- *Corresponding Author:
- Yi Li
Department of Pulmonary and Critical Care Medicine, Haining People’s Hospital, Jiaxing, Zhejiang 314400, China
E-mail: jinsely3469@163.com
| This article was originally published in a special issue,“Exploring the Role of Biomedicine in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(1) Spl Issue “96-103” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Radiotherapy is one of the most effective treatment strategies for lung cancer. However, radioresistance is a main limitation for lung cancer therapy. It is required to identify novel target to improve the radiotherapy efficacy of lung cancer. This study aimed to investigate the role of long non-coding ribonucleic acid lung cancer associated transcript 1 in the radioresistance of lung cancer. The correlation between lung cancer associated transcript 1 and patient survival was analyzed by using the cancer genome Atlas database. The expression of lung cancer associated transcript 1 in cells and lung cancer tissues was measured by real time polymerase chain reaction assay. Cell counting kit-8 assay and colony formation assay were used to determine the sensitivity of cells to ionizing radiation. Western blotting assay was performed to detect protein expressions and bioinformatic strategy was used to predict the target competing endogenous ribonucleic acids of lung cancer associated transcript 1. In the present study, we showed that long non-coding ribonucleic acid lung cancer associated transcript 1 is dramatically elevated in lung cancer cells and tissues, and high expression of lung cancer associated transcript 1 was negatively correlated with poor outcome in terms of overall survival. Moreover, knockdown of this long non-coding ribonucleic acid inhibited cell viability and increased cell apoptosis after irradiation, while lung cancer associated transcript 1 overexpression showed opposite effects. Mechanistically, knockdown of lung cancer associated transcript 1 elevated the activation of apoptosis factors B-cell lymphoma 2-associated X protein and cleaved-caspase 3. And lung cancer associated transcript 1 functions through binding with micro ribonucleic acid-199a-5p to regulate radiation response. In conclusion, we identified lung cancer associated transcript 1 as a factor promoting radioresistance in lung cancer, which provide novel target for cancer therapy.
Keywords
Long non-coding ribonucleic acid lung cancer associated transcript 1, radiotherapy, lung cancer, micro ribonucleic acid-199a-5p
Rather than transcriptional noise, long non-coding Ribonucleic Acids (lncRNAs) were identified as non-coding RNAs (larger than 200 nucleotides (nt)) and were found to play critical role in cancer development, progression and metastasis[1,2]. Dysregulation of lncRNAs were also observed in most types of cancer. Along with the progress of sequencing techniques, the genomic analysis have revealed numerous lncRNAs up-regulated or downregulated in cancer tissues, or in serum or wrapped in exosomes, which provide diagnostic advantages of diseases[3,4]. Despite cancer biomarkers, lncRNA were also demonstrated to exert oncogenic or tumor suppressive functions. There are also lncRNAs, which were related to chemoresistance or radioresistance of cancer[5-7]. However, the role of most lncRNAs in cancer resistance remains largely unknown.
Lung cancer is the most common cause of mortality and the overall survival of lung cancer is really poor[8]. Non-small cell lung cancer constitute 80 %-85 % of all lung cancer patients and the current treatment strategies mainly falls into surgery, chemotherapy as well as radiotherapy[8-10]. However, surgery can only be applied in early stages, while cancer resistance is an important obstacle for both chemotherapy and radiotherapy[11,12]. So uncovering the underlying mechanism of cancer resistance is critical for improving the therapeutic effects.
Recently, lncRNAs were reported to play key roles in lung cancer progression and therapy, which provide novel target for cancer therapy. For instance, lncRNA HOX Transcript Antisense RNA (HOTAIR) was upregulated in lung cancer and correlated with poor outcomes of patients[13]. LncRNA-Urothelial Carcinoma Associated 1 (UCA1) regulated lung cancer progression and could be a possible biomarker in plasma[14]. Upregulation of lncRNA Small Nucleolar RNA Host Gene 1 (SNHG1) contribute to lung cancer progression through interacting with microRNA (miR)-101-3p, which targets SRY-Box transcription factor 9 (SOX9) and beta-catenin pathway[15]. Particularly, lncRNA Lung Cancer Associated Transcript 1 (LUCAT1) was proved to be associated in patient survival in lung cancer through repressing p25 and p57[16]. LUCAT1 was also found to promote cell proliferation and tumorigenesis in esophageal squamous cell carcinoma, renal cell carcinoma, glioma etc[17-19]. However, whether LUCAT1 was involved in cancer resistance including chemotherapy or radiotherapy has not been reported. In the present study, we measured the expression of LUCAT1 in lung cancer tissues and cells, and determined the cell proliferation and survival in LUCAT1 knockdown and Overexpressing (OE) cells. Finally, LUCAT1 was proved to be sponging with miR-199a-5p to regulate radiation sensitivity.
