- *Corresponding Author:
- Chenwei Li
Department of Clinical Laboratory, The People’s Hospital of Yubei District of Chongqing, Yubei, Chongqing 401120, China
E-mail: 285452208@qq.com
| Date of Received | 17 June 2024 |
| Date of Revision | 09 August 2024 |
| Date of Acceptance | 11 September 2024 |
| Indian J Pharm Sci 2024;86(5):1678-1689 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Breast cancer, the most common malignant tumor among women worldwide, remains an incurable disease once it has spread to distant organs. The human epidermal growth factor receptor 2-positive and triple-negative molecular subtypes are associated with an increased risk of developing breast cancer. This study aimed to investigate the potential impact of crucial neutrophil extracellular traps-related genes on the survival of patients with human epidermal growth factor receptor 2-positive breast cancer and triple-negative breast cancer. 69 neutrophil extracellular traps-related genes were selected as the study subjects to their association with survival, immune infiltration impacts, interaction networks, and potential drugs for human epidermal growth factor receptor 2-positive breast cancer/triple-negative breast cancer. Using least absolute shrinkage and selection operator regression models based on 69 candidate neutrophil extracellular traps-related genes for predicting overall survival and progressionfree survival, we identified 16 key human epidermal growth factor receptor 2 neutrophil extracellular trap genes and 4 key triple-negative breast cancer genes. We identified four core neutrophil extracellular traps-related genes: Alkaline phosphatase, biomineralization associated, autophagy related 7, colony stimulating factor 3 receptor, and cathelicidin antimicrobial peptide responsive element binding protein 5, which may influence breast cancer progression through mechanisms associated with neutrophil extracellular traps. The key neutrophil extracellular traps-related genes were closely correlated with neutrophils, myeloid dendritic cells macrophage, clusters of differentiation 4+ and clusters of differentiation 8+ T cells. Moreover, potential sensitizing drugs for human epidermal growth factor receptor 2-positive breast cancer and triple-negative breast cancer treatment identified using the genomics of therapeutics response portal and genomics of drug sensitivity in cancer databases may include; SB52334, 17-alkyladenine DNA glycosylase, bromodomain containing-K51490254, CAY10618, GMX-1778, daporinad, serdemetan, CIL70 and panobinostat. In summary, the key neutrophil extracellular traps-related genes may play important roles in the growth and development of human epidermal growth factor receptor 2-positive breast cancer and triple-negative breast cancer. They can impact different immune cells, tumor immune microenvironment and predict overall survival and progression-free survival of breast cancer.
Keywords
Triple negative breast cancer, human epidermal growth factor receptor 2-positive breast cancer, bioinformatics, immune infiltration, drug sensitivity
In general, there are four molecular subtypes of Breast Cancer (BC), namely luminal A (Estrogen Receptor (ER) and Progesterone Receptor (PR) positive and negative Human Epidermal Growth Factor Receptor 2 (HER2)), luminal B (ER, PR and HER2 all positive), HER2-positive ((HER2+), ER and PR negative), and Triple-Negative BC (TNBC). Among these types, luminal A BC proliferation is slow and usually has a good prognosis; luminal B BC is sensitive to targeted drugs, and has a relatively satisfactory therapeutic effect. These two types have a lower degree of malignancy. Relatively speaking, HER2BC and TNBC have poor prognosis and are prone to Drug Resistance (DR)[1]. HER2-positive BC is prone to recurrence or metastasis and its prognosis is significantly worse than luminal A and luminal B; and TNBC has rapid metastasis and the worst prognosis. Once the disease progresses to metastasize, the median survival time is very short. Commonly, TNBC is regarded as the most malignant subtype of BC[2,3]. The vast majority of TNBCs are categorized as overlapping with the Basal-Like BC (BLBC) subtype.
Neutrophils play an important role in innate immunity. They constantly patrol organisms to prevent microbial infections and invading pathogens. Recently, it has been progressively recognized that Tumor Associated Neutrophils (TANs) play an important role in the development of cancer and progression of tumor in the microenvironment, and one of the important mechanisms is the Neutrophil Extracellular Trap (NET)[4]. NET was first reported in 2004[5]. Specifically, after stimulation, neutrophils may lose the integrity of their intracellular membrane, chromatin begins to stretch and depolymerize, followed by disintegration of the nuclear membrane and neutrophils may release of Deoxyribonucleic Acid (DNA) and histones. In the process of the constructing extracellular traps, they include a variety of antimicrobial peptides and enzymes, such as neutrophil elastase, histone G, and Myeloperoxidase (MPO), etc., which immobilize and kill invading microorganisms. The scaffolds of NETs are composed of chromatin fibres with a diameter of 15-17 nm; DNA and histones are the main components of NETs. The above process is usually called NETosis. NETosis was induced by stimulation of neutrophils in contact with pathogens (fungi, bacteria, protozoa, parasites), bacterial Lipopolysaccharide (LPS), Interleukin-8 (IL-8) or chemical stimulation with Protein Kinase C (PKC) activators[6]. The effect of NETs on tumor development is dual. As an important step in the innate and adaptive immune responses, which can be triggered by both infectious and sterile stimuli, the role of NETosis in tumorigenesis and development remains controversial. From the perspective of antitumor immunity, NETs may inhibit tumor growth by activating the immune system. NET itself has both anti-infective and pro-inflammatory effects. On the one hand, it can enhance the tumor-killing toxicity through a highly inflammatory and high-immune level microenvironment, including Reactive Oxygen Species (ROS) released by neutrophil granules, neutrophil elastase, and MPO. On the other hand, tumors may also induce NETosis in neutrophils to promote metastasis. Multiple stimuli, including the hypoxic environment, inflammatory cytokines and activated platelets may trigger the formation of NETs in the tumor microenvironment. Tumors may interact with neutrophils through various pathways to affect the formation of NETs. Several recent studies have shown that tumor cells can induce the formation of NETs, including BC, ovarian cancer, colon cancer, oesophagogastric adenocarcinoma and lung cancer[7]. Naturally, NETs can lead to an increase in the malignancy of tumors. For example, IL-8 released by tumor cells via exosomes can activate the release of NETs, ovarian cancer promotes the formation of NETs through the Cathepsin C (CTSC)-Proteinase 3 (PR3)-Interleukin-1Beta (IL-1β) axis, and colorectal cancer cells can transfer mutant Kirsten Rat Sarcoma (KRAS) viral oncogene homologue to neutrophils via exosomes, which promotes the formation of NETs by mediating the up-regulation of IL-8, ultimately leading to the progression of colorectal cancer[8-10].
So far, few studies have focused on the association between NETosis and BC DR/progression (as well as the treatment outcomes, immunogenicity, and immunotherapy responses). Given the duality of NETs, their role in BC DR/progression remains controversial. Besides its toxicity to BC cells, it is also reasonable to hypothesize that tumor-driven NETosis can drive metastasis. For example, NETs have been shown to activate dormant single BC cells in mouse lungs, which can lead to metastasis[11]. The activation of cancer cell dormancy is believed to occur through the remodelling of extracellular matrix induced by NET-associated NE and is further facilitated by Granulocyte-Colony Stimulating Factor (G-CSF). Moreover, Deoxyribonuclease I (DNase I) treatment can degrade NETs, and studies have shown that it can lead to the loss of the reticular structure and a decrease in the metastasis ability[12]. MPO and Citrullinated Histone H3 (H3Cit), the typical biomarker of NETs, were present in primary BC and liver metastasis[13]. In addition, NETs were reported to promote metastasis through endothelial cell damage[14]. Therefore, it is worthwhile to exploring the role of NETs, and in particular, how the NETs associated genes affect BCs that are prone to DR and progression.
