- *Corresponding Author:
- Qinying Feng
Central Laboratory, Beijing Jishuitan Hospital Guizhou Hospital, 1Department of Clinical Examination, Guiyang Nanming District Maternal and Child Health Hospital, Guiyang, Guizhou, 550000, China
E-mail: fqy13985563892@163.com
| Date of Received | 29 December 2023 |
| Date of Revision | 07 August 2024 |
| Date of Acceptance | 03 September 2024 |
| Indian J Pharm Sci 2024;86(5):1611-1620 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study aims to uncover the regulatory mechanism behind the up-regulation of dual-specificity phosphatase 28 in hepatocellular carcinoma. The differential expression and survival analyses of studied genes were carried out by the Encyclopedia of Ribonucleic Acid Interactomes platform. The GeneMANIA and biological general repository for interaction datasets were applied to obtain the regulatory network of dual-specificity phosphatase 28. Western blot analyzed the protein expression levels of dual-specificity phosphatase 28 and Josephin domain containing 2 in clinical samples collected from hepatocellular carcinoma individuals. To study the functions of dual-specificity phosphatase 28 and Josephin domain containing 2, transfection was performed in hepatocellular carcinoma cells, of which efficiency was determined by quantitative real-time polymerase chain reaction and Western blot. Cell proliferation was evaluated by colony formation and ethynyl deoxyuridine assays. Both dual-specificity phosphatase 28 and Josephin domain containing 2 were found to be highly expressed in hepatocellular carcinoma. The overexpression of dual-specificity phosphatase 28 exerts a promotion role in the cell proliferation and migration of human hepatocellular carcinoma cells, whereas the deletion of dual-specificity phosphatase 28 exhibited the opposite role. Bioinformatics tools found that Josephin domain containing 2 is an interacted gene of dual-specificity phosphatase 28, which was verified by co-immunoprecipitation. Josephin domain containing 2 regulates dual-specificity phosphatase 28 stability through deubiquitination. Furthermore, overexpressing dual-specificity phosphatase 28 reversed both the proliferation and migration of hepatocellular carcinoma cells inhibited by Josephin domain containing 2 silencing. Josephin domain containing 2 interacts with and deubiquitinates dual-specificity phosphatase 28 to improve its protein stability, thereby promoting the proliferation and migration of hepatocellular carcinoma cells.
Keywords
Hepatocellular carcinoma, dual-specificity phosphatase 28, Josephin domain containing 2, ubiquitination, Western blot
Liver cancer worldwide, of which incidence rates consistently increased in the past decades[1]. Despite various research projects suggesting that the incidence and mortality rates of liver cancer are expected to decrease by 2030 in China, the liver cancer burden in the country remains serious[2,3]. Approximately 90 % of all liver cancer cases are diagnosed as Hepatocellular Carcinoma (HCC). Although advances have been made in the prevention and treatment of HCC in the last few decades, the prognosis and over survival of HCC individuals are still poor. Currently, have little curative effect on HCC individuals[4]. Sorafenib, the first and only systemic agent approved by the Food and Drug Administration (FDA) for the treatment of advanced HCC, has been shown to prolong patient survival by only a few months[5]. Therefore, it’s urgent to exploit new effective therapies for HCC. Elucidating the underlying mechanism behind is critical in developing novel therapeutics for HCC.
Dual-Specificity Phosphatases (DUSPs) represent a group of heterogeneous protein phosphatases that can dephosphorylate both phospho-Tyr and phospho-Ser residues, resulting in conformational changes in proteins[6]. DUSPs are primarily classified into typical and atypical DUSPs (aDUSPs). Typical DUSPs can specifically inactivate Mitogen-Activated Protein Kinases (MAPKs), which are also named as MAPK Phosphatases (MKPs). Given the critical role of MAPKs in various pathological conditions, MKPs are emerging as attractive targets for drug development in several diseases, including diabetes[7], depression[8], and cancers[9]. According to previous studies, the substrate specificities and physiological roles of aDUSPs are greatly different from MKPs[10,11]. Several aDUSPs have been proven to be closely related to the oncogenesis or malignant progression in diverse cancers, including HCC. A previous study indicated that DUSP26 could inhibit p53 to prevent Doxorubicin-induced cell death in neuroblastoma. Recently, Jacques et al.[12] indicated the crucial role of DUSP3 in the initiation and progression of HCC by using a mouse DUSP3 knockout model. Wang et al.[13], the overexpression of DUSP28 contributed to HCC progression by promoting the proliferation and colony formation of HCC cells. This finding suggested the possibility of DUSP28 as a promising target for anti-HCC therapy. This research also indicated that the expression of DUSP28 was over-activated in HCC tissues and cell lines compared with the normal liver[13]. However, the underlying mechanism involving the up-regulation of DUSP28 in HCC is little known. Herein, we confirmed that is up-regulated in HCC based on the Encyclopedia of Ribonucleic Acid (RNA) Interactomes (ENCORI) database and the analysis of HCC clinical samples. The effect of DUSP28 in facilitating the proliferation and migration of HCC cells was proven by overexpressing or silencing DUSP28. Then, we found that Josephin Domain Containing 2 (JOSD2), a gene-targeted DUSP28 predicted by 2 online databases, was highly expressed in HCC samples compared with normal liver, which could regulate to enhance the proliferation and migration of HCC cells.