Materials and Methods
Cells and treatments:
Human lung cancer cell lines like Adenocarcinomic human alveolar basal epithelial cells (A549), Human lung cancer cell line (H460) and Human non-small cell lung cancer cell line (H358) cells, as well as normal cells like Human Bronchial Epithelial cell line (BEAS-2B) were purchased from American Type Culture Collection, Virginia, United States of America (USA). All cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10 % fetal bovine serum (Gibco) and incubated at 37° in a humid condition. Cells were transfected with lipofectamineTM 3000 reagents to introduce cell knockdown or OE plasmids. After different treatments, cells were exposed to Cobalt-60 (60Co) gamma irradiation and used for next experiments.
Reagents and samples:
All this study was approved by the Ethics Committee of Haining People’s Hospital (Zhejiang, China). Samples of lung cancer patients were collected from 30 patients with lung cancer surgery in Haining People's Hospital. Written informed consent was obtained from each patient. After the isolation of samples, samples were stored at -80° for further detection. All procedures performed in studies involving human participants were approved by the ethics committee of Haining People’s Hospital in accordance with the ethical standards of the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Real time Polymerase Chain Reaction (PCR) assay:
Total RNA was extracted from frozen tissues as well as treated cell lines by using a TRIzol reagent according to the manufacturer’s instructions. The complementary Deoxyribonucleic Acid (cDNA) was synthesized by using a Reverse Transcription (RT) kit (Yeasen Co., Shanghai, China). Then the expressing LUCAT1 OE was detected with a SYBR green real time PCR kit (Yeasen Co. Shanghai, China). For lncRNA expression, all results were normalized for the expression of Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH). For miRNA, U6 was used as internal control.
Short hairpin RNA (shRNA) and OE vectors transfection:
Lentiviral transfection vector of LUCAT1 knockdown and OE was constructed by Genepharma Co. (Shanghai, China), by the way, empty vectors were used as Negative Control (NC). LipofectamineTM 3000 was used to transfect these plasmids into 293T cells, from which the virus was collected at 48 h and 72 h. A549 cells were transfected with either NC/ knockdown virus or NC/OE virus. After 48 h, cells were irradiated and used for next experiments.
Luciferase assay:
A549 cells were seeded in 96 well plates at the concentration of 5000 cells per well. 24 h later, cells were transfected with wild type and mutant LUCAT1 reporter plasmid and miR-199a-5p mimics. At 48 h, the relative luciferase activity was assessed with dual-luciferase reporter assay system (Promega, USA).
Western blotting assay:
The proteins were extracted from cells with a Radio- Immunoprecipitation Assay (RIPA) lysis buffer and subjected to Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE). After transfer, proteins were blocked with milk and probed with the following primary antibodies like B-cell lymphoma 2 (Bcl2) (Cell Signaling Technology (CST), 1:1000); Bcl-2-associated X protein (Bax) (CST, 1:1000); caspase 3 (Abcam, 1:1000); p53 (Abcam, 1:1000); p21 (CST, 1:1000), while GAPDH (CST, 1:1000) at 4° overnight. Then the membrane were incubated with secondary antibodies and exposed with an Enhanced Chemiluminescence (ECL) kit (Thermal Fisher).
Cell proliferation assay:
A Cell Counting Kit-8 (CCK-8) kit (Beyotime, China) was used for cell viability and proliferation determination in different groups. The measurements were strictly performed according to the manufacturer’s instructions.
Apoptosis assay:
At 24 h after irradiation, cells were washed with Phosphate Buffered Saline (PBS) and trypsinized and collected. Then cells were incubated with an Annexin V-Fluorescein Isothiocyanate (FITC) and Propidium Iodide (PI) double staining kit (Beyotime, Haimen, China), according the manufacturer’s instructions. Then the cells were analyzed with flow cytometry equipment (BD, USA).