Given that HER2BC and TNBC are more malignant subtypes of BC, the influence of NETs-related genes in these types are particularly interesting, and new therapeutic strategies may be discovered based on targeting the key NETs-related genes in HER2BC and TNBC. This study aimed to observe the potential impact of important NETs-related genes on the survival of HER2BC/TNBC patients and to screen the key NETs-related genes in DR/progression of HER2BC/TNBC through bioinformatics analysis and we explored their associated molecular mechanisms, immune infiltration mechanisms and potential therapeutic agents.
Materials and Methods
Selection of NETs-related genes:
As described by Zhang et al.[15], we searched for NETsinitial biomarkers in different studies. In the progression of NETosis in immunity and various diseases, 30 genes (including ligands and receptors) stimulate the formation of NETs, and 39 genes were identified as the downstream signals adhere to the NETs framework of NETs. A total of 69 genes were used as NETs-related genes.
NETs-related genes in the progression of HER2BC and TNBC:
Next, we used the 69 NETs-related genes to establish models for survival prediction by Least Absolute Shrinkage and Selection Operator (LASSO) regression, including Overall Survival (OS) and Progression-Free Survival (PFS). In the process, the LASSO regression algorithm was applied to feature selection and ensure the simplicity of the model (using the R package glmnet and survival). After dimensionality reduction, the most import features (genes) that impact the survival were selected. And multivariate Cox regression analysis was used to construct a prognostic model (using the R package survival). Ribonucleic Acid (RNA)- sequencing expression profiles and corresponding clinical information for HER2BC and TNBC were downloaded from The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.com). The counts data were converted to Transcript Per Million (TPM) and log2 TPM1 was calculated. Logarithmic-rank test was used to compare the survival differences between these groups. In the Kaplan-Meier curves, the p-values and Hazard Ratio (HR) with 95 % Confidence Interval (CI) were generated through log-rank tests and univariate Cox proportional hazards regression. The key factors validated in the prognostic models were considered as HER2BC-NETs key related genes and TNBC-NETs key related genes.
Core genes in HER2BC and TNBC NETs:
Among the key genes acquired by the LASSO regression model, we tested whether there was a strong correlation between the gene expression and survival (OS or PFS) rate. We downloaded RNA-sequencing data of 185 HER2BC samples and 140 TNBC samples from TCGA (https://portal.gdc.cancer.gov/). For each key genes, samples were divided into two groups (low expression and high expression); the OS and PFS in two groups were compared by the Kaplan-Meier method. The key genes associated with significant differences in survival rates were selected as the core genes, which may be highly likely to influence the DR/progression of HER2BC and TNBC.
Key gene interaction networks:
There were a total of 19 key genes associated with HER2BC-NETs and TNBC-NETs. The interaction between these genes were analysed by the STRING and GeneMANIA online tools. First, the overall Protein- Protein Interaction (PPI) network was generated by STRING (confidence >0.15, for all interactions except for text mining). Then, following specific interactions were explored by the GeneMANIA tool: Genetic interaction (two genes are functionally associated if the effects of perturbing one gene were found to be modified by perturbations to a second gene. These data were collected from primary studies and BioGRID), protein physical interaction (according to BioGRID and pathway commons, if two gene products were found to interact in PPI studies) and pathways (two gene products were in the same pathway).
Enrichment analysis:
Based on 19 key BC-NETs-related genes, we used the Metascape tool to explore the enriched Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and reactome pathways. Firstly, for pathway and process enrichment analysis, we identified all statistically enriched terms, accumulative hypergeometric p-values and enrichment factors were calculated and used them for filtering. The first top 10 enrichment terms were represented as bars. The representative terms were converted into a network layout. More specifically, each term was represented by a circular node, whose size is proportional to the number of input genes fall under that term, and whose colour represented its cluster identity. Terms with a similarity score >0.3 were connected by an edge (the thickness of the edge represents the similarity score). The network was visualized with Cytoscape with a “force-directed” layout and clear edge binding. One term from each cluster was selected to display its term description as a label. Next, gene list enrichments was determined in the following ontology categories: DisGeNET, Transcription Regulatory Relationships Unraveled Sentence-based Text mining (TRRUST) and transcription factor targets. All genes in the genome have been used as the enrichment background. Terms with a p<0.05, a minimum count of 3, and an enrichment factor >1.5 (the enrichment factor was the ratio between the observed counts and the random expected counts) were collected and grouped into clusters based on their membership similarities.
Tumor immune infiltration levels associated with key genes:
The RNA expression profiles of 185 HER2BC and TNBC samples in TCGA dataset were normalized to log2 (TPM1). The immune score evaluation was analysed by the Tumor Immune Estimation Resource (TIMER) algorithm in the immuneeconv R software package. Visualization was performed by the R software ggClusterNet package. The heatmaps were generated to show the correlations between gene expression and immune cell expression (Spearman’s correlation analysis). In the heatmaps, red represents positive correlation, green represents negative correlation, the more red or blue colour means the greater correlation, also the larger circle means the stronger correlation. The red line in the diagram represents the negative correlation between the gene expression and immune cell score, while the green line means the positive correlation.
Potential drugs for HER2BC and TNBC based on KEYnet database:
The gene set cancer analysis tool was implemented to identify potential drugs for TNBC based on Key- TNBC-CRGs. Two databases were referenced to acquire the Half-Maximal Inhibitory Concentration (IC50) value of drugs; Genomics of Drug Sensitivity in Cancer (GDSC) and Genomics of Therapeutics Response Portal (CTRP). The messenger RNA (mRNA) expression data and drug sensitivity data were merged. Pearson correlation analysis was performed to obtain the correlation between gene mRNA expression and drug IC50. The p-value was adjusted by the False Discovery Rate (FDR) value. In the generated bubble plots, we summarized the correlations between key HER2BC-NET genes or TNBC-NET genes and drugs. Blue bubbles represent negative correlations, red bubbles represent positive correlations, the darker colour, and the higher of the correlation. Bubble size is positively correlated with the FDR significance. The black outline border indicates FDR≤0.05.The drugs are ranked by the integrated level of correlation coefficient and FDR, and only the top 30 ranked drugs were listed.