Materials and Methods
Reagents and antibodies:
Dulbecco’s Modified Eagle’s Medium (DMEM), Fetal Bovine Serum (FBS), penicillin-streptomycin, and 0.25 % trypsin were purchased from Gibco (New York, United States of America (USA)). The transfection reagent Lipofectamine® 2000 (#11668019) was purchased from Invitrogen (California, USA). GenElute™ total RNA purification kit (#RNB100), first strand copy Deoxyribonucleic Acid (cDNA) synthesis kit (#GE27-9261-01), and SYBR® green quantitative Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) kit (#QR0100) were obtained from Sigma-Aldrich (Missouri, USA). Ethynyl deoxyuridine (EdU) staining proliferation kit (#ab219801), anti-Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) antibody (#ab8245), goat anti-human IgG Fc Horseradish Peroxidase (HRP) preadsorbed (#ab98624), and Enhanced Chemiluminescence (ECL) Western blotting substrate kit (#ab65623) were supplied by Abcam (Massachusetts, USA). Enhanced Bicinchoninic Acid (BCA) protein assay kit (#P0009), Western and Immunoprecipitation (IP) lysis buffer (#P0013) were purchased from Beyotime (Jiangsu, China). The information on antibodies used in this study was listed in Table 1.
| Antibodies | Manufacturer | Cat. # |
|---|---|---|
| DUSP28 | MyBioSource | MBS832311 |
| JOSD2 | MyBioSource | MBS9202885 |
| GAPDH | Beyotime | AF0006 |
| Flag tag | Beyotime | AF0036 |
| HA tag | Beyotime | AF2858-50 μl |
| OTUB2 | MyBioSource | MBS9204470 |
| Goat anti-mouse IgG | Beyotime | A0216 |
| Goat anti-rabbit IgG | Beyotime | A0208 |
Table 1: The Antibodies used in this Study
Patient samples collection:
Clinical cancerous and non-cancerous liver samples were collected from the resected tissues of three HCC individuals, and stored at -80° before use.This research obtained written informed consent from all participants and was reviewed and approved by the Ethics Committee of Beijing Jishuitan Hospital Guizhou Hospital.
Bioinformatics online tools:
The pan-cancer analysis platform of ENCORI (http://starbase.sysu.edu.cn/panCancer.php)[14] was applied to perform the survival and differential expression analysis of studied genes in HCC. The Human Protein Atlas (HPA) (https://www.proteinatlas.org) provided the protein expression of DUSP28 in HCC and normal liver tissues. The GeneMANIA (http://genemania.org)[15] and Biological General Repository for Interaction Datasets (BioGRID) (https://thebiogrid.org)[16] were used to predict genes that potentially interacted with DUSP28.
Cell lines and culture:
Human HCC cell lines SNU449 cells and SNU423 cells obtained from ATCC (Virginia, USA) were cultured in DMEM containing 10 % FBS and 1 % penicillin-streptomycin in a humidified environment of 5 % CO2 at 37°. After cell confluency reached 80 %-90 %, cultured cells were digested with 0.25 % trypsin, followed by subcultured at a ratio of 1:3.
Cell transfection:
The cells with good growth were divided into diverse groups to receive different transfections: a sh-NC group with cells transfected with irrelevant nucleotides to act as a negative control; a sh-DUSP28-1 group with cells transfected with shRNA#1 specifically targeting DUSP28; a sh-DUSP28-2 group with cells transfected with shRNA#2 specifically targeting DUSP28; a vector group with cells transfected with an empty vector; a DUSP28 group with cells transfected with lentivirus vectors for DUSP28 overexpression; a JOSD2 group with cells transfected with lentivirus vectors for JOSD2 overexpression; a sh-JOSD2-1 group with cells transfected with shRNA#1 specifically targeting JOSD2; a sh-JOSD2-2 group with cells transfected with shRNA#2 specifically targeting JOSD2, and a sh-JOSD2-1+DUSP28 group with cells co-transfected with sh-JOSD2-1 and DUSP28 overexpression.
The transfection procedure was conducted as previously described by Cui et al.[17]. In brief, both SNU449 and SNU423 cells were transfected using Lipofectamine® 2000 according to the manufacturer’s instructions, and then harvested 48 hours post-transfection for various experiments. The sequences of shRNAs used in this study were listed in Table 2.
| Groups | Directionality | Sequence |
|---|---|---|
| sh-DUSP28-1 | 5’-3’ | CCGGATTAGATGTTGCTATAT |
| sh-DUSP28-2 | 5’-3’ | TGGTCTCAGCTCCAGAAGTAT |
| sh-JOSD2-1 | 5’-3’ | CGATGAGATCTGCAAGAGGTT |
| sh-JOSD2-2 | 5’-3’ | CAACTATGATGTCAATGTGAT |
Table 2: Sequences of shRNA against Specific Targets
Colony formation assay:
The transfected cells were seeded into 6-well plates (Corning, New York, USA) at a density of 2.5×102 cells and subsequently cultured in a 5 % CO2 incubator at 37°. After cultivation for 2 w, cell colonies were fixed with 4 % Paraformaldehyde (PFA), followed by staining with 1 % crystal violet for 10 min. The dishes were gently washed with distilled water, photographed, and counted under a BX51 microscope (Olympus, Tokyo, Japan).
EdU assay:
According to protocols provided by manufacturer, EdU staining proliferation kit was utilized to assess the proliferation of transfected cells in different groups. By using a BX51 microscope, images were obtained to observe and calculate EdU-positive cells.