Statistical analysis:
All the experiments were repeated for three independent times and the data were expressed as mean±Standard Error of Mean (SEM). Data was analyzed with one way Analysis of Variance (ANOVA) followed by student t test, p<0.05 was considered statistically significant.
Results and Discussion
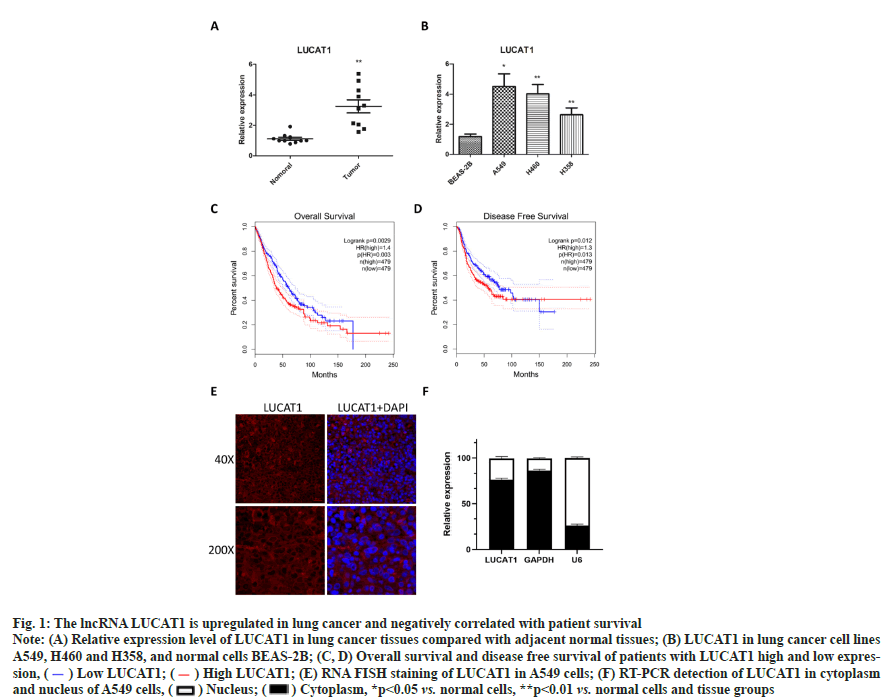
The lncRNA LUCAT1 is up-regulated in lung cancer tissues and cell lines as shown in fig. 1. To check the expression of lncRNA LUCAT1 in lung cancer and cells, total RNA was extracted and transcribed into cDNA, from which LUCAT1 was amplified and measured. We found that the expression of LUCAT1 was significantly increased in lung cancer tissues, compared with normal lung tissues (fig. 1A). LUCAT1 was also observed to be upregulated in lung cancer cells A549, H460 and H358, compared with BEAS-2B (fig. 1B). Through a bioinformatics analysis, we found that high expression of LUCAT1 was negatively correlated with overall survival and disease free survival in lung cancer patients (fig. 1C and fig. 1D). These data indicated that LUCAT1 may play key role in lung cancer. To determine the cellular localization of LUCAT1, we performed RNA Fluorescence In Situ Hybridization (FISH) assay as well RT-PCR in cytoplasm and nucleus. It was observed that most of the LUCAT1 was located outside nucleus, which was also confirmed in the RTPCR assay (fig. 1D and fig. 1E).