Results and Discussion
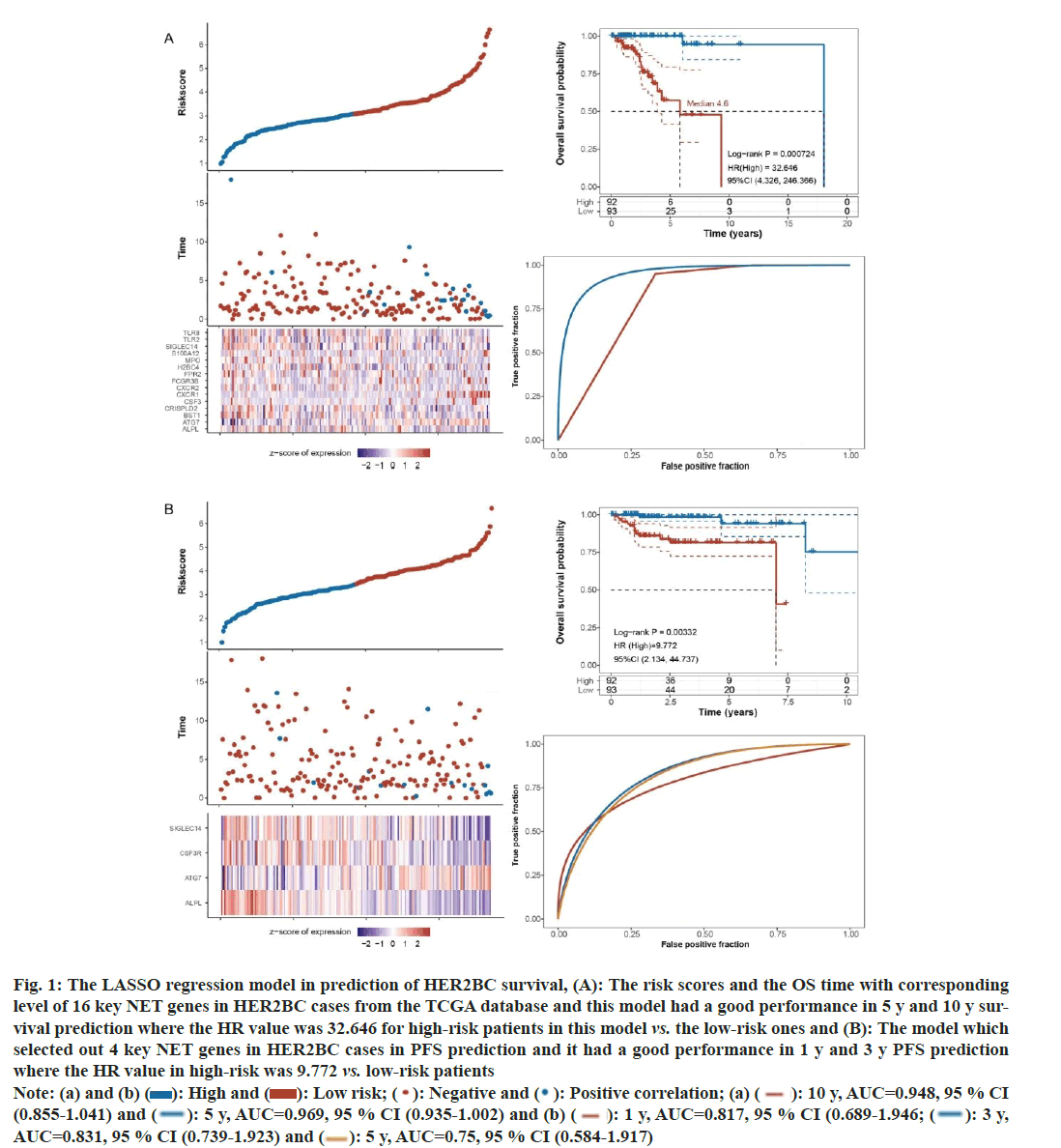
For OS prediction in HER2BC, the model included following 16 NET-associated genes; Toll-Like Receptor 8 (TLR8), Autophagy Related 7 (ATG7), Bone marrow stromal cell antigen 1 (BST1), Cysteine Rich Secretory Protein LCCL Domain containing 2 (CRISPLD2), Colony Stimulating Factor 3 (CSF3), CSF3 Receptor (CSF3R), C-X-C Motif Chemokine Receptor 2 (CXCR2), Fc Gamma Receptor 3B (FCGR3B), Formyl Peptide Receptor 2 (FPR2), H2B Clustered Histone (4H2BC4), MPO, S100 Calcium Binding Protein A12 (S100A12), Sialic Acid Binding Ig Like Lectin 14 (SIGLEC14), TLR2, Alkaline Phosphatase (ALPL), biomineralization associated and CSF3R. This model had a good performance in 5 y and 10 y survival prediction (5 y, Area Under the Curve (AUC)=0.969, 95 % CI: 0.935-1.002; 10 y, AUC=0.948, 95 % CI: 0.855-1.041). The High-Risk (HR) value was 32.646 for high-risk patients in this model compared to the low-risk ones, log-rank p<0.01 (fig. 1A).
For PFS prediction in HER2BC, the LASSO regression model found four import features; SIGLEC14, CSF3R, ATG7, and ALPL. This model performed well in 1 y and 3 y PFS prediction (1 y, AUC=0.817, 95 % CI: 0.689-0.946; 3 y, AUC=0.831, 95 % CI: 0.739-0.923). The HR value was 9.772 (95 % CI: 2.134, 44.737) in this model, log-rank p<0.01 (fig. 1B).
Fig. 1: The LASSO regression model in prediction of HER2BC survival, (A): The risk scores and the OS time with corresponding
level of 16 key NET genes in HER2BC cases from the TCGA database and this model had a good performance in 5 y and 10 y survival
prediction where the HR value was 32.646 for high-risk patients in this model vs. the low-risk ones and (B): The model which
selected out 4 key NET genes in HER2BC cases in PFS prediction and it had a good performance in 1 y and 3 y PFS prediction
where the HR value in high-risk was 9.772 vs. low-risk patients
Note: (a) and (b) ( ): High and (
): High and ( ): Low risk; (
): Low risk; (  ): Negative and (
): Negative and (  ): Positive correlation; (a) (
): Positive correlation; (a) (  ): 10 y, AUC=0.948, 95 % CI
(0.855-1.041) and (
): 10 y, AUC=0.948, 95 % CI
(0.855-1.041) and ( ): 5 y, AUC=0.969, 95 % CI (0.935-1.002) and (b) (
): 5 y, AUC=0.969, 95 % CI (0.935-1.002) and (b) ( ): 1 y, AUC=0.817, 95 % CI (0.689-1.946; (
): 1 y, AUC=0.817, 95 % CI (0.689-1.946; (  ): 3 y,
AUC=0.831, 95 % CI (0.739-1.923) and (
): 3 y,
AUC=0.831, 95 % CI (0.739-1.923) and ( ): 5 y, AUC=0.75, 95 % CI (0.584-1.917)
): 5 y, AUC=0.75, 95 % CI (0.584-1.917)
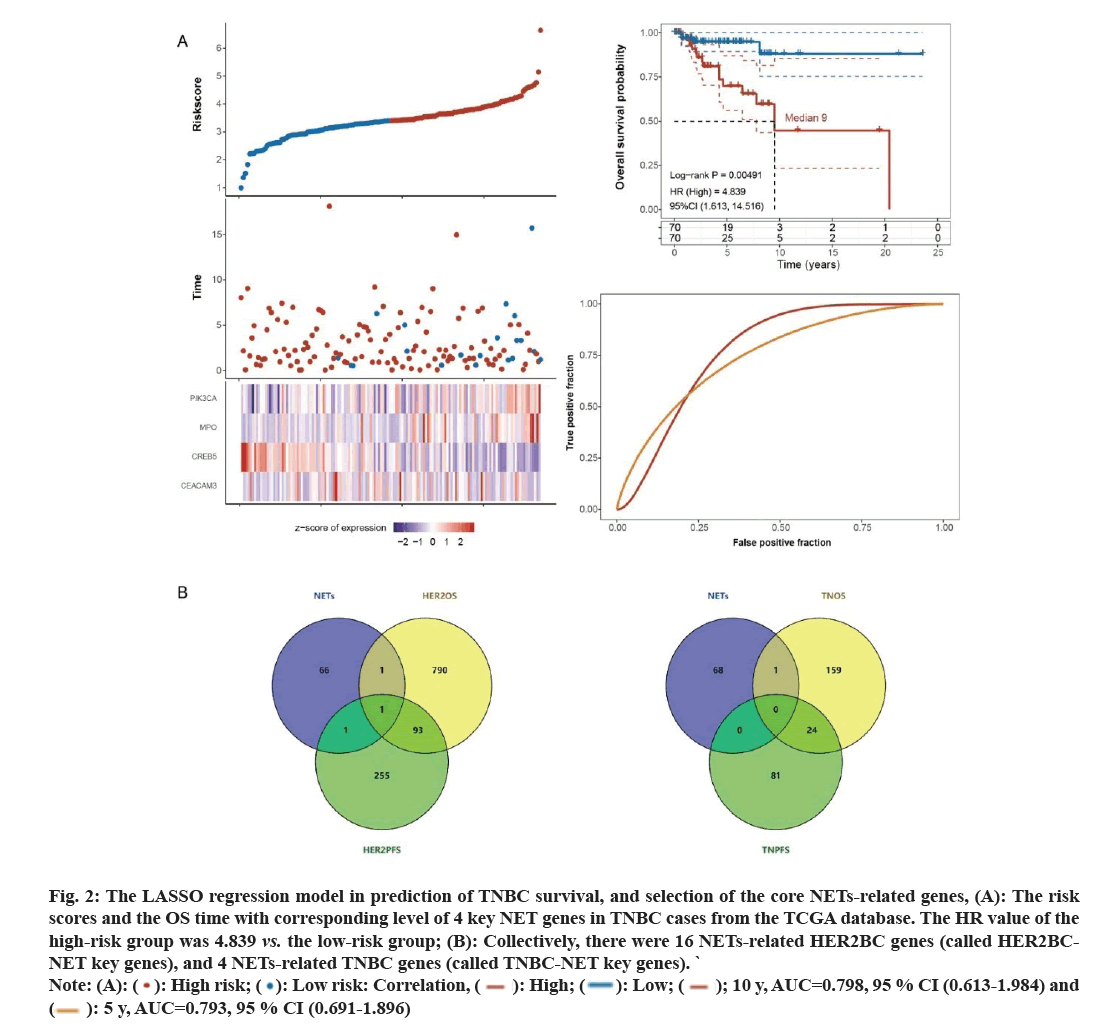
For OS prediction in TNBC (fig. 2A), dimensionality reduction was conducted, and four genes were focused on; Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic subunit Alpha (PIK3CA), MPO, Cathelicidin Antimicrobial Peptide (CAMP) Responsive Element Binding protein 5 (CREB5) and CEA Cell Adhesion Molecule 3 (CEACAM3). The performance of 5 y and 10 y survival prediction was as follows; 5 y, AUC=0.793, 95 % CI: 0.691-0.896; 10 y, AUC=0.798, 95 % CI: 0.613- 0.984. The HR value of the high-risk group was 4.839 compared to the low-risk group (log-rank p<0.01).
