP 1L1, a novel associating partner with vinculin, modulates cellular morphology and motility, and promotes the progression of colorectal cancers. Cancer Med 2014;3(4):759-74.
[Crossref] [Google Scholar] [PubMed]