Wound healing assay:
A density of 2×105 transfected cells/well was seeded into 6-well plates and cultured until a monolayer of cells had formed. The 200 μl pipette tip was used to create a similar size of scratches in the cell layer for each group. Then, scratched cells were removed by gently rinsing Phosphate Buffered Saline (PBS) thrice; the remaining cells were cultured in DMEM in a 5 % CO2 incubator at 37° for 24 h. A BX51 microscope was utilized to photograph the same position of scratches at 0 and 24 h after scratching.
Transwell migration assay:
The migration of transfected cells was also evaluated by using transwell chambers (Corning, New York, USA). Briefly, transfected cells were seeded into the upper chambers containing 200 μl DMEM. Simultaneously, 700 μl DMEM with serum was added to the lower chamber. After incubation for 24 h, cells still in the upper chamber were wiped using a cotton swab, while cells traversing the membranes to the lower chamber were fixed in 4 % PFA and stained with 0.1 % crystal violet for 15 min. The stained cells were imaged and counted in 5 random visual fields under a BX51 microscope.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) analysis:
The extraction of total RNA and subsequent reverse transcription was conducted in line with standard protocols. Afterward, qRT-PCR analysis was performed with SYBR green reagent on the 7500 RT-PCR system. This study used GAPDH as an internal reference gene to quantitate messenger RNA (mRNA) expression of DUSP28 based on the 2−ΔΔCt method[18]. Primer sequences were presented as follows: DUSP28 (forward: 5’-GCGCCTGCCTAGTCTACTG-3’, reverse: 5’-CGGGTTCGGTTCTGCTACC-3’) and GAPDH (forward: 5’- GGAGCGAGATCCCTCCAAAAT-3’, reverse: 5’- GGCTGTTGTCATACTTCTCATGG-3’).
Western Blot (WB) analysis:
Transfected cells were lysed with Radioimmunoprecipitation Assay (RIPA) buffer on ice and for 20 min at 4° to acquire the total protein. After determining the concentration of total protein, equivalent amounts of protein were separated on 10 % Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDSPAGE) gels and subsequently transferred onto Poly(Vinylidene Fluoride) (PVDF) membranes. Membranes were blocked with skimmed milk, followed by incubated with primary antibodies. Next, the membranes were rinsed thrice with Tuberculosis antigen-Based Skin Tests (TBST) prior to incubating with a secondary antibody. Finally, by using an ECL kit, the protein bands were visualized, of which intensities were measured by Image J 6.0 software.
Co-IP:
Transfected cells were lysed and incubated with bead-conjugated FLAG® or the specific antibody with gentle rotation overnight at 4°, followed by incubation with incubated with antibodyconjugated beads for 2 h. Then, beads coupling immuno-complexes were washed four times with lysis buffer before eluting precipitated proteins by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) buffer. Thereafter, eluted proteins were separated by SDS-PAGE gels. The interacting proteins were detected by WB analysis.
