- *Corresponding Author:
- J. Zheng
Department of Pathology, Taizhou Hospital, Wenzhou Medical University, Linhai 317000, China
E-mail: zhengjingmin@enzemed.com
| Date of Received | 15 June 2022 |
| Date of Revision | 19 February 2023 |
| Date of Acceptance | 21 June 2023 |
| Indian J Pharm Sci 2023;85(3):709-720 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Calponin is an actin filament-associated regulatory protein that can inhibit the activity of myosin- ATPase and stabilize the dynamics of the actin cytoskeleton. Although calponin 2 has been reported to play roles in several cancers, whether it takes part in the progression of esophageal cancer still remains unknown. To explore the pathologic significance of calponin 2 in esophageal squamous cell carcinoma, the expression level of calponin 2 proteins in the tumor tissue of 190 esophageal squamous cell carcinoma patients was examined with immunohistochemistry while the expression level of calponin 2 messenger ribonucleic acid was analyzed by using the data from The Cancer Genome Atlas database. Both the calponin 2 messenger ribonucleic acid and protein level were increasingly expressed in the tumor tissues of esophageal squamous cell carcinoma patients compared with the adjacent non-tumor tissue and correlated negatively with the tumor grade. Patients with higher calponin 2 in the tumor tissue were found to have longer overall survival time. Calponin 2 was shown to be an independent factor influencing the overall survival of the esophageal squamous cell carcinoma patients. Methylation analysis based on MethSurv database revealed a methylation site in the body of calponin 2 gene, which was associated with a better prognosis. Further, in esophageal cancer tumor tissue, calponin 2 gene was found to co-express with genes associated with tight junction and the expression level of calponin 2 was observed to correlate significantly with the number of infiltrating immune cells. These results supported the idea that calponin 2 is involved in esophageal cancer and may function as a tumor inhibitor probably through modulating cancer cells tight junction and tumor immunity.
Keywords
Clinicopathological feature, calponin 2, esophageal cancer, prognosis
Esophageal Cancer (ESCA), one of the top ten most commonly diagnosed cancers worldwide, has caused 604 000 new diagnosed cases in 2020 and the number will continue to increase in 2021[1]. ESCA cancer has two most common histologic subtypes; Esophageal Squamous Cell Carcinoma (ESCC) and esophageal adenocarcinoma[2,3]. In China, more than 90 % ESCA cases belong to ESCC[4]. Because of the difficulty of early diagnosis, ESCC is frequently found at an advanced stage[5]. Although surgical treatment with adjuvant chemo-radiotherapy, targeted therapy and immunotherapy have been used in patients with this disease, the long-term survival is still disappointing[6,7]. The poor prognosis is generally related to loco-regional recurrence, regional lymphnode invasion, and distant metastases[8,9]. However, the underlying mechanism is far from clear. Thus, further exploring the mechanism, finding new disease relative markers, and developing novel therapies to improve the prognosis of this malignancy have become an urgent need.
Calponin is a group of actin filament-associated regulatory proteins. The family includes three members, calponin 1, calponin 2, and calponin 3 which are encoded by CNN1, CNN2, and CNN3 gene respectively[10,11]. Calponin has been reported to be associated with lots of biologic functions, such as cell proliferation, adhesion, migration, cytokinesis, and so on[10,12,13].
In Oncology, calponin has been reported to be involved in the pathogenesis of some cancers[14-18]. In ESCC, in a previous protein profile analysis study, Deng et al.[19] examined the protein profiles of three ESCC patients and observed that CNN2 was differentially expressed. Except for this, no study has reported the significance of calponin in ESCC. To explore the possible pathological significance of calponin in ESCC, in this study, we first examined the expression of calponin isoforms in the tumor tissue of ESCC patients and further explored their association with the disease.
Materials and Methods
Patients:
There were two cohorts of ESCC patients included in the present study, TCGA cohort and TZYY ESCC cohort. The TCGA cohort included 80 ESCC patients, including 68 males and 12 females with a median age of 57.5 y (ranging from 36 to 90 y). The data of tumor tissue samples from 80 ESCC patients and 10 normal oesophagus samples and clinicopathological data were downloaded from The Cancer Genome Atlas (TCGA) database (TCGA-ESCA). The ESCC cohort contained 190 ESCC patients, including 116 males and 74 females with a median age of 62 y (ranging from 42 to 82 y). The patients were hospitalized at the Cardiothoracic Surgery Department of Taizhou Hospital affiliated to Wenzhou Medical University between 2003 to 2018 and were histologically diagnosed as ESCC after radicalsurgery. The cases were pathologically staged based on the 8th edition of the Tumor Nodes and Metastases (TNM) classification for esophagus cancer. We excluded the following cases with history of other malignancy or anticancer therapy before surgery; mixed histological types; developed other cancers during follow-up. The clinical and pathological data of the TZYY ESCC cohort patients were obtained from medical records and follow-up records. The Overall Survival time (OS) was defined as the time interval between surgery and death or the date of last visit in our study.
Immunohistochemistry and Masson’s trichrome staining:
The sections (2 mm thick) were deparaffinised in xylene and rehydrated with graded ethanols. After autoclaved for antigen repair, the sections were treated with 3 % hydrogen peroxide solution for 10 min to inactivate the endogenous peroxidase, blocked for 30 min in 10 % fetal calf serum, and incubated with rabbit anti-human CNN2 antibody (NBP2-13848, Novus Biologicals, CO, USA, dilution 1:1000) at 4° overnight. Then, the sections were washed with PBS, incubated with horseradish peroxidase-conjugated secondary antibody for 30 min at 37°, washed again and developed with diaminobenzidine. Finally, the sections were counterstained with haematoxylin. Hematoxylin and eosin (H&E) and Masson’s trichrome staining were performed routinely.