Fig. 1: The lncRNA LUCAT1 is upregulated in lung cancer and negatively correlated with patient survival
Note: (A) Relative expression level of LUCAT1 in lung cancer tissues compared with adjacent normal tissues; (B) LUCAT1 in lung cancer cell lines A549, H460 and H358, and normal cells BEAS-2B; (C, D) Overall survival and disease free survival of patients with LUCAT1 high and low expression, ( ) Low LUCAT1; (
) Low LUCAT1; ( ) High LUCAT1; (E) RNA FISH staining of LUCAT1 in A549 cells; (F) RT-PCR detection of LUCAT1 in cytoplasm and nucleus of A549 cells, (
) High LUCAT1; (E) RNA FISH staining of LUCAT1 in A549 cells; (F) RT-PCR detection of LUCAT1 in cytoplasm and nucleus of A549 cells, ( ) Nucleus; (
) Nucleus; ( ) Cytoplasm, *p<0.05 vs. normal cells, **p<0.01 vs. normal cells and tissue groups
) Cytoplasm, *p<0.05 vs. normal cells, **p<0.01 vs. normal cells and tissue groups
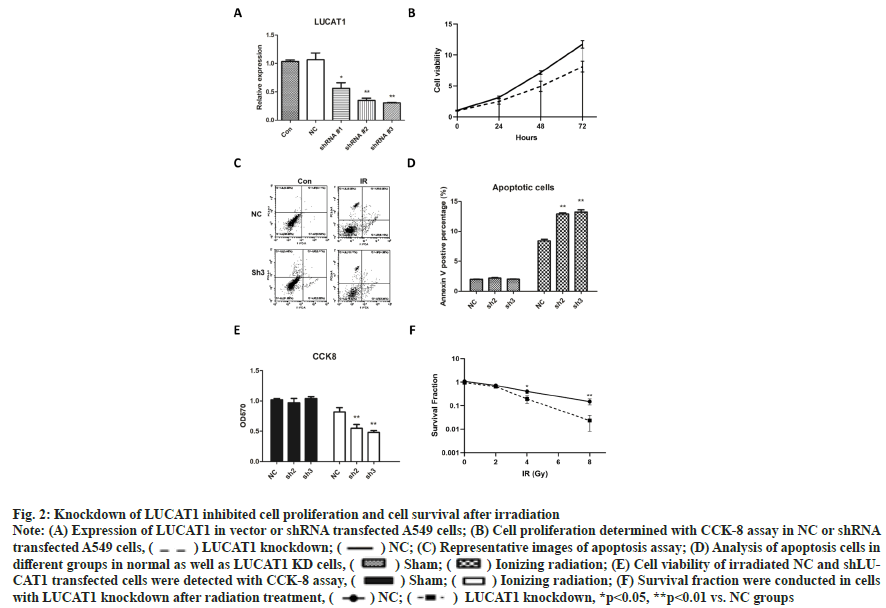
Knockdown of LUCAT1 inhibited cell proliferation and cell survival after irradiation was shown in fig. 2. We used small hairpin RNA to knockdown LUCAT1 in A549 cells and we found that shRNA2 and shRNA3 have good knockdown capacity (fig. 2A). Then cell proliferation was determined with CCK-8 assay at 24 h, 48 h and 72 h, from which we found that knockdown of LUCAT1 significantly, suppressed cell proliferation of A549 cells (fig. 2B). Then, we performed flow cytometry analysis to detect cell apoptosis in different groups. Knockdown of LUCAT1 resulted in more apoptotic cells when combined with ionizing radiation (fig. 2C) and the difference was significant (fig. 2D). Next, CCK- 8 assay was used to measure the cell viability and it was shown that irradiation induced a decrease in cell viability. In two LUCAT knockdown groups, cell viability reduced significantly than the single radiation groups (fig. 2E). In colony formation experiments, significantly fewer colonies survived in LUCAT1 knockdown groups after 4 and 8 Gy irradiation as shown in fig. 2F.