Collectively, there were 16 NETs-related HER2BC genes (called HER2BC-NET key genes), and 4 NETsrelated TNBC genes (called TNBC-NET key genes). Between them, a common gene was MPO, and the union set has 19 key genes.
Next, we explored which of these 19 key BC-NET genes the core genes were in. In 185 cases of HER2BC, we found 885 genes associated with OS and 350 genes associated with PFS and their intersection with 69 NETs-related genes were analysed. As shown in fig. 2B, ATG7 was shared by the NETs and HER2OS sets; CSF3R was shared by the HER2PFS and NETs sets; and ALPL was shared by three sets. Consistently, ALPL, ATG7 and CSF3R were all involved in the 16 NETs-related HER2BC genes. Moreover, we identified 184 OS associated genes and 105 PFS associated genes in 140 TNBC samples. Only one gene, CREB5, was shared between NETs and TNOS sets (fig. 2C). CREB5 was also involved in the 4 NETs-related TNBC genes. Meanwhile, among the 19 key genes, ALPL, ATG7, CSF3R, and CREB5 were the core genes which may have effect on progression of BC through NETs associated mechanisms.
Fig. 2: The LASSO regression model in prediction of TNBC survival, and selection of the core NETs-related genes, (A): The risk
scores and the OS time with corresponding level of 4 key NET genes in TNBC cases from the TCGA database. The HR value of the
high-risk group was 4.839 vs. the low-risk group; (B): Collectively, there were 16 NETs-related HER2BC genes (called HER2BCNET
key genes), and 4 NETs-related TNBC genes (called TNBC-NET key genes). `
Note: (A): (  ): High risk; (
): High risk; ( ): Low risk: Correlation, (
): Low risk: Correlation, ( ): High; (
): High; (  ): Low; (
): Low; ( ); 10 y, AUC=0.798, 95 % CI (0.613-1.984) and (
); 10 y, AUC=0.798, 95 % CI (0.613-1.984) and (  ): 5 y, AUC=0.793, 95 % CI (0.691-1.896)
): 5 y, AUC=0.793, 95 % CI (0.691-1.896)
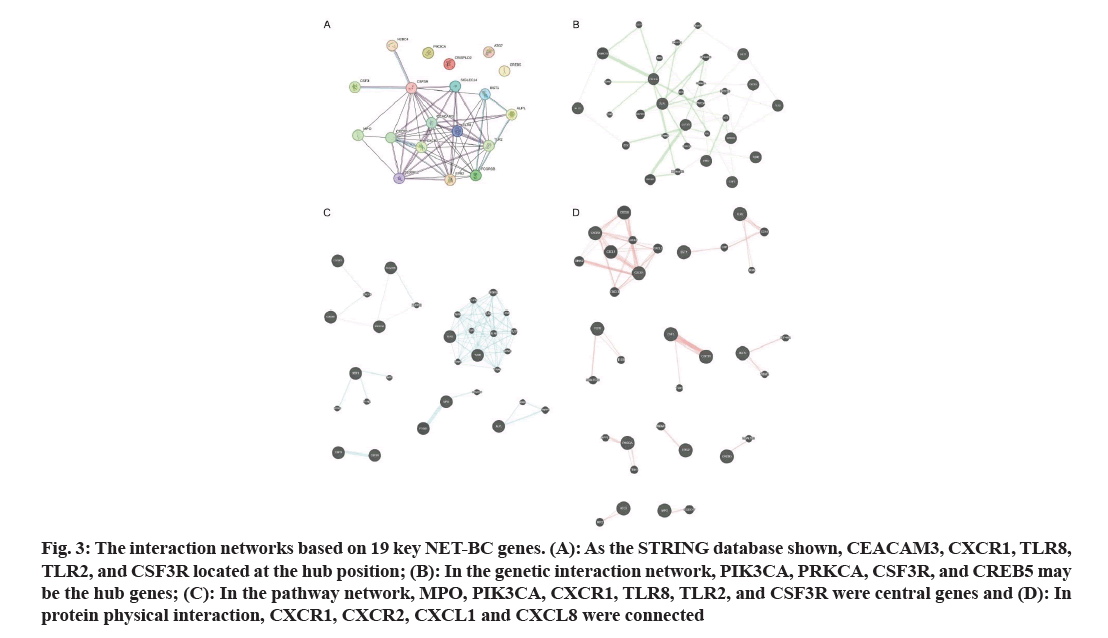
Based on the 19 key genes, interaction networks are shown in fig. 3. As the STRING database shown, CEACAM3, CXCR1, TLR8, TLR2, and CSF3R located at the central position (fig. 3A). In the genetic interaction network, PIK3CA, PRKCA, CSF3R, and CREB5 may be the hub genes (fig. 3B). In the pathway network, PIK3CA, CXCR1, TLR8, TLR2, and CSF3R are also important hub genes, as is MPO (fig. 3C). In protein physical interaction, CXCR1, CXCR2, CXCL1 and CXCL8 are connected through a network (fig. 3D).