Cycloheximide (CHX) half-life assay:
After transfection post 48 h, CHX, (20 μg/ml) was added to the cell medium. At the designed time points (0 h, 2 h, 4 h and 6 h), cells were collected and lysed to detect the protein levels by WB analysis.
Statistical analysis:
All statistical analysis was conducted on GraphPad Prism 8.0.1 software. All experiments were repeated at least 3 times, and all data were presented as mean±Standard Deviation (SD). Students’ test or one-way Analysis of Variance (ANOVA) with post-hoc test (Bonferroni) was performed to analyze the difference among the groups in the present study. It is considered to be statistically different if p<0.05.
Results and Discussion
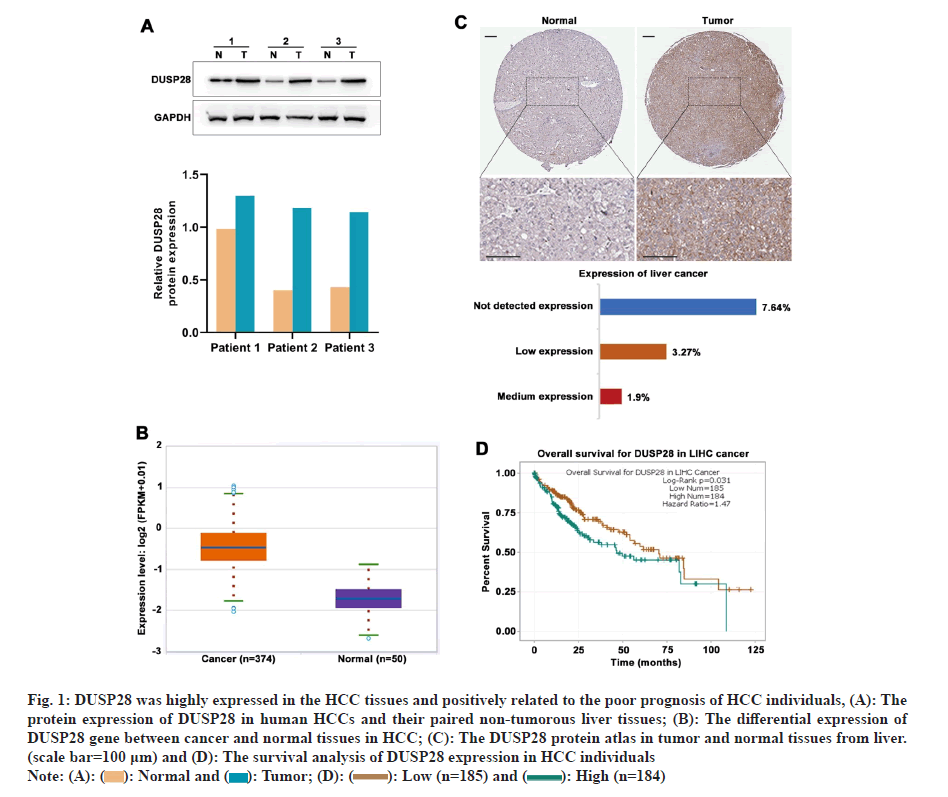
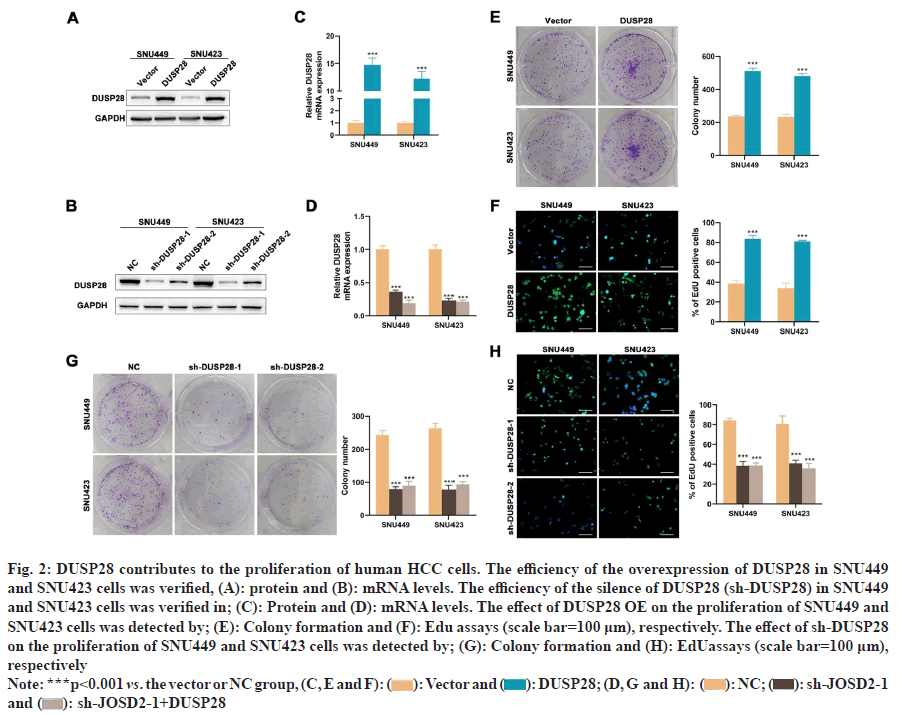
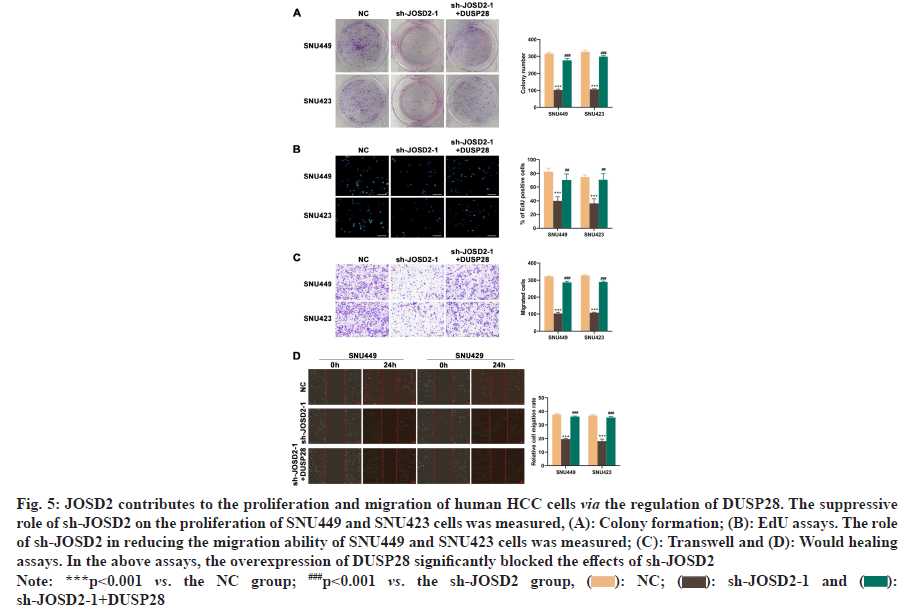
To understand the expression profile of DUSP28 in HCC, WB analysis was performed to determine the expression levels of DUSP28 in 3 pairs of cancerous and non-cancerous liver samples. As depicted in fig. 1A, the expression of DUSP28 in HCC tissues was significantly higher than that in non-cancerous tissues. A similar result was found in the data downloaded from HPA, showing that the protein expression of DUSP28 was generally up-regulated in HCC tissues compared with normal tissues. Based on the ENCORI database, we found that the mRNA expression of DUSP28 in HCC tissues was also significantly increased in comparison of normal tissues (fig. 1B and fig. 1C). Additionally, survival analysis showed that the Over-Survival (OS) of HCC individuals with the low expression of DUSP28 was obviously longer than that with the high expression of DUSP28, revealing that the expression of DUSP28 was positively associated with the poor prognosis of HCC individuals (fig. 1D). These data suggested that the high expression of DUSP28 is related to the development of HCC. To study the roles of DUSP28 on the proliferation of human HCC cells, colony formation and EdU assays were performed in SNU449 and SNU423 cells after overexpressing or silencing DUSP28 by transfection. As shown in fig. 2A and fig. 2B, both WB and qRT-PCR results confirmed the efficiency of overexpressing DUSP28 in SNU449 and SNU423 cell lines. Simultaneously, we have also confirmed the silencing efficiency of sh- DUSP28-1 and sh-DUSP28-2 (fig. 2C and fig. 2D). According to colony formation and EdU assays, we found that the proliferation of SNU449 and SNU423 cells was significantly enhanced after overexpressing DUSP28 (fig. 2E and fig. 2F). Conversely, silencing DUSP28 expression could observably reduce the proliferation of SNU449 and SNU423 cells (fig. 2G and fig. 2H). Our results demonstrated that DUSP28 is capable of promoting the proliferation of human HCC cells.