Fig. 2: Knockdown of LUCAT1 inhibited cell proliferation and cell survival after irradiation
Note: (A) Expression of LUCAT1 in vector or shRNA transfected A549 cells; (B) Cell proliferation determined with CCK-8 assay in NC or shRNA transfected A549 cells, (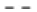 ) LUCAT1 knockdown; (
) LUCAT1 knockdown; ( ) NC; (C) Representative images of apoptosis assay; (D) Analysis of apoptosis cells in different groups in normal as well as LUCAT1 KD cells, (
) NC; (C) Representative images of apoptosis assay; (D) Analysis of apoptosis cells in different groups in normal as well as LUCAT1 KD cells, ( ) Sham; (
) Sham; ( ) Ionizing radiation; (E) Cell viability of irradiated NC and shLUCAT1 transfected cells were detected with CCK-8 assay, (
) Ionizing radiation; (E) Cell viability of irradiated NC and shLUCAT1 transfected cells were detected with CCK-8 assay, ( ) Sham; (
) Sham; ( ) Ionizing radiation; (F) Survival fraction were conducted in cells with LUCAT1 knockdown after radiation treatment, (
) Ionizing radiation; (F) Survival fraction were conducted in cells with LUCAT1 knockdown after radiation treatment, ( ) NC; (
) NC; ( ) LUCAT1 knockdown, *p<0.05, **p<0.01 vs. NC groups
) LUCAT1 knockdown, *p<0.05, **p<0.01 vs. NC groups
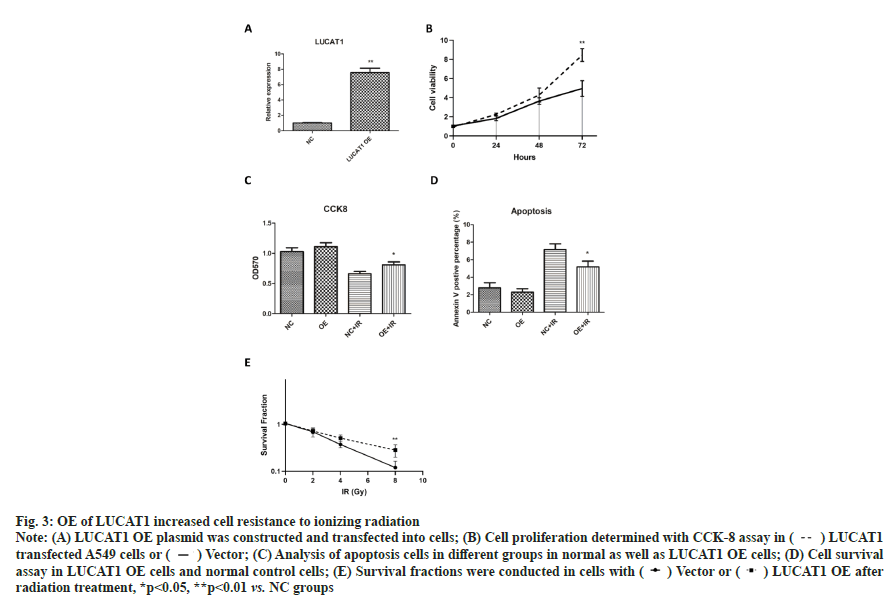
OE of LUCAT1 increased cell resistance to ionizing radiation as shown in fig. 3. To further confirm the role of LUCAT1 in lung cancer radiation sensitivity, we constructed a LUCAT1 OE cell with lentiviral vectors. The upregulation of LUCAT1 was confirmed with real-time PCR assay (fig. 3A). For cell proliferation, we found that OE of LUCAT1 significantly promoted cell proliferation of A549 cells (fig. 3B). After irradiation, less apoptosis cells were detected in LUCAT1 OE cells compared with single radiation group (fig. 3C). From the CCK-8 data, we found that cells with LUCAT1 high expression was significantly resistant to ionizing radiation (fig. 3D). Significantly more colonies were found in LUCAT1 OE groups after irradiation (fig. 3E). These data suggest that high level of LUCAT1 promotes cell resistance to radiotherapy.
Fig. 3: OE of LUCAT1 increased cell resistance to ionizing radiation
Note: (A) LUCAT1 OE plasmid was constructed and transfected int cells; (B) Cell proliferation determined with CCK-8 assay in ( ) LUCAT1 transfected A549 cells or (
) LUCAT1 transfected A549 cells or ( ) Vector; (C) Analysis of apoptosis cells in different groups in normal as well as LUCAT1 OE cells; (D) Cell survival assay in LUCAT1 OE cells and normal control cells; (E) Survival fractions were conducted in cells with (
) Vector; (C) Analysis of apoptosis cells in different groups in normal as well as LUCAT1 OE cells; (D) Cell survival assay in LUCAT1 OE cells and normal control cells; (E) Survival fractions were conducted in cells with ( ) Vector or (
) Vector or ( ) LUCAT1 OE after radiation treatment, *p<0.05, **p<0.01 vs. NC groups
) LUCAT1 OE after radiation treatment, *p<0.05, **p<0.01 vs. NC groups
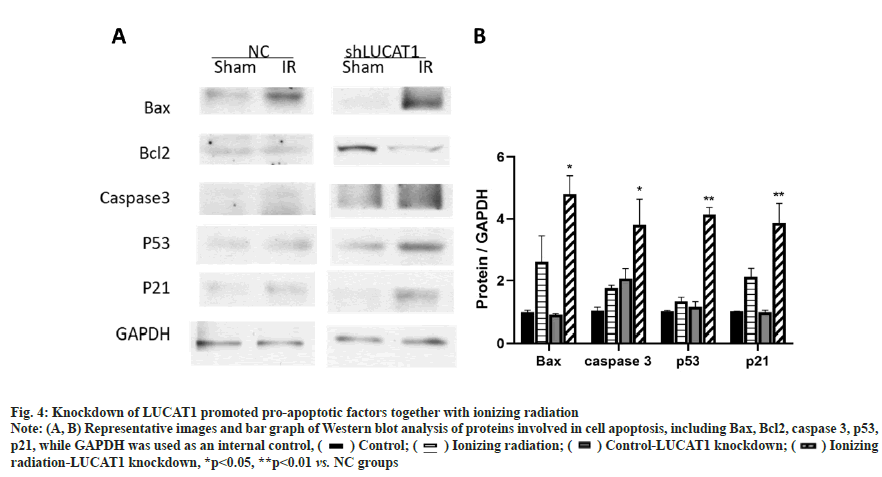