Two independent researchers scored the level of CNN2 in each section according to the immunostaining intensity in a blind manner. 1 weak; 2 mild; 3 medium; 4 strong. The slides with different scores obtained by the two researchers were reviewed again until the agreed scores were made. For survival analysis based on the level of CNN2 protein, patients were divided into low CNN2 protein group (patients scored as 1 and 2) and high CNN2 protein group (patients scored as 3 and 4).
DNA methylation information of CNN2:
The MethSurv database (https://biit.cs.ut.ee/ methsurv/) is a web portal that provides survival analysis based on DNA methylation biomarkers using TCGA data.DNA methylation of CNN2 at CpG sites and the prognostic value of these CpG sites in ESCA were analyzed by MethSurv.
Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genome (KEGG) analyses:
The cBioPortal database (https://www.cbioportal. org/) integrated data from multiple databases including TCGA and the International Cancer Genome Gonsorium (ICGC), providing cancer genome data, comprehensive analysis of genome data and clinical data. GO and KEGG analysis were performed on co-expressed genes with the package R "clusterProfiler" to explore possible biological functions and signaling pathways. In our study, cBioPortal database was used to obtain co-expression information of CNN2. To explore the potential molecular biological functions of CNN2, the 500 transcriptionally associated genes obtained were performed for the GO/KEGG pathway analyses.
Immune infiltration analysis of CNN2:
The Transcripts Per Million (TPM) normalized RNA-seq data of ESCA were downloaded from TCGA. Single-sample Gene Set Enrichment Analysis (ssGSEA) was analyzed with R package GSVA to assess the tumour sample’s immune infiltration level and calculate the responding immune infiltration score in ESCA. Six immune cells of which the number was highly correlated (both positive and negative) with the expression level of CNN2 were selected for Spearman’s correlation analysis.
Statistical analysis:
SPSS software (version 17, IBM, CHI, USA) was used to analyze the data. Independent sample T test was used to compare the CNN1, CNN2, and CNN3 messenger ribonucleic acid (mRNA) levels in the tumor tissue of ESCC patients with those of the normal control group. The Mann-Whitney U test was used to compare other differences between two groups. The chi-square test was used to explore the correlation between CNN2 mRNA and protein levels (expressed as scores) and clinicopathological parameters. The Kaplan-Meier method and log-rank test were used in survival analysis. The Univariable and Multivariable Cox regression model was used to determine which variables affect survival. Missing values were excluded from the analysis. All statistical tests were two tailed, and p values <0.05 were considered significant.
Results and Discussion
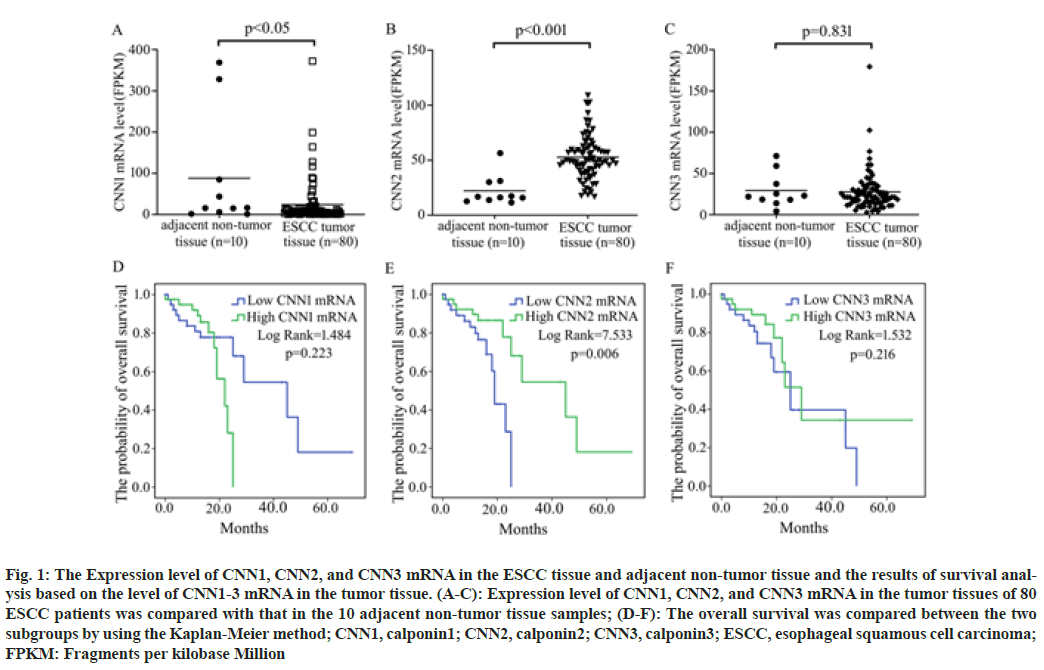
According to the data from TCGA database, CNN1 mRNA was significantly decreased while CNN2 mRNA was significantly increased in ESCC tumor tissues compared with the controls. No significant difference in the expression level of CNN3 mRNA in the tumor tissue of ESCC patients was observed compared with that in the normal esophageal tissues (fig. 1A- fig. 1C).