Fig 1: DUSP28 was highly expressed in the HCC tissues and positively related to the poor prognosis of HCC individuals, (A): The
protein expression of DUSP28 in human HCCs and their paired non-tumorous liver tissues; (B): The differential expression of
DUSP28 gene between cancer and normal tissues in HCC; (C): The DUSP28 protein atlas in tumor and normal tissues from liver.
(scale bar=100 μm) and (D): The survival analysis of DUSP28 expression in HCC individuals 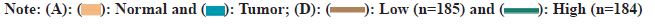
Fig 2: DUSP28 contributes to the proliferation of human HCC cells. The efficiency of the overexpression of DUSP28 in SNU449
and SNU423 cells was verified, (A): protein and (B): mRNA levels. The efficiency of the silence of DUSP28 (sh-DUSP28) in SNU449
and SNU423 cells was verified in; (C): Protein and (D): mRNA levels. The effect of DUSP28 OE on the proliferation of SNU449 and
SNU423 cells was detected by; (E): Colony formation and (F): Edu assays (scale bar=100 μm), respectively. The effect of sh-DUSP28
on the proliferation of SNU449 and SNU423 cells was detected by; (G): Colony formation and (H): EdUassays (scale bar=100 μm),
respectively 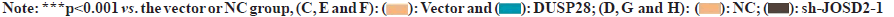
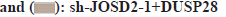
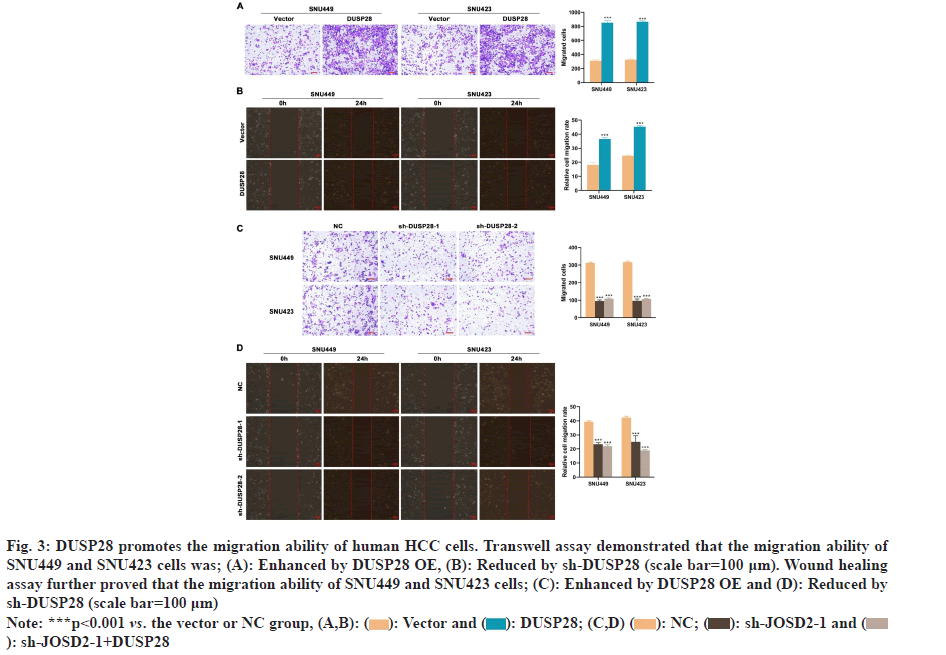
Fig 3: DUSP28 promotes the migration ability of human HCC cells. Transwell assay demonstrated that the migration ability of
SNU449 and SNU423 cells was; (A): Enhanced by DUSP28 OE, (B): Reduced by sh-DUSP28 (scale bar=100 μm). Wound healing
assay further proved that the migration ability of SNU449 and SNU423 cells; (C): Enhanced by DUSP28 OE and (D): Reduced by
sh-DUSP28 (scale bar=100 μm)
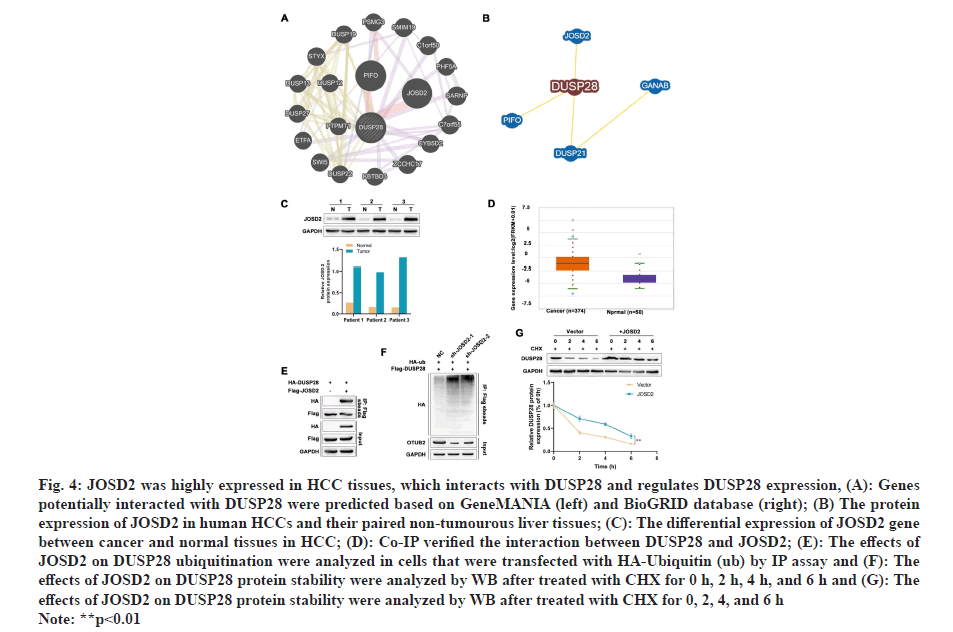
Next, the effects of DUSP28 on the migration of human HCC cells were also explored in our study. In the transwell assay, we found that the migration ability of SNU449 and SNU423 cells could be partly strengthened by overexpressing DUSP28 (fig. 3A), whereas weakened by silencing DUSP28 (fig. 3B). The results of the wound healing assay were similar to those of the transwell assay (fig. 3C and fig. 3D), which further consolidated that DUSP28 could promote the migration of human HCC cells. To find out genes involved in the up-regulation of DUSP28 in HCC, the genes had potential interaction with DUSP28 were predicted based on GeneMANIA and BioGRID database, of which results were shown in fig. 4A. After intersecting the results from these two