Knockdown of LUCAT1 promoted pro-apoptotic factors together with ionizing radiations as shown in fig. 4. The above data showed that LUCAT1 was related to cell killing which resulted from radiotherapy. We checked the expression of apoptosis related changes and found that LUCAT1 downregulated cells. Apoptosis promoted molecules such as Bax and caspase 3, which was increased when LUCAT1 was down-regulated as shown in fig. 4A and fig. 4B. This data proved that the apoptosis signaling pathway was further activated in LUCAT1 knockdown cells compared with normal control groups.
Fig. 4: Knockdown of LUCAT1 promoted pro-apoptotic factors together with ionizing radiation
Note: (A, B) Representative images and bar graph of Western blot analysis of proteins involved in cell apoptosis, including Bax, Bcl2, caspase 3, p53, p21, while GAPDH was used as an internal control, ( ) Control; (
) Control; ( ) Ionizing radiation; (
) Ionizing radiation; ( ) Control-LUCAT1 knockdown; (
) Control-LUCAT1 knockdown; ( ) Ionizing radiation-LUCAT1 knockdown, *p<0.05, **p<0.01 vs. NC groups
) Ionizing radiation-LUCAT1 knockdown, *p<0.05, **p<0.01 vs. NC groups
LUCAT1 interacts with miR-199a-5p to regulate apoptosis after irradiation was explained in fig. 5. The lncRNA often function via sponging with miRNAs to reverse the inhibitory effects of miRNA on its target genes. Through a bio-informatics tool (miRBase database), we predicted that miR-199a-5p as a potential target of LUCAT1 (fig. 5A). Further, we used a luciferase assay to check the binding property of LUCAT1 and miR-199a-5p. We found that the luciferase activity reduced significantly in miR-199a-5p transfected group, while not in the LUCAT1 mutant groups (fig. 5B and fig. 5C). We also found that miR-199a-5p expression was negatively correlated with LUCAT1 expression in patient samples (fig. 5D). To determine whether the miR- 199a-5p is related to the radioresistance-promoting effect of LUCAT1, we transfected the miR-199a-5p inhibitor into LUCAT1 knockdown cells and then determined cell survival and apoptosis. Our data showed that miR-199a-5p inhibitor reversed the radio sensitizing effects of LUCAT1 on lung cancer cells (fig. 5E and fig. 5F).
Fig. 5: LUCAT1 interacts with miR-199a-5p to regulate apoptosis after irradiation
Note: (A) Bioinformatics prediction of miR-199a-5p to interact with LUCAT1; (B, C) Luciferase activity assay of miR-199a-5p binding with wild type LUCAT1 instead of mutant LUCAT1; (D) Expression of miR-199a-5p is negatively correlated with LUCAT1; (E) Cell survival was measured with colony formation assay in irradiated cells transfected with LUCAT1 shRNA and/or miR-199a-5p inhibitor; (F) Cell apoptosis was detected with
flow cytometry assay in cells transfected with LUCAT1 shRNA and/or miR-199a-5p inhibitor, ( ) DMSO; (
) DMSO; ( ) Ionizing radiation, **p<0.01 vs. NC groups
) Ionizing radiation, **p<0.01 vs. NC groups
In the present study, we demonstrated that lncRNA LUCAT1 promote cell proliferation and radiation resistance through sponging miR-199a-5p and knockdown of LCUAT1 reduced the cell viability while promoting cell death under radiation treatment. We also found that LUCAT1 was related to patient survival in lung cancer patients and negatively correlated with miR-199a-5p level. Our findings provide novel potential target for lung cancer therapy for adjuvant radiotherapy.