Fig. 1: The Expression level of CNN1, CNN2, and CNN3 mRNA in the ESCC tissue and adjacent non-tumor tissue and the results of survival analysis based on the level of CNN1-3 mRNA in the tumor tissue. (A-C): Expression level of CNN1, CNN2, and CNN3 mRNA in the tumor tissues of 80 ESCC patients was compared with that in the 10 adjacent non-tumor tissue samples; (D-F): The overall survival was compared between the two subgroups by using the Kaplan-Meier method; CNN1, calponin1; CNN2, calponin2; CNN3, calponin3; ESCC, esophageal squamous cell carcinoma; FPKM: Fragments per kilobase Million
To investigate the association of the expression level of CNN1, CNN2, and CNN3 with patients’ survival time, patients were respectively divided into two subgroups; high expression subgroup (mRNA level>median) and low expression subgroup (mRNA level≤median). Analysis based on the Kaplan-Meier method revealed that patients with higher CNN2 mRNA in the tumor tissue had a longer OS time than patients with lower one (p=0.006), while the expression level of CNN1 (p=0.223) and CNN3 (p=0.216) was not associated with the survival of ESCC patients in fig.1D-fig. 1F.
To determine whether CNN2 is an independent risk factor for the survival of ESCC patients, both Univariable and Multivariable Cox regression were performed. The expression level of CNN2 mRNA was observed to be an independent risk factor for patients’ OS along with the TNM stage in both Univariable and Multivariable Cox regression, while age was shown to be an independent risk factor for patients’ OS only in Multivariable Cox regression (Table 1).
| Variable | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR (95 % CI) | p | HR (95 % CI) | p | |
| Age (years) (<63 vs. ≥63) | 1.69(0.71-4.03) | 0.241 | 2.81(1.05-7.50) | 0.039 |
| Gender (Male vs. Female) | 0.03(0.000-1.62) | 0.083 | 0.000(0.000-2.43E149) | 0.945 |
| Drinking (Yes vs. No) | 0.38(0.09-1.62) | 0.189 | 0.23(0.05-1.07) | 0.061 |
| TNM stage (I-II vs. III-IV) | 2.71(1.12-6.55) | 0.028 | 2.69(1.05-6.87) | 0.039 |
| CNN2 level (Lower vs. Higher) | 0.29(0.11-0.74) | 0.01 | 0.22(0.08-0.64) | 0.005 |
Note: CNN2: Calponin2; HR: Hazard Ratio; CI: Confidence Interval; p: p value
Table 1: Univariable and Multivariable Cox Regression Analysis for Overall Survival
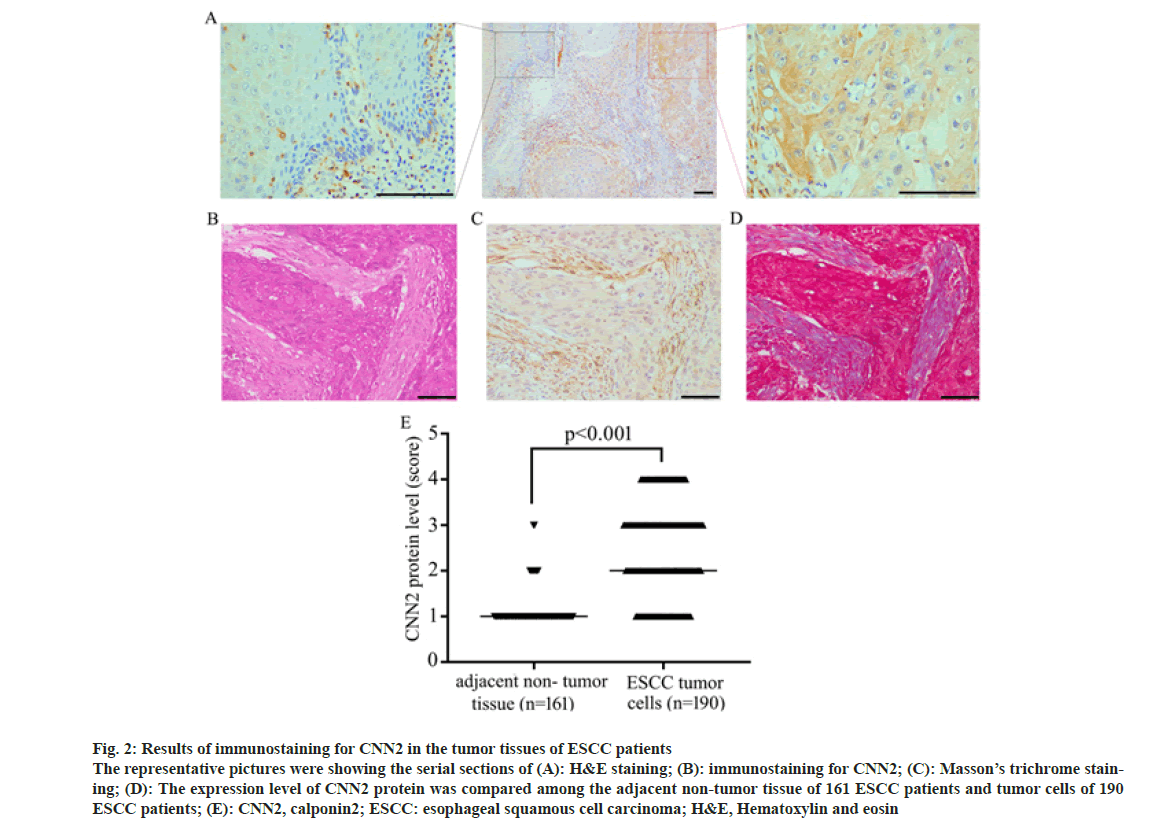
Immunohistochemical staining was performed on the tumor tissue of 190 ESCC patients of TZYY ESCC cohort. CNN2 was observed to distribute mainly in the cytoplasm of cells, express higher in the tumor tissue rather than adjacent normal esophagus tissue. Some tumor stromal also expressed CNN2, which was later proved to be Fibroblasts by Masson’s trichrome (fig. 2).