Fig. 3: The interaction networks based on 19 key NET-BC genes. (A): As the STRING database shown, CEACAM3, CXCR1, TLR8, TLR2, and CSF3R located at the hub position; (B): In the genetic interaction network, PIK3CA, PRKCA, CSF3R, and CREB5 may be the hub genes; (C): In the pathway network, MPO, PIK3CA, CXCR1, TLR8, TLR2, and CSF3R were central genes and (D): In protein physical interaction, CXCR1, CXCR2, CXCL1 and CXCL8 were connected
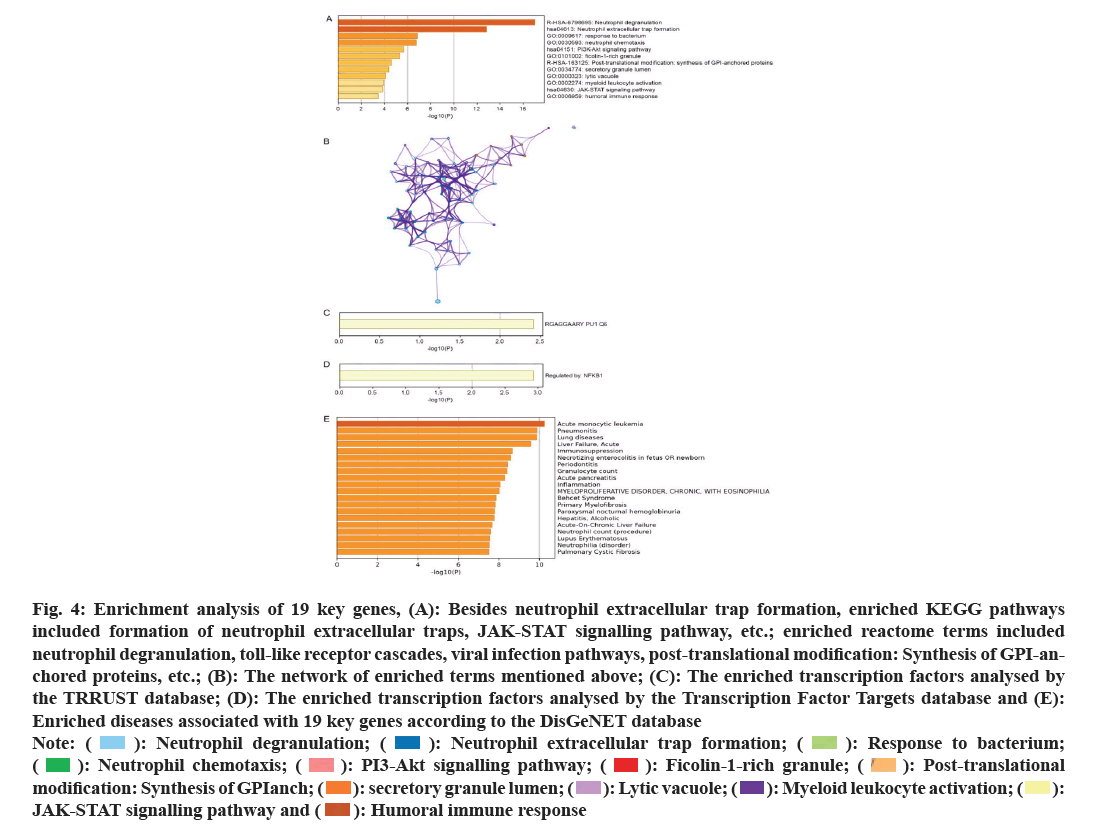
In GO enrichment analysis, the following terms were enriched: Response to bacteria, neutrophil chemotaxis, ficolin-1-rich granule, secretory granule lumen, lytic vacuole, myeloid leukocyte activation, etc., (fig. 4A). In addition to NET formation, enriched KEGG pathways include Janus Kinase/Signal Transducers and Activators of Transcription (JAK/STAT) signalling pathway, etc., (fig. 4A). And enriched reactome terms include neutrophil degranulation, toll-like receptor cascades, viral infection pathways, post-translational modification: Synthesis of GPI-anchored proteins, etc. The network of above enriched terms was shown in fig. 4B. The enriched transcription factors may include PU1Q6 (fig. 4C) and NF-kappaB1 (fig. 4D). Moreover, according to the DisGeNET database, enriched diseases associated with 19 key genes include: Acute monocytic leukemia, pneumonitis, lung diseases, etc., (fig. 4E).
Fig. 4: Enrichment analysis of 19 key genes, (A): Besides neutrophil extracellular trap formation, enriched KEGG pathways
included formation of neutrophil extracellular traps, JAK-STAT signalling pathway, etc.; enriched reactome terms included
neutrophil degranulation, toll-like receptor cascades, viral infection pathways, post-translational modification: Synthesis of GPI-anchored
proteins, etc.; (B): The network of enriched terms mentioned above; (C): The enriched transcription factors analysed by
the TRRUST database; (D): The enriched transcription factors analysed by the Transcription Factor Targets database and (E):
Enriched diseases associated with 19 key genes according to the DisGeNET database
Note: (  ): Neutrophil degranulation; (
): Neutrophil degranulation; ( ): Neutrophil extracellular trap formation; (
): Neutrophil extracellular trap formation; (  ): Response to bacterium; (
): Response to bacterium; (  ): Neutrophil chemotaxis; (
): Neutrophil chemotaxis; ( ): PI3-Akt signalling pathway; (
): PI3-Akt signalling pathway; (  ): Ficolin-1-rich granule; (
): Ficolin-1-rich granule; ( ): Post-translational modification: Synthesis of GPIanch; (
): Post-translational modification: Synthesis of GPIanch; ( ): secretory granule lumen; (
): secretory granule lumen; (  ): Lytic vacuole; (
): Lytic vacuole; ( ): Myeloid leukocyte activation; (
): Myeloid leukocyte activation; ( ): JAK-STAT signalling pathway and (
): JAK-STAT signalling pathway and (  ): Humoral immune response
): Humoral immune response
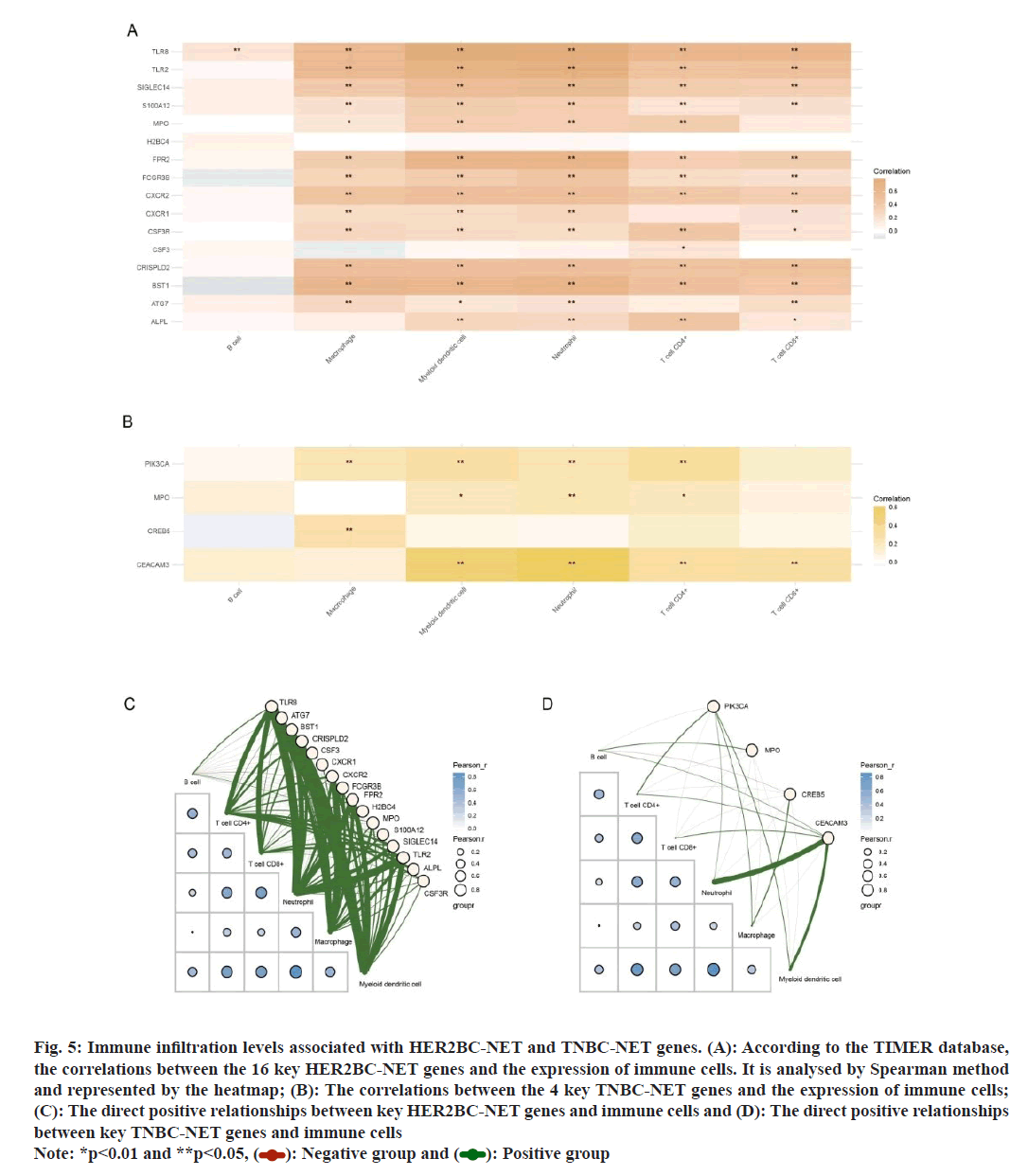
According to the TIMER database, the correlations between the 16 key HER2BC-NET genes and immune cell was analysed by Spearman method, as shown in the heat-map in fig. 5A. Except for H2BC4 and CSF3, all these genes were positively correlated with neutrophils and myeloid dendritic cells and most of these genes were positively correlated with macrophage, Clusters of Differentiation 4+ m(CD4+) and CD8+ T cells. Similarly, among the 4 key TNBC-NET genes, all genes except CREB5 were positively correlated with neutrophils, CD4+ T cells and myeloid dendritic cells (fig. 5B). The direct positive correlation between the key NETs-related genes and immune cells is shown in fig. 5C and fig. 5D.