databases, we identified two genes, JOSD2 and Primary cilia Formation Protein (PIFO), as the most likely candidates to interact with DUSP28. Since JOSD2 encodes a Josephin domain-containing protein involved in deubiquitination, we selected JOSD2 as our focus and explored its regulatory role in the expression of DUSP28. The protein and mRNA expression profiles of JOSD2 in HCC were investigated based on clinical samples and the ENCORI database, respectively. As shown in fig. 4B and fig. 4C, we found a similar tendency of JOSD2 expression for mRNA and protein levels to DUSP28 expression, which let us speculate that JOSD2 plays a positive regulation on the expression of DUSP28. Next, Co- IP assay was performed to verify whether there is a protein interaction between JOSD2 and DUSP28. The result showed that the DUSP28 expression in the JOSD2 precipitates was enhanced after overexpressing JOSD2 (fig. 4D), indicating that DUSP28 protein interacts with JOSD2. Then, using IP assay, we further observed the effect of JOSD2 on the ubiquitination of DUSP28 protein. After silencing JOSD2 expression, the ubiquitin levels in DUSP28 precipitates were dramatically increased compared with the control (fig. 4E). Furthermore, the CHX half-life assay showed that the overexpression of JOSD2 significantly reduced the degradation of DUSP28 protein induced by CHX, which suggested that JOSD2 promotes the stability of DUSP28 protein (fig. 4F and fig. 4G). To clarify the specific role of JOSD2 in HCC, the proliferation and migration ability of sh-JOSD2 transfected cells were evaluated. In the meantime, the cells were also co-transfected with sh-JOSD2 and DUSP28 OE to further determine whether the effect of JOSD2 on HCC progression and metastasis via the up-regulation of DUSP28. As shown in fig. 5A and fig. 5B, the proliferation of both SNU449 and SNU423 cells were significantly reduced by silencing of JOSD2 in comparison of the control. However, the suppressive role of sh- JOSD2 in SNU449 and SNU423 cells was countered by the overexpression of DUSP28 (fig. 5A and fig. 5B). Both the Transwell and wound healing assays demonstrated that the deletion of JOSD2 significantly decreased the migration ability of human HCC cells, while the overexpression of DUSP28 rescued this phenotype (fig. 5C and fig. 5D). These results hinted that knocking down JOSD2 suppresses the proliferation and migration of human HCC cells by reducing the expression of DUSP28.