Fig. 2: Results of immunostaining for CNN2 in the tumor tissues of ESCC patients The representative pictures were showing the serial sections of (A): H&E staining; (B): immunostaining for CNN2; (C): Masson’s trichrome staining; (D): The expression level of CNN2 protein was compared among the adjacent non-tumor tissue of 161 ESCC patients and tumor cells of 190 ESCC patients; (E): CNN2, calponin2; ESCC: esophageal squamous cell carcinoma; H&E, Hematoxylin and eosin
According to the score of immunostaining intensity for CNN2, the association between the expression level of CNN2 protein in the tumor tissue of ESCC patients and patients’ clinicopathological parameters was analyzed. CNN2 protein level in the tumor cells was observed to be significantly associated with tumor differentiation, T, N, pTNM stage, and hypertension status (Table 2). There was no significant difference in the expression level of CNN2 protein in patients with different ages, gender, tumor length, and diabetes status.
| Variable | CNN2 expression level in tumor cells (score) | ||||||
|---|---|---|---|---|---|---|---|
| n | 1(%) | 2 (%) | 3 (%) | 4 (%) | p | ||
| Age (years) | <63 | 95 | 22(23.2) | 25(26.3) | 28(29.5) | 20(21.1) | 0.557 |
| ≥63 | 95 | 19(20.0) | 31(32.6) | 31(32.6) | 14(14.7) | ||
| Gender | Male | 116 | 22(19.0) | 37(31.9) | 31(26.7) | 26(22.4) | 0.081 |
| Female | 74 | 19(25.7) | 25.7(19) | 28(37.8) | 8(10.8) | ||
| Tumor length (cm) | <4 | 95 | 18(18.9) | 31(32.6) | 29(30.5) | 17(17.9) | 0.613 |
| ≥4 | 83 | 22(26.5) | 23(27.7) | 26(31.3) | 12(14.5) | ||
| Differentiation | Poor | 42 | 12(28.6) | 9(21.4) | 18(42.9) | 3(7.1) | 0.036 |
| Moderate/Well | 145 | 28(19.3) | 46(31.7) | 40(27.6) | 31(21.4) | ||
| T stage | T1-2 | 78 | 16(20.5) | 15(19.2) | 33(42.3) | 14(17.9) | 0.021 |
| T3-4 | 111 | 25(22.5) | 40(36.0) | 26(23.4) | 20(18.0) | ||
| N stage | N0 | 99 | 14(14.1) | 30(30.3) | 39(39.4) | 16(16.2) | 0.017 |
| N1-3 | 90 | 27(30.0) | 26(28.9) | 20(22.2) | 17(18.9) | ||
| pTNM stage | I-II | 99 | 15(15.2) | 28(28.3) | 38(38.4) | 18(18.2) | 0.033 |
| III-IV | 86 | 26(30.2) | 26(30.2) | 19(22.1) | 15(17.4) | ||
| Diabetes | Yes | 17 | 5(29.4) | 5(29.4) | 6(35.3) | 1(5.9) | 0.528 |
| No | 172 | 35(20.3) | 51(29.7) | 53(30.8) | 33(19.2) | ||
| Hypertension | Yes | 54 | 11(20.4) | 16(29.6) | 23(42.6) | 4(7.4) | 0.048 |
| No | 136 | 30(22.1) | 40(29.4) | 36(26.5) | 30(22.1) | ||
Note: CNN2: Calponin2; n: Number of patients and p: p value
Table 2: Association between Cnn2 Expression Level and Clinicopathological Parameters
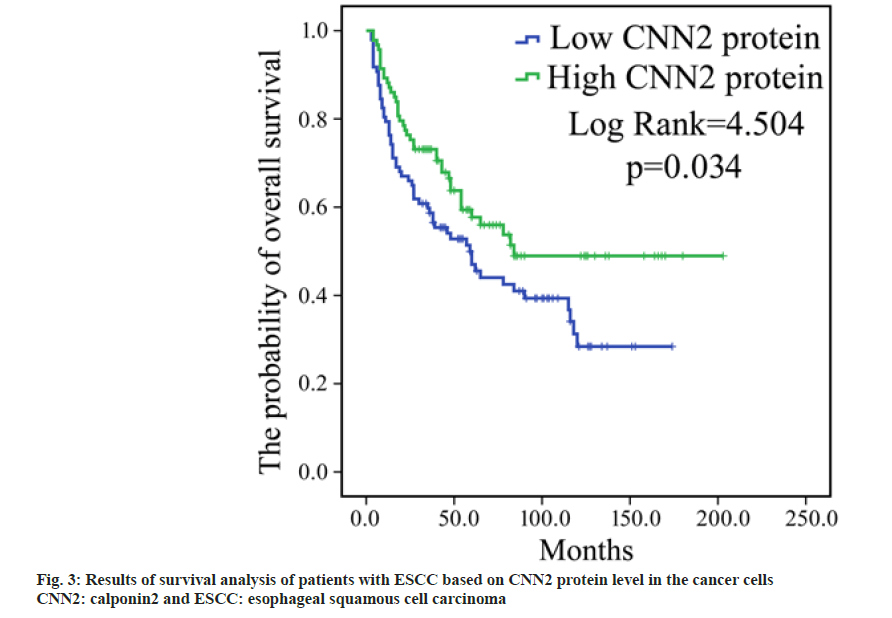
To determine whether the expression level of CNN2 protein in the tumor cells is associated with the survival, according to the level of CNN2 in cancer cells, patients were divided into two subgroups; high CNN2 subgroup (with immunostaining score for CNN2 being 3 or 4) and low CNN2 subgroup (with immunostaining score for CNN2 being 1 or 2). The Kaplan-Meier survival analysis revealed that those patients who expressed stronger CNN2 level had a longer OS than patients expressed weaker CNN2 level (p=0.034, fig. 3).