LUCAT1 had been found to be up-regulated in several types of cancer, including breast cancer, esophageal squamous cell carcinoma, glioma and colorectal cancer[18,20-22]. And the present findings suggest a oncogene role of LUCAT1 because its cancer promotion property[23]. In our study, we found that LUCAT1 was significantly upregulated in lung cancer cells as well as tissues and LUCAT1 was also correlated with survival in lung cancer patients. Furthermore, the possible role of LUCAT1 in cancer resistance was investigated. It has been shown in literature that knockdown of LUCAT1 inhibits cell proliferation[17]. In our model, we found that knockdown of LUCAT1 together with ionizing radiation (radiotherapy) produced an additional effect of cell killing, less proliferation while more apoptosis was observed in LUCAT1 knockdown cells. Apoptosis signaling pathway was also affected by LUCAT1 knockdown, which was consistent with the phenotype. These data showed that LUCAT1 is a possible target for cancer treatment.
The lncRNA regulated biological processes mostly through a competing endogenous RNA (ceRNA) mechanism, in which miRNA was often found to be bound[24-26]. It has been proved that LUCAT1 can be sponged with several miRNAs in different disease models. For instance, LUCAT1 regulates methotrexate resistance in osteosarcoma via miR- 200c/ATP Binding Cassette subfamily B member 1 ABCB1 axis[27]. LUCAT1 bind with miR-5582-3p and regulates stemness of cancer stem cells[22]. In glioma, LUCAT1 sponge with miR-375 and promote cell survival and metastasis[17]. In this study, we identified miR-199a-5p as a potential target through bioinformatic prediction, which was further validated in luciferase assay. Surprisingly, miR-199a-5p was found to be a cancer suppressive factor in lung cancer and other types of cancer[28,29]. These data showed novel mechanism of LUCAT1 in lung cancer.
In conclusion, our findings identified a novel lncRNA LUCAT1 which was related to lung cancer survival and the therapeutic effects of radiotherapy. Knockdown of LUCAT1 inhibited cell proliferation and increased cell killing effects combined with radiotherapy. We demonstrated that LUCAT1-miR- 199a-5p as a potential signaling pathway contributing to radiation resistance and cancer malignancy. Intervention of LUCAT1-miR-199a-5p was also critical for the improvement of cancer therapy.
Acknowledgements:
We appreciate for the help of Genepharma Co. (Shanghai, China) to construct the plasmids in this study.
Conflict of interests:
The authors have no conflict of interest to disclose.
References
- Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: Orchestrating the epithelial-mesenchymal plasticity. Oncotarget 2017;8(7):12472.
[Crossref] [Google scholar] [PubMed]
- Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta Gene Regul Mech 2014;1839(11):1097-109.
[Crossref] [Google scholar] [PubMed]
- Huang C, Liu S, Wang H, Zhang Z, Yang Q, Gao F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res 2016;8(11):5025-34.
[Google scholar] [PubMed]
- Zhang K, Luo Z, Zhang YI, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark 2016;17(2):187-94.
[Crossref] [Google scholar] [PubMed]
- Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:1-3.
[Crossref] [Google scholar] [PubMed]
- Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016;35(21):2746-55.
[Crossref] [Google scholar] [PubMed]
- Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 2013;8(5):e65309.
[Crossref] [Google scholar] [PubMed]
- Bunn Jr PA. Worldwide overview of the current status of lung cancer diagnosis and treatment. Arch Pathol Lab Med 2012;136(12):1478-81.
[Crossref] [Google scholar] [PubMed]
- Rahman SJ, Gonzalez AL, Li M, Seeley EH, Zimmerman LJ, Zhang XJ, et al. Lung cancer diagnosis from proteomic analysis of pre invasive lesions. Cancer Res 2011;71(8):3009-17.
[Crossref] [Google scholar] [PubMed]
- Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis and therapy. Curr Cancer Drug Targets 2009;9(4):572-94.