Fig 4: JOSD2 was highly expressed in HCC tissues, which interacts with DUSP28 and regulates DUSP28 expression, (A): Genes
potentially interacted with DUSP28 were predicted based on GeneMANIA (left) and BioGRID database (right); (B) The protein
expression of JOSD2 in human HCCs and their paired non-tumourous liver tissues; (C): The differential expression of JOSD2 gene
between cancer and normal tissues in HCC; (D): Co-IP verified the interaction between DUSP28 and JOSD2; (E): The effects of
JOSD2 on DUSP28 ubiquitination were analyzed in cells that were transfected with HA-Ubiquitin (ub) by IP assay and (F): The
effects of JOSD2 on DUSP28 protein stability were analyzed by WB after treated with CHX for 0 h, 2 h, 4 h, and 6 h and (G): The
effects of JOSD2 on DUSP28 protein stability were analyzed by WB after treated with CHX for 0, 2, 4, and 6 h
Note: **p<0.01
Fig 5: JOSD2 contributes to the proliferation and migration of human HCC cells via the regulation of DUSP28. The suppressive
role of sh-JOSD2 on the proliferation of SNU449 and SNU423 cells was measured, (A): Colony formation; (B): EdU assays. The role
of sh-JOSD2 in reducing the migration ability of SNU449 and SNU423 cells was measured; (C): Transwell and (D): Would healing
assays. In the above assays, the overexpression of DUSP28 significantly blocked the effects of sh-JOSD2
Note: ***p<0.001 vs. the NC group; ###p<0.001 vs. the sh-JOSD2 group, sh-JOSD2-1+DUSP28
sh-JOSD2-1+DUSP28
The research of the physiological roles of DUSPs in cancers has been increasing over a few decades. The overexpression of DUSP1 has been observed in diverse cancers, which enhances the dephosphorylation of Extracellular signal Regulated Kinase (ERK) and c-JUN N-terminal Kinase (JNK) to exert a carcinogenesis role in several malignancies[19]. DUSP2 could be as a tumor suppressor, of which down-regulation induced by hypoxia is crucial for the promotion of cancer stemness in colorectal cancer[20]. Several publications in pancreatic cancer revealed that DUSP28 is closely related to the malignant behaviors of tumor[21,22]. Besides, DUSP28 is a potential HCC-related gene selected by a genomewide approach[13]. However, its roles and the related regulatory mechanism in HCC progression remain elusive. Our study demonstrated that the promotion role of DUSP28 in HCC progression and metastasis is modulated by the deubiquitination effect of JOSD.
In clinical samples collected from HCC individuals, we found that DUSP28 expression levels in cancerous tissues were obviously higher than their adjacent normal samples, which was corroborated by the differential expression analysis in the ENCORI database. Infinite proliferation and metastasis are accounting for the tumor developing into malignancy. Therefore, we performed a series of in vitro experiments, which included colony formation assay, EdU assay, Transwell assay, as well as wound healing assay to study the functions of DUSP28 on human HCC cells. Consistent with a previous study[13], our results showed that the overexpression of DUSP28 promoted the proliferation of human HCC cells whereas the deletion of DUSP28 led to the converse effects. In addition, our study also revealed that the expression of DUSP28 was positively associated with the migration ability of HCC cells. Besides, in the ENCORI database, the expression of DUSP28 showed a positive correlation with the overall survival of HCC individuals. All the above data imply that DUSP28 is a tumor promoter in HCC. Then, we further explored the related regulatory mechanism of DUSP28 in HCC. By using bioinformatics tools, we predicted a potential interacted gene of DUSP28, JOSD2, for further analysis. Both the ENCORI database and clinical samples revealed that the expression levels of JOSD2 were increased in HCC when compared with the normal. JOSD2 is a deubiquitinase containing Josephin domain, which probably functions through deubiquitylating target substrates. A growing body of studies documented that deubiquitinases exert critical functions in the development of various cancers. DUSP5 promotes the growth and chemo-resistance of colorectal cancer cells[23]. In a recent study, DUSP21 played a deubiquitination role to stabilize YY1, a wellknown oncogene, leading to the promotion of cell proliferation, migration, and invasion, as well as in vivo tumor growth in non-small cell lung cancer[24]. JOSD2 has recently been considered a positive regulator of cancer cell proliferation[25]. As the relatively short half-lives (<1 h) of most DUSPs, protein levels of DUSPs are strictly modulated by post-translational modifications, including phosphorylation, ubiquitination, and methylation[26]. Accordingly, we hypothesized that JOSD2 directly reverses the ubiquitination of DUSP28 to enhance DUSP28 expression, thereby promoting the development of HCC. As expected, the interaction between DUSP28 and JOSD2 was confirmed by Co-IP. Moreover, IP and CHX assays further verified our hypothesis that JOSD2 could stabilize DUSP28 by decreasing the ubiquitination of DUSP28. To further verify whether the effect of JOSD2 on DUSP28 participates the development and metastasis of HCC, we transfected sh-JOSD2 alone or in combination with DUSP28 OE into human HCC cells. Unsurprisingly, knocking down JOSD2 suppressed not only the proliferation but also the migration of human HCC cells, which can be blocked by the overexpression of DUSP28.
In summary, our study is the first to demonstrate that DUSP28 is a substrate of JOSD2. JOSD2 interacts with and deubiquitinates DUSP28, stabilizing its protein expression and thereby promoting the proliferation and migration of HCC cells. Further exploration of the precise mechanism of JOSD2/DUSP28 in driving HCC development is required. Moreover, the present study lacks in vivo experiment, which might reduce the credibility of our findings in this study. Therefore, a further investigation involving in vivo studies would be our next research direction. Our study revealed that JOSD2 functions as deubiquitinates for DUSP28 to reduce its degradation, thus enhancing the proliferation and migration of human HCC cells. Such findings provide scientific information of the modulating mechanism of DUSPs by posttranslational modifications in HCC, which has further implications in the development of a novel therapeutic strategy for HCC.
Funding:
The Guizhou Science and Technology Projects (Guizhou Science and Technology Cooperation Achievement-LC[2024]002, and Guizhou Science and Technology Cooperation Achievement- LC[2024]103).