We performed Univariable and Multivariable Cox regression analyses in order to determine whether CNN2 level is an independent risk factor for the survival of the patients. As shown in Table 3, results of Univariable Cox regression analysis showed that low CNN2 level was significantly associated with increased risk of death (HR=0.65, p=0.037). However, the significance of the association between the expression level of CNN2 and patients’ OS disappeared due to the close association of CNN2 with the tumor stage in Multivariable Cox regression analysis (HR=0.37, p =0.097).
| Variable | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR (95 % CI) | p | HR (95 % CI) | p | |
| Age (years) (<63 vs. ≥63) | 1.34(0.90-1.99) | 0.152 | 1.31(0.87-1.97) | 0.194 |
| Gender (Male vs. Female) | 0.57(0.37-0.88) | 0.011 | 0.54(0.35-0.85) | 0.008 |
| Differentiation (poor vs. moderate/well) | 1.07(0.67-1.72) | 0.779 | 1.23(0.75-2.00) | 0.416 |
| pTNM stage (I-II vs. III-IV) | 3.25(2.13-4.97) | <0.001 | 2.70(1.75-4.17) | <0.001 |
| Hypertension (Yes vs. No) | 1.79(1.18-2.72) | 0.007 | 1.94(1.25-3.01) | 0.003 |
| CNN2 level (Lower vs. Higher) | 0.65(0.44-0.97) | 0.037 | 0.70(0.46-1.07) | 0.097 |
Note: CNN2: Calponin2; HR: Hazard Ratio; CI: Confidence Interval; p: p value
Table 3: Univariable and Multivariable Cox Regression Analysis for Overall Survival
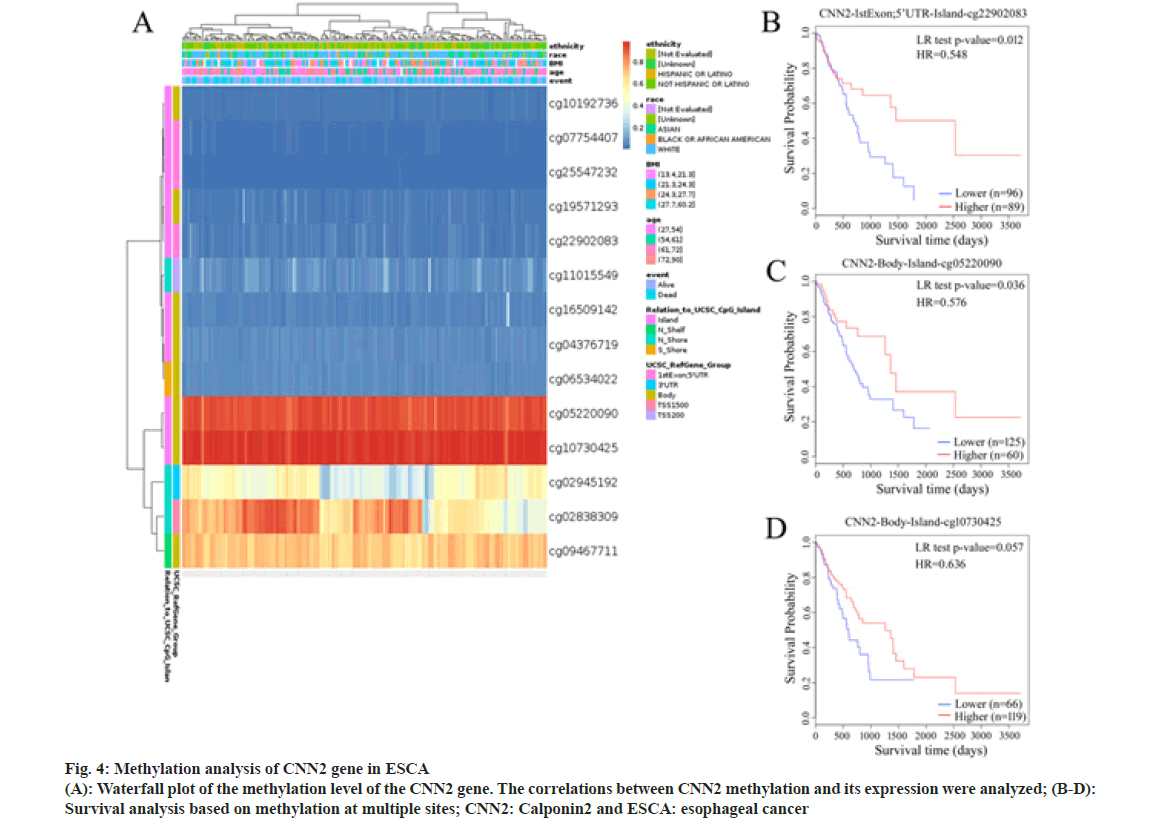
DNA methylation plays an important role in the regulation of gene expression. To determine whether DNA methylation also plays a role in CNN2 expression regulation, we examined the DNA methylation site in CNN2 gene in ESCA patients by using the MethSurv tool. A total of 14 methylation sites were found (Table 4). Among them, cg22902083 had a significant correlation with the prognosis of ESCA patients (HR=0.573, p=0.022). In fig. 4, results of survival analysis also showed that higher DNA methylation level of CpG at cite cg22902083 correlated with better OS. The highest frequency of methylation was observed at cg05220090 and cg10730425 site. Although no significant meaning was observed between the two sites and OS, the result showed the same trend as cg22902083.