Fig. 5: Immune infiltration levels associated with HER2BC-NET and TNBC-NET genes. (A): According to the TIMER database,
the correlations between the 16 key HER2BC-NET genes and the expression of immune cells. It is analysed by Spearman method
and represented by the heatmap; (B): The correlations between the 4 key TNBC-NET genes and the expression of immune cells;
(C): The direct positive relationships between key HER2BC-NET genes and immune cells and (D): The direct positive relationships
between key TNBC-NET genes and immune cells
Note: *p<0.01 and **p<0.05, ( ): Negative group and (
): Negative group and (  ): Positive group
): Positive group
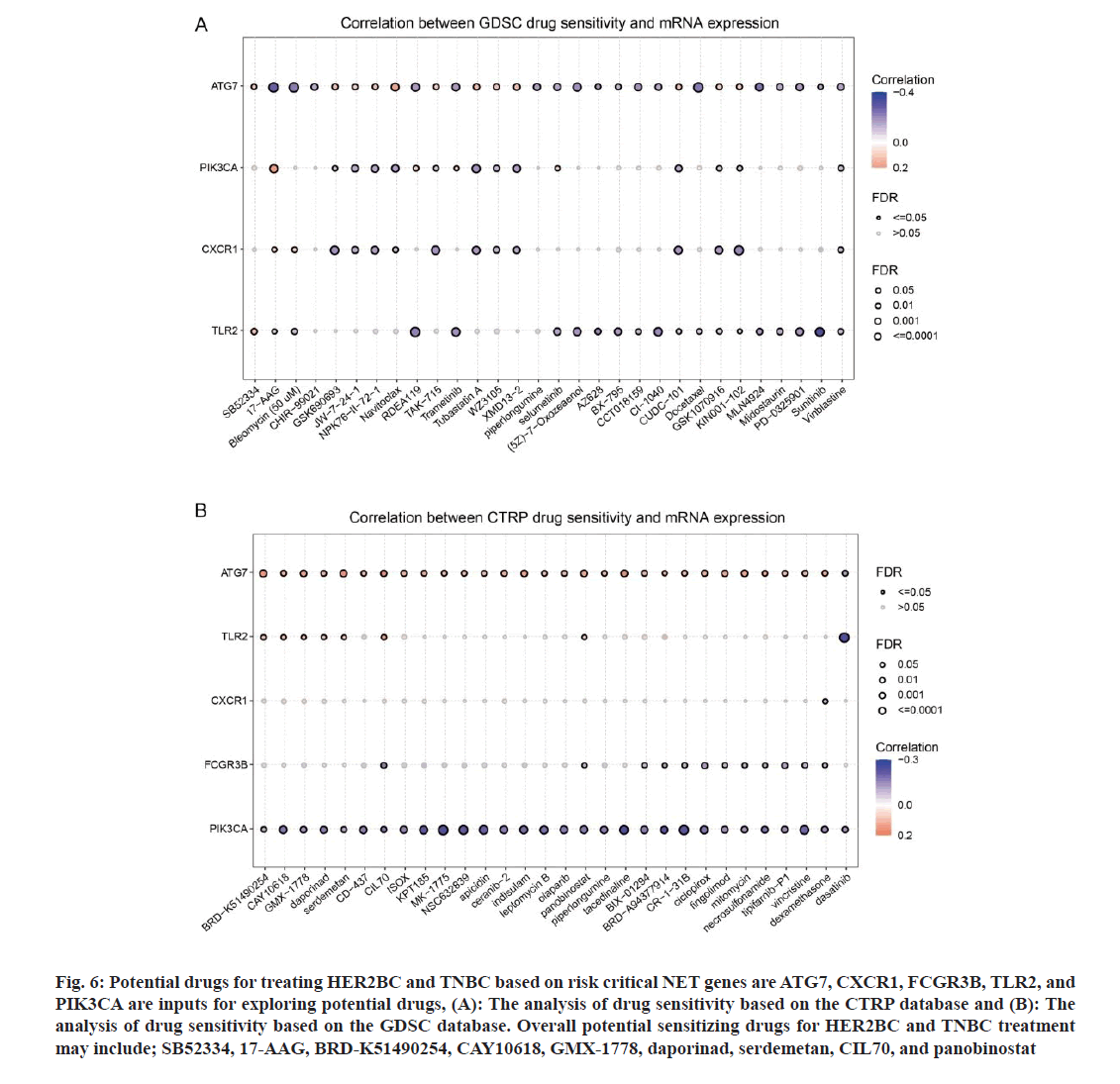
Finally, we aimed to identify potential drugs for improving HER2BC and TNBC progression based on the strategy of targeting high-risk genes among the 19 key genes based on the CTRP and GDSC databases. According to fig. 1A, following genes were associated with low survival (high risk) of HER2BC: ATG7, CXCR1, FCGR3B, and TLR2 and ATG7 was also a risk factor in the PFS model (fig. 1B). In the TNBC OS model, MPO only showed weak risk, while PIK3CA was a relatively strong risk factor. Therefore, we input ATG7, CXCR1, FCGR3B, TLR2, and PIK3CA to explore the potential drugs. The correlations between drug sensitivity and mRNA expression were shown in fig. 6A and fig. 6B. Overall, the potential sensitizing drugs for HER2BC and TNBC treatment may include SB52334, 17-Alkyladenine DNA Glycosylase (AAG), Bromodomain Containing (BRD)-K51490254, CAY10618, GMX-1778, daporinad, serdemetan, CIL70, and panobinostat. The future work can evaluate the performance of above mentioned drugs (or drug combinations) in the treatment of DR BC.
Fig. 6: Potential drugs for treating HER2BC and TNBC based on risk critical NET genes are ATG7, CXCR1, FCGR3B, TLR2, and PIK3CA are inputs for exploring potential drugs, (A): The analysis of drug sensitivity based on the CTRP database and (B): The analysis of drug sensitivity based on the GDSC database. Overall potential sensitizing drugs for HER2BC and TNBC treatment may include; SB52334, 17-AAG, BRD-K51490254, CAY10618, GMX-1778, daporinad, serdemetan, CIL70, and panobinostat
In this study, we first used the LASSO regression method to identify 16 HER2BC-NET OS genes, 4 HER2BCNET PFS genes and 4 TNBC-NET OS genes. Among them, MPO was a common gene. MPO is one of the most abundant proteins in neutrophils. It is stored in the azurophilic granules and secreted when stimulated. MPO contributed to DNA de-condensation, binding to DNA, and catalysing oxidative reactions. It is necessary for autonomous cells to promote the formation of NETs. Generally speaking, MPO is considered as a representative biomarker of NETosis. The role of MPO in the progression of HER2BC and TNBC has not been fully understood. In the OS prognostic model of HER2BC, MPO was a protective factor; in the OS prognostic model of TNBC, it was a very weak risk factor. In summary, NETosis represented by MPO may play an anti-tumor role in the progression of BC. It has been reported that MPO variants, related to reduce the generation of ROS, are associated with decreased BC risk[16]. In consistency with our results, a study in 2019 performed immunohistochemical staining on MPO on multiple tissue microarrays comprising a total of 928 BC samples, and they found that infiltration of MPOpositive cells indicated significantly better performance of OS in the luminal B/HER2-negative subtype cases (p=0.005), the HER2 enriched subtype cases (p=0.011), and the triple negative subtype cases (p<0.001); and multivariate analysis showed that MPO expression was an independent prognostic factor for improving OS (p<0.001)[17].