Conflict of interests:
The authors declared no conflict of interests.
References
- Dasgupta P, Henshaw C, Youlden DR, Clark PJ, Aitken JF, Baade PD. Global trends in incidence rates of primary adult liver cancers: A systematic review and meta-analysis. Front Oncol 2020;10:171.
[Crossref] [Google Scholar] [PubMed]
- Valery PC, Laversanne M, Clark PJ, Petrick JL, McGlynn KA, Bray F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology 2018;67(2):600-11.
[Crossref] [Google Scholar] [PubMed]
- Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu X, et al. Liver cancer incidence and mortality in China: temporal trends and projections to 2030. Chin J Cancer Res 2018 Dec;30(6):571.
[Crossref] [Google Scholar] [PubMed]
- Ingle PV, Samsudin SZ, Chan PQ, Ng MK, Heng LX, Yap SC, et al. Development and novel therapeutics in hepatocellular carcinoma: A review. Ther Clin Risk Manag 2016:445-55.
[Crossref] [Google Scholar] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359(4):378-90.
[Crossref] [Google Scholar] [PubMed]
- Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov 2007;6(5):391-403.
[Crossref] [Google Scholar] [PubMed]
- Wei Q, Pu X, Zhang L, Xu Y, Duan M, Wang Y. Expression of dual-specificity phosphatase 9 in placenta and its relationship with gestational diabetes mellitus. J Diabetes Res 2019;2019(1):1963178.
[Crossref] [Google Scholar] [PubMed]
- An N, Bassil K, Al Jowf GI, Steinbusch HW, Rothermel M, de Nijs L, et al. Dual-specificity phosphatases in mental and neurological disorders. Prog Neurobiol 2021;198:101906.
[Crossref] [Google Scholar] [PubMed]
- Vogt A, McDonald PR, Tamewitz A, Sikorski RP, Wipf P, Skoko JJ, et al. A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol Cancer Ther 2008;7(2):330-40.
[Crossref] [Google Scholar] [PubMed]
- Li JP, Fu YN, Chen YR, Tan TH. JNK pathway-associated phosphatase dephosphorylates focal adhesion kinase and suppresses cell migration. J Biol Chem 2010;285(8):5472-8.
[Crossref] [Google Scholar] [PubMed]
- Shang X, Vasudevan SA, Yu Y, Ge N, Ludwig AD, Wesson CL, et al. Dual-specificity phosphatase 26 is a novel p53 phosphatase and inhibits p53 tumor suppressor functions in human neuroblastoma. Oncogene 2010;29(35):4938-46.
[Crossref] [Google Scholar] [PubMed]
- Jacques S, Arjomand A, Perée H, Collins P, Mayer A, Lavergne A, et al. Dual-specificity phosphatase 3 deletion promotes obesity, non-alcoholic steatohepatitis and hepatocellular carcinoma. Sci Rep 2021;11(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Wang D, Han S, Peng R, Jiao C, Wang X, Han Z, et al. DUSP28 contributes to human hepatocellular carcinoma via regulation of the p38 MAPK signaling. Int J Oncol 2014;45(6):2596-604.
[Crossref] [Google Scholar] [PubMed]
- Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42(D1):D92-7.
[Crossref] [Google Scholar] [PubMed]
- Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46(W1):W60-4.
[Crossref] [Google Scholar] [PubMed]
- Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 2019 Jan 8;47(D1):D529-41.
[Crossref] [Google Scholar] [PubMed]
- Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut 2017;66(12):2149-59.
[Crossref] [Google Scholar] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. 2001;25(4):402-8.
[Crossref] [Google Scholar] [PubMed]
- Shen J, Zhang Y, Yu H, Shen B, Liang Y, Jin R, et al. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med 2016;5(8):2061-8.
[Crossref] [Google Scholar] [PubMed]
- Hou PC, Li YH, Lin SC, Lin SC, Lee JC, Lin BW, et al. Hypoxia-induced downregulation of DUSP-2 phosphatase drives colon cancer stemness. Cancer Res 2017;77(16):4305-16.
[Crossref] [Google Scholar] [PubMed]
- Lee J, Hun YJ, Lee J, Choi C, Hoon KJ. Blockade of dual-specificity phosphatase 28 decreases chemo-resistance and migration in human pancreatic cancer cells. Sci Rep 201527;5(1):12296.
[Crossref] [Google Scholar] [PubMed]
- Lee J, Lee J, Yun JH, Choi C, Cho S, Kim SJ, et al. Autocrine DUSP28 signaling mediates pancreatic cancer malignancy via regulation of PDGF-A. Sci Rep 2017;7(1):12760.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics 2019;9(14):4208.
[Crossref] [Google Scholar] [PubMed]
- Xu P, Xiao H, Yang Q, Hu R, Jiang L, Bi R, et al. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp Mol Med 2020;52(1):41-55.
[Crossref] [Google Scholar] [PubMed]
- Krassikova L, Zhang B, Nagarajan D, Queiroz AL, Kacal M, Samakidis E, et al. The deubiquitinase JOSD2 is a positive regulator of glucose metabolism. Cell Death Differ 2021;28(3):1091-109.
[Crossref] [Google Scholar] [PubMed]
- Huang CY, Tan TH. DUSPs, to MAP kinases and beyond. Cell Biosci 2012;2:1-0.
[Crossref] [Google Scholar] [PubMed]