| CpG | HR | p |
|---|---|---|
| cg22902083 | 0.573 | 0.022 |
| cg10730425 | 0.636 | 0.053 |
| cg07754407 | 1.555 | 0.062 |
| cg05220090 | 0.667 | 0.078 |
| cg11015549 | 0.71 | 0.184 |
| cg02945192 | 1.372 | 0.248 |
| cg16509142 | 0.743 | 0.256 |
| cg09467711 | 0.771 | 0.257 |
| cg25547232 | 1.336 | 0.294 |
| cg02838309 | 1.261 | 0.316 |
| cg04376719 | 0.799 | 0.332 |
| cg10192736 | 0.8 | 0.336 |
| cg06534022 | 1.195 | 0.498 |
| cg19571293 | 0.857 | 0.528 |
Note: CNN2: calponin3; ESCA: esophageal cancer; HR: hazard ratio; p: p value
Table 4: The Influence of Hyper Cnn2 Methylation Level on Prognosis in Esca
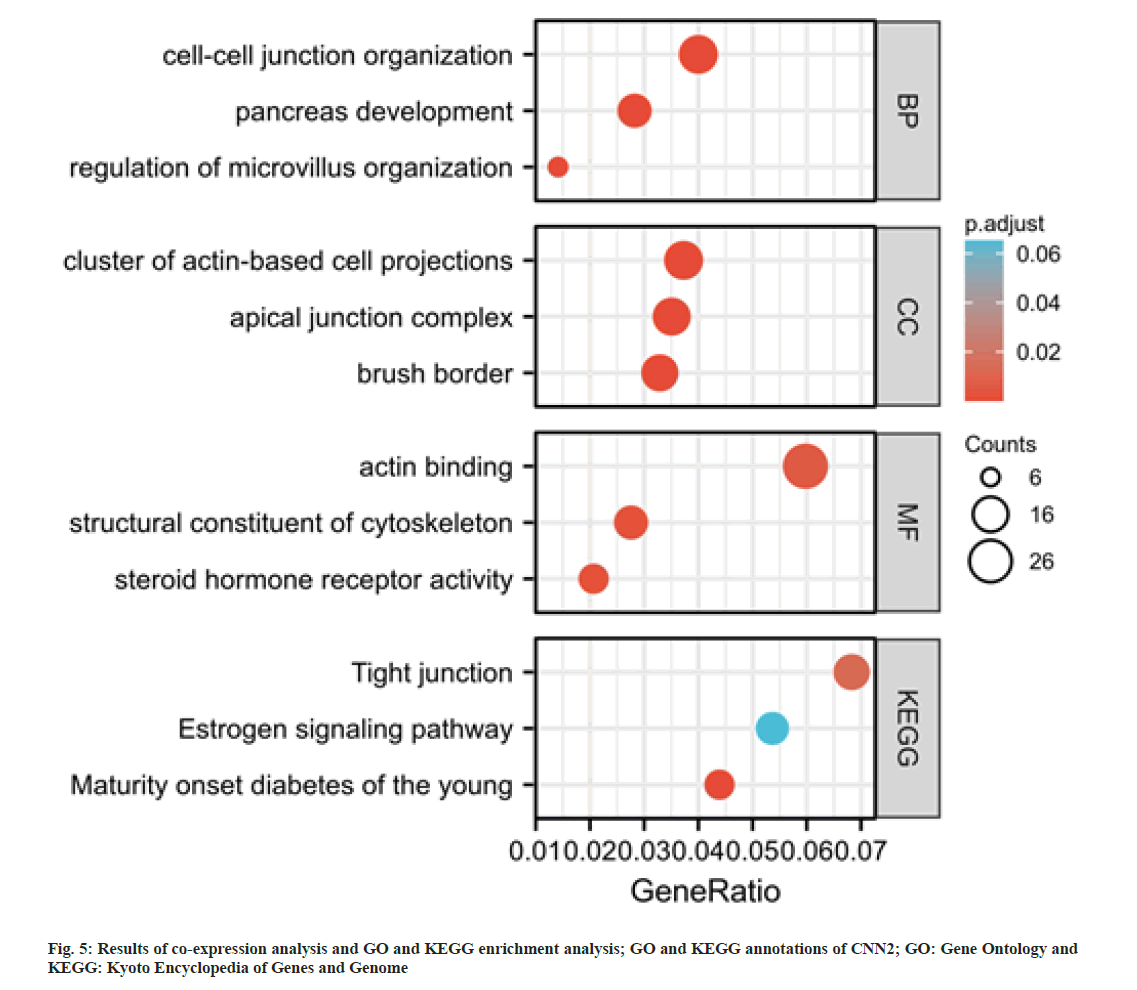
To further explore the possible roles of CNN2 gene in ESCA, a co-expression analysis was performed by using cBioPortal database. A total of 500 transcriptionally associated genes were selected (|r|>0.6, p<0.05). Based on these genes, GO and KEGG analysis were performed. As shown in fig. 5, these genes were enriched in “cell-cell junction organization”, “cluster of actin-based cell projections”, “apical junction complex”, “actin binding”, and “structural constituent of cytoskeleton”. The KEGG pathway analysis revealed that CNN2 co-expressed genes were highly enriched in tight junction.