Among the key NET genes, we identified 4 core genes; ALPL, ATG7, CSF3R, and CREB5. In the both OS and PFS prognostic models (fig. 1 and fig. 2), ALPL showed as a protective factor for survival. In line with our research results, downregulation of ALPL (alkaline phosphatase, biomineralization associated) has been reported to be a potential biomarker gene for bone metastasis progression of invasive BC[18]. ATG7 (the autophagy gene) was both positively related to the low survival rates in OS and PFS prognostic models. As a support, in vivo experiments have confirmed ATG7 was crucial for autophagy activation; mechanistically, inhibition of the autophagic flux in dormant BC cells leads to the accumulation of damaged mitochondria and ROS, resulting in cell apoptosis[19]. Our result implied that, in the NET microenvironment, BC may develop into DR and evade neutrophil toxicity through ATG7 signals (such as autophagy activation). In the PFS prognostic model of HER2BC, CSF3R is a slight protective factor. It has been noted that tumorderived G-CSF can remodel the vascular endothelium in a hematopoietic cell-independent manner through CSF3R; and targeting of the metastatic niche by blocking G-CSF receptor may inhibit pathological vascular remodelling and reduce the burden of bone metastasis[20]. Additionally, CREB5 was a protective factor for OS in the TNBC prognostic model. The role of CREB5 in BC DR/progression has not been fully clear. A study in 2020 constructed a circularRNA-microRNAmRNA regulatory network and generated a prognostic model based on 7 signatures (including CREB5) of BC (all subtypes)[21]. Similarly, they found that the negative correlation between CREB5 expression and risk score. However, the exact mechanism by which CREB5 inhibits BC development remains unclear. Our results findings suggest that it may exert an anti-tumor effects through NETosis.
In immune infiltration analysis, except for H2BC4 and CSF3, all NET genes were positively correlated with neutrophils and myeloid dendritic cells; most of them were positively correlated with macrophage, CD4+ and CD8+ T cells; except for CREB5, all 4 key TNBCNET genes were positively correlated with neutrophils, CD4+ T cells and myeloid dendritic cells. This result implied that, in the NET microenvironment, key NET genes may regulate the network of neutrophils/myeloid dendritic cells/CD4+/CD8+ T cells to affect DR and progression of BC.
Finally, we suggested that the high expression of five key NET genes (ATG7, CXCR1, FCGR3B, TLR2, and PIK3CA) may contribute to the progression of BC and proposed some potential targeted therapeutic strategies based on pharmacological sensitivity analysis. The mechanism of ATG7 has been discussed above. It is known that CXCR1 blockade may selectively target human BC stem cells in vitro and in xenografts[22]. IL-8/CXCR1/2 paracrine activation induces BC cells to acquire migration and invasive features[23]. TLR2 signalling may lead to a chemotherapyresistant phenotype[24]; and unprecedented use of TLR2 inhibitors in vivo may reduce tumor growth and potentiate doxorubicin[24]. PIK3CA is a widely known oncogene, and the mutation (causing signalling activation or increased copies) of PIK3CA is an important driving force of BC metastasis[25-33]. Thereby, we proposed that following drugs may be effective in sensitizing HER2BC and TNBC treatment based on CTRP and GDSC databases; SB52334, 17-AAG, BRD-K51490254, CAY10618, GMX-1778, daporinad, serdemetan, CIL70, and panobinostat. As far as we know, the efficacy of SB52334, BRD-K51490254, CAY10618, GMX-1778, daporinad, and CIL70 on BC has not been extensively explored. There has been some supportive evidence for 17-Allylamino-17-demethoxy Geldanamycin (17-AAG), an inhibitor of heat shock protein 90), which has been used in combination therapies in BC[34,35]. Nanogels co-encapsulated with doxorubicin and 17-AAG have potent anti-tumor activity in HER2BC models[36]. It strongly inhibits BC growth and the HIF-1α/VEGF mediated angiogenesis in vitro and in vivo[37,38]. Panobinostat (LBH589) is an oral histone deacetylase inhibitor. In 2012, a study reported that panobinostat has significant toxicity to TNBC cells in vitro and decreases tumorigenesis in vivo. Additionally, treatment of up-regulating anti-proliferative, tumor suppressor, and epithelial marker genes in MDA-MB-231 cells and initiating a partial reversal of the epithelial-to-mesenchymal transition was reported in the research[39]. Moreover, panobinostat inhibits Wnt/β-catenin signalling pathway by upregulating the expression of APCL in BC[40]. We here demonstrated that panobinostat may also sensitize HER2BC and TNBC by targeting ATG7 and TLR2 through regulating NETosis. In summary, these potential drugs for improving the progression of HER2BC and TNBC can be preferentially studied.
In conclusion, the key NETs-associated genes have shown to play important roles in the development of HER2BC and TNBC. They can impact different immune cells and tumor immune microenvironment. The prognostic model including key NETs-associated genes had a robust performance in prediction of OS and PFS. Finally, based on the risk NET genes, some new drugs are potentially effective in the treatment of HER2BC and TNBC.
Funding:
This study was supported by Chongqing Natural Science Foundation (Grant No: cstc2021jcyjmsxmX0851) and Yongchuan District Technology Innovation and Application Development Project (Grant No: 2023yc-cxfz30007).
Author’s contributions:
Chenwei Li and Ke Wang have contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat 2009;115:423-8.
[Crossref] [Google Scholar] [PubMed]
- Lin WX, Xie YN, Chen YK, Cai JH, Zou J, Zheng JH, et al. Nomogram for predicting overall survival in Chinese triple-negative breast cancer patients after surgery. World J Clin Cases 2022;10(31):11338.
[Crossref] [Google Scholar] [PubMed]
- Zheng YZ, Liu Y, Deng ZH, Liu GW, Xie N. Determining prognostic factors and optimal surgical intervention for early-onset triple-negative breast cancer. Front Oncol 2022;12:910765.
[Crossref] [Google Scholar] [PubMed]
- Huang H, Zhang H, Onuma AE, Tsung A. Neutrophil elastase and neutrophil extracellular traps in the tumor microenvironment. Adv Exp Med Biol 2020;1263:13-23.
[Crossref] [Google Scholar] [PubMed]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science 2004;303(5663):1532-5.
[Crossref] [Google Scholar] [PubMed]
- Mamtimin M, Pinarci A, Han C, Braun A, Anders HJ, Gudermann T, et al. Extracellular DNA traps: Origin, function and implications for anti-cancer therapies. Front Oncol 2022;12:869706.
[Crossref] [Google Scholar] [PubMed]
- Chen Q, Zhang L, Li X, Zhuo W. Neutrophil extracellular traps in tumor metastasis: pathological functions and clinical applications. Cancers 2021;13(11):2832.
[Crossref] [Google Scholar] [PubMed]
- Cao TM, King MR. Supercharged eGFP-TRAIL decorated NETs to ensnare and kill disseminated tumor cells. Cell Mol Bioeng 2020;13:359-67.
- Lee W, Ko SY, Mohamed MS, Kenny HA, Lengyel E, Naora H. Neutrophils facilitate ovarian cancer premetastatic niche formation in the omentum. J Exp Med 2019;216(1):176-94.