[Crossref] [Google scholar] [PubMed]
- Bergsagel DE, Jenkin RD, Pringle JF, White DM, Fetterly JC, Klaassen DJ, et al. Lung cancer: Clinical trial of radiotherapy alone vs. radiotherapy plus cyclophosphamide. Cancer 1972;30(3):621-7.
- Koh MS, Tee A, Wong P, Antippa P, Irving LB. Advances in lung cancer diagnosis and staging: Endobronchial ultrasound. Intern Med J 2008;38(2):85-9.
[Crossref] [Google scholar] [PubMed]
- Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med 2015;8(7):11824-30.
[Google scholar] [PubMed]
- Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget 2017;8(11):17785.
[Crossref] [Google scholar] [PubMed]
- Sun Y, Jin SD, Zhu Q, Han L, Feng J, Lu XY, et al. Long non-coding RNA LUCAT1 is associated with poor prognosis in human non-small cell lung cancer and regulates cell proliferation via epigenetically repressing p21 and p57 expression. Oncotarget 2017;8(17):28297-311.
[Crossref] [Google scholar] [PubMed]
- Gao YS, Liu XZ, Zhang YG, Liu XJ, Li LZ. Knockdown of long noncoding RNA LUCAT1 inhibits cell viability and invasion by regulating miR-375 in glioma. Oncol Res 2018;26(2):307.
[Crossref] [Google scholar] [PubMed]
- Yoon JH, You BH, Park CH, Kim YJ, Nam JW, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett 2018;417:47-57.
[Crossref] [Google scholar] [PubMed]
- Zheng Z, Zhao F, Zhu D, Han J, Chen H, Cai Y, et al. Long non-coding RNA LUCAT1 promotes proliferation and invasion in clear cell renal cell carcinoma through AKT/GSK-3β signaling pathway. Cell Physiol Biochem 2018;48(3):891-904.
[Crossref] [Google scholar] [PubMed]
- Chen Y, Yu X, Xu Y, Shen H. Identification of dysregulated lncRNAs profiling and metastasis?associated lncRNAs in colorectal cancer by genome?wide analysis. Cancer Med 2017;6(10):2321-30.
[Crossref] [Google scholar] [PubMed]
- Xiao H, Bao L, Xiao W, Ruan H, Song Z, Qu Y, et al. Long non-coding RNA Lucat1 is a poor prognostic factor and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Oncotarget 2017;8(69):113622-34.
[Crossref] [Google scholar] [PubMed]
- Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, et al. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J Exp Clin Cancer Res 2019;38(1):1-4.
[Crossref] [Google scholar] [PubMed]
- Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun Y, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40?MDM 2?p53 pathway through binding with UBA 52. Cancer Sci 2019;110(4):1194-207.
[Crossref] [Google scholar] [PubMed]
- Xie J, Guo B, Ding Z, Kang J, Deng X, Wu B, et al. Microarray analysis of lncRNAs and mRNAs co-expression network and lncRNA function as ceRNA in papillary thyroid carcinoma. J Biomater Tissue Eng 2015;5(11):872-80.
- Li R, Yang YE, Jin J, Zhang MY, Liu X, Liu XX, et al. Identification of lncRNA biomarkers in lung squamous cell carcinoma using comprehensive analysis of lncRNA mediated ceRNA network. Artif Cells Nanomed Biotechnol 2019;47(1):3246-58.
[Crossref] [Google scholar] [PubMed]
- Song X, Cao G, Jing L, Lin S, Wang X, Zhang J, et al. Analysing the relationship between lncRNA and protein?coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med 2014;18(6):991-1003.
[Crossref] [Google scholar] [PubMed]
- Han Z, Shi L. Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem Biophys Res Commun 2018;495(1):947-53.
[Crossref] [Google scholar] [PubMed]
- Ahmadi A, Khansarinejad B, Hosseinkhani S, Ghanei M, Mowla SJ. miR-199a-5p and miR-495 target GRP78 within UPR pathway of lung cancer. Gene 2017;620:15-22.
[Crossref] [Google scholar] [PubMed]
- Li W, Wang H, Zhang J, Zhai L, Chen W, Zhao C. miR?199a?5p regulates β1 integrin through Ets?1 to suppress invasion in breast cancer. Cancer Sci 2016;107(7):916-23.
[Crossref] [Google scholar] [PubMed]