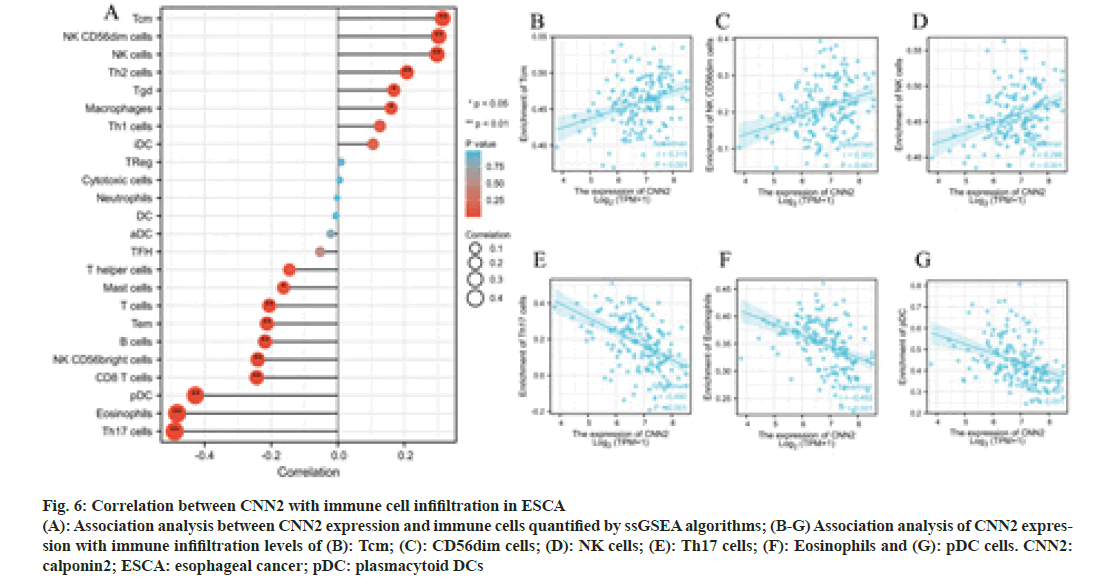
To further explore the possible role of CNN2, the correlation analysis between CNN2 expression and immune cells quantified was performed. The lollipop diagram in fig. 6A showed the correlation between CNN2 expression levels and 24 immune cells in ESCA quantified by ssGSEA algorithms. The expression level of CNN2 was positively correlated with the enrichment of Tcm, NK CD56dim cells, NK cells, Th2 cells, Tgd, and macrophages, and negatively correlated with the enrichment of Th17 cells, eosinophils, pDC, CD8 T cells, NK CD56bright cells, B cells, Tem, T cells, and mast cells. Six immune cells with the strongest positive or negative correlation with CNN2 expression level were selected for further Spearman’s correlation analysis (fig. 6B-fig. 6G).
Fig. 6: Correlation between CNN2 with immune cell infifiltration in ESCA (A): Association analysis between CNN2 expression and immune cells quantified by ssGSEA algorithms; (B-G) Association analysis of CNN2 expression with immune infifiltration levels of (B): Tcm; (C): CD56dim cells; (D): NK cells; (E): Th17 cells; (F): Eosinophils and (G): pDC cells. CNN2: calponin2; ESCA: esophageal cancer; pDC: plasmacytoid DCs
In the present study, for the first time, we investigated the expression of calponin in the tumor tissue of ESCC patients and associated them with the clinicopathological parameters. According to the TCGA database, compared with that in the adjacent non-tumor esophagus tissues, the expression level of CNN1 in the tumor tissues was much lower while the expression level of CNN2 in the tumor tissues was much higher. The expression level of CNN3 showed no significant change in the tumor tissues compared with adjacent non-tumor esophagus tissues. Patients with higher level of CNN2 mRNA were observed to have longer OS time. The expression level of CNN1 and CNN3 mRNA did not seem to correlate with the survival of the patients. Considering that genes work primarily through the proteins they encoded rather than mRNA, we further examined the expression level of CNN2 protein in the ESCC cancer cells by immunohistochemistry. Much higher CNN2 protein was found in the tumor tissues compared with the adjacent non-tumor esophagus tissues, which was in accordance with the results of mRNA expression. The increased CNN2 was observed mainly in the cancer cells and interstitial fibroblast. Furthermore, the expression level of CNN2 in the cancer cells was found to correlate with tumor stage and patients’ OS. All these results suggested that CNN2 might be involved in the pathogenesis of ESCC and might act as a tumor-inhibiting factor.
DNA methylation plays an important role in the regulation of gene expression and thus participates in the pathogenesis of a variety of human diseases. Aberrant DNA methylation has been reported in various cancers[20,21]. Increased DNA methylation within the promoter often results in silencing of a gene, whereas a high level of intragenic methylation (gene body, 5’UTR, and 3’UTR) is linked to increased gene expression[22-24]. To determine whether DNA methylation also plays a role in CNN2 expression regulation, three intragenic methylation sites were found in CNN2 gene in ESCA patients in our study. Among them, two highly methylated sites (cg05220090 and cg10730425) were at the body of the gene, another site (cg22902083) was at the 5’UTR of the gene, suggesting that DNA methylation in these sites might contribute to the increased expression of CNN2 in ESCA patients. Interestingly, the level of methylation at another CpG site in 5’UTR (cg22902083) was significantly associated with prognosis, patients with higher frequency of methylation at this site were found to have longer OS, which emphasized that CNN2 might be involved in the pathological process of ESCA and further supported our idea that CNN2 might play a role in ESCA and work as an inhibitor.
Cytoskeleton is the basic structure of a cell, playing key roles in cell morphology, movement, and invasion[25-29]. Cell-cell junctions included Adherens Junctions (AJs) and tight junctions (TJs)[30,31]. When tumor cells lose their intercellular connection, they would become invasive and spread from the primary site, then tumor metastases occur[32-34]. GO enrichment analysis showed that CNN2 and interacted genes participate in the cell-cell junction organization and structural constituent of cytoskeleton with 500 transcriptionally associated genes. The KEGG pathway analysis revealed that CNN2 co-expressed genes were highly enriched in tight junction. Thus, we considered that the high expression of CNN2 and its co-expression genes might influence tumor proliferation and metastasis in ESCA via regulating cytoskeleton, cell-cell junctions and other relevant mechanisms.