[Crossref] [Google Scholar] [PubMed]
- Shang A, Gu C, Zhou C, Yang Y, Chen C, Zeng B, et al. Exosomal KRAS mutation promotes the formation of tumor-associated neutrophil extracellular traps and causes deterioration of colorectal cancer by inducing IL-8 expression. Cell Commun Signal 2020;18:1-4.
[Crossref] [Google Scholar] [PubMed]
- Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 2018;361(6409):eaao4227.
[Crossref] [Google Scholar] [PubMed]
- Leal AC, Mizurini DM, Gomes T, Rochael NC, Saraiva EM, Dias MS, et al. Tumor-derived exosomes induce the formation of neutrophil extracellular traps: Implications for the establishment of cancer-associated thrombosis. Sci Rep 2017;7(1):6438.
[Crossref] [Google Scholar] [PubMed]
- Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 2020;583(7814):133-8.
[Crossref] [Google Scholar] [PubMed]
- Snoderly HT, Boone BA, Bennewitz MF. Neutrophil extracellular traps in breast cancer and beyond: Current perspectives on NET stimuli, thrombosis and metastasis, and clinical utility for diagnosis and treatment. Breast Cancer Res 2019;21(1):145.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Guo L, Dai Q, Shang B, Xiao T, Di X, et al. A signature for pan-cancer prognosis based on neutrophil extracellular traps. J Immunother Cancer 2022;10(6).
[Crossref] [Google Scholar] [PubMed]
- Ahn J, Gammon MD, Santella RM, Gaudet MM, Britton JA, Teitelbaum SL, et al. Myeloperoxidase genotype, fruit and vegetable consumption, and breast cancer risk. Cancer Res 2004;64(20):7634-9.
[Crossref] [Google Scholar] [PubMed]
- Zeindler J, Angehrn F, Droeser R, Däster S, Piscuoglio S, Ng CK, et al. Infiltration by myeloperoxidase-positive neutrophils is an independent prognostic factor in breast cancer. Breast Cancer Res Treat 2019;177(3):581-9.
[Crossref] [Google Scholar] [PubMed]
- Tayubi IA, Madar IH. Biomineralization associated alkaline phosphatase as a potential marker of bone metastasis in the patients with invasive breast cancer. Saudi J Biol Sci 2022;29(8):103340.
[Crossref] [Google Scholar] [PubMed]
- Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9(1):1944.
- Yip RK, Rimes JS, Capaldo BD, Vaillant F, Mouchemore KA, Pal B, et al. Mammary tumour cells remodel the bone marrow vascular microenvironment to support metastasis. Nat Commun 2021;12(1):6920.
[Crossref] [Google Scholar] [PubMed]
- Song H, Sun J, Kong W, Ji Y, Xu D, Wang J. Construction of a circRNA-related ceRNA prognostic regulatory network in breast cancer. OncoTargets Ther 2020:8347-58.
[Crossref] [Google Scholar] [PubMed]
- Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest 2010;120(2):485-97.
[Crossref] [Google Scholar] [PubMed]
- Santolla MF, Talia M, Cirillo F, Scordamaglia D, De Rosis S, Spinelli A, et al. The AGEs/RAGE transduction signaling prompts IL-8/CXCR1/2-mediated interaction between Cancer-Associated Fibroblasts (CAFs) and breast cancer cells. Cells 2022;11(15):2402.
[Crossref] [Google Scholar] [PubMed]
- Di Lorenzo A, Bolli E, Ruiu R, Ferrauto G, Di Gregorio E, Avalle L, et al. Toll-like receptor 2 promotes breast cancer progression and resistance to chemotherapy. Oncoimmunology 2022;11(1):2086752.
[Crossref] [Google Scholar] [PubMed]
- André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor–positive advanced breast cancer. New Engl J Med 2019;380(20):1929-40.
[Crossref] [Google Scholar] [PubMed]
- Xing Y, Lin NU, Maurer MA, Chen H, Mahvash A, Sahin A, et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res 2019;21:1-2.
[Crossref] [Google Scholar] [PubMed]
- Martínez-Sáez O, Chic N, Pascual T, Adamo B, Vidal M, González-Farré B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 2020;22(1):1-45.
[Crossref] [Google Scholar] [PubMed]
- Mosele F, Stefanovska B, Lusque A, Dien AT, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol 2020;31(3):377-86.
[Crossref] [Google Scholar] [PubMed]
- André F, Ciruelos EM, Juric D, Loibl S, Campone M, Mayer IA, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2–negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann Oncol 2021;32(2):208-17.
[Crossref] [Google Scholar] [PubMed]
- Cai Y, Xu G, Wu F, Michelini F, Chan C, Qu X, et al. Genomic alterations in PIK3CA-mutated breast cancer result in mTORC1 activation and limit the sensitivity to PI3Kα inhibitors. Cancer Res 2021;81(9):2470-80.
[Crossref] [Google Scholar] [PubMed]
- Fusco N, Malapelle U, Fassan M, Marchiò C, Buglioni S, Zupo S, et al. PIK3CA mutations as a molecular target for hormone receptor-positive, HER2-negative metastatic breast cancer. Front Oncol 2021;11:644737.
[Crossref] [Google Scholar] [PubMed]
- Rugo HS, Lerebours F, Ciruelos E, Drullinsky P, Ruiz-Borrego M, Neven P, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): One cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol 2021;22(4):489-98.
[Crossref] [Google Scholar] [PubMed]
- Donzelli S, Cioce M, Sacconi A, Zanconato F, Daralioti T, Goeman F, et al. A PIK3CA-mutant breast cancer metastatic patient-derived organoid approach to evaluate alpelisib treatment for multiple secondary lesions. Mol Cancer 2022;21(1):152.
- Saxena V, Naguib Y, Hussain MD. Folate receptor targeted 17-allylamino-17-demethoxygeldanamycin (17-AAG) loaded polymeric nanoparticles for breast cancer. Colloids Surf B Biointerfaces 2012;94:274-80.
[Crossref] [Google Scholar] [PubMed]
- Ghadban T, Jessen A, Reeh M, Dibbern JL, Mahner S, Mueller V, et al. In vitro study comparing the efficacy of the water-soluble HSP90 inhibitors, 17-AEPGA and 17-DMAG, with that of the non‑water-soluble HSP90 inhibitor, 17-AAG, in breast cancer cell lines. Int J Mol Med 2016;38(4):1296-302.
[Google Scholar] [PubMed]
- Desale SS, Raja SM, Kim JO, Mohapatra B, Soni KS, Luan H, et al. Polypeptide-based nanogels co-encapsulating a synergistic combination of doxorubicin with 17-AAG show potent anti-tumor activity in ErbB2-driven breast cancer models. J Control Release 2015;208:59-66.
[Crossref] [Google Scholar] [PubMed]
- Zhang PC, Liu X, Li MM, Ma YY, Sun HT, Tian XY, et al. AT-533, a novel Hsp90 inhibitor, inhibits breast cancer growth and HIF-1α/VEGF/VEGFR-2-mediated angiogenesis in vitro and in vivo. Biochem Pharmacol 2020;172:113771.
[Crossref] [Google Scholar] [PubMed]
- Zuo Y, Xu H, Chen Z, Xiong F, Zhang B, Chen K, et al. 17‑AAG synergizes with Belinostat to exhibit a negative effect on the proliferation and invasion of MDA‑MB‑231 breast cancer cells. Oncol Rep 2020;43(6):1928-44.
[Google Scholar] [PubMed]
- Tate CR, Rhodes LV, Segar HC, Driver JL, Pounder FN, Burow ME, et al. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res 2012;14(3):R79.
[Crossref] [Google Scholar] [PubMed]
- Qin G, Li Y, Xu X, Wang X, Zhang K, Tang Y, et al. Panobinostat (LBH589) inhibits Wnt/β-catenin signaling pathway via upregulating APCL expression in breast cancer. Cell Signal 2019;59:62-75.
[Crossref] [Google Scholar] [PubMed]