Tumor development and growth typically requires an appropriate microenvironment, which consists of a complex network of extracellular matrix components and distinct cell types containing infiltrating immune cells[35-37]. In our research, we also investigated the relationship between the expression level of CNN2 and infiltrating level of immune cells by ssGSEA. The result showed that the CNN2 expression level was positively correlated with the infifiltrating level of Tcm cells, NK CD56dim cells, and NK cells, and negatively correlated with the infifiltrating level of Th17 cells, Eosinophils, and pDC. Among them, Tcm cells, CD56dim NK cells and NK cells play important roles in killing cancer cells,whileTh17 cells, Eosinophils, and pDCs are associated with chronic inflammation or angiogenesis, which could drive tumor development further more[36,38- 49]. The expression level of CNN2 was positive correlated with the infifiltrating level of tumor inhibition cells and negatively correlated with the infifiltrating level of those cells related to chronic inflammation or angiogenesis.The cell surface receptors, which expressed in immune cells are immune checkpoints and Programmed cell Death protein 1 (PD-1) is one of the most concerned immune checkpoints[50,51]. PDL1 was mainly expressed in human cancers and contributes to cancer immune evasion by engaging with PD-1 on immune cells[52]. PD-PDL1 in different cells can be induced by several signaling pathways, like MEK, MAPK, PI3K, and AKT, and several transcriptional factors, including HIF-1α, STAT3, and NF-κB[51]. It’s worth mentioning that in hepatocellular carcinoma, CNN2 could affect tumor progression by regulating the phosphorylation level of ERK1/2 in the MAPK pathway, while in pancreatic ductal adenocarcinoma, CNN2 affected tumor progression by inhibiting PI3K/ AKT and NF-κB pathways[15,18]. In the ESCA, therefore, we speculated that CNN2 might could via some pathways to regulate PD-L1 expression in cancer cells and then influence the engagement of PD-1, and then affect immune invasion. There are some limitations to our study. Firstly, some clinical information is missing due to the retrospective nature of data collection which might impact on data analysis. Secondly, since this is a study based on tumor specimens, there may be deviations due to specimen selection. Thirdly, the present study has not explored the concrete mechanism, further mechanism research is essential to explore how CNN2 exerts its roles in ESCA.
In summary, for the first time, the present study investigated the expression of CNN2 in the tumor tissues and combined that with clinicopathological parameters of ESCC patients. CNN2 was increasingly expressed in the cancer cells and correlated negatively with the tumor grade and positively with the survival of the patients. Furthermore, DNA methylation of CNN2 was related to the prognosis of ESCA. CNN2 was co-expressed with genes associated with tight junction and the expression level of CNN2 was observed to correlate significantly with infiltrating level of immune cells.CNN2 may not only participate in the occurrence and progression but also the immune regulation of ESCA. Of all, over-expressed CNN2 might play as a tumor inhibitor in ESCA. Thus, the present study may help us obtain deeper insights into therapeutic targeting for ESCA. Further experiments are still needed to confirm these ideas.
Author Contributions:
M. GAN and YUHUI XIA contributed equally to this work.
Acknowledgments:
This work was supported by the Open Project Program of Key Laboratory of Minimally Invasive Techniques & Rapid Rehabilitation of Digestive System Tumor of Zhejiang Province (21SZDSYS08).
Conflict of interests:
The authors declared no conflict of interests.References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Short MW, Burgers K, Fry V. Esophageal cancer. Am Fam Physician 2017;95(1):22-8. [Crossref]
[Google Scholar] [PubMed]
- Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P. Oesophageal cancer. Nat Rev Dis Primers 2017;3:17048.
[Crossref] [Google Scholar] [PubMedsss]
- Wang X, Li K, Cheng M, Wang G, Han H, Chen F, et al. Bmi1 severs as a potential tumor-initiating cell marker and therapeutic target in esophageal squamous cell carcinoma. Stem Cells Int 2020;2020:1-9.
[Crossref] [Google Scholar] [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2018;41(3):210-5.
[Crossref] [Google Scholar] [PubMed]
- Zhao Q, Yu J, Meng X. A good start of immunotherapy in esophageal cancer. Cancer Med 2019;8(10):4519-26.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Yang T, Xu Z, Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother 2016;82:406-12.
[Crossref] [Google Scholar] [PubMed]
- Yu WW, Fu XL, Cai XW, Sun MH, Guo YM. Identification of differentially expressed proteins in the locoregional recurrent esophageal squamous cell carcinoma by quantitative proteomics. J Gastrointest Oncol 2021;12(3):991-1006.
[Crossref] [Google Scholar] [PubMed]
- Lin XF, Zhang CQ, Dong BR. MiR-421 expression independently predicts unfavorable overall survival in patients with esophageal adenocarcinoma. Eur Rev Med Pharmacol Sci 2019;23(9):3790-8.
[Crossref] [Google Scholar] [PubMed]
- Liu R, Jin JP. Calponin isoforms CNN1, CNN2 and CNN3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene 2016;585(1):143-53.
[Crossref] [Google Scholar] [PubMed]
- Plazyo O, Liu R, Hossain MM, Jin JP. Deletion of calponin 2 attenuates the development of calcific aortic valve disease in ApoE−/− mice. J Mol Cell Cardiol 2018;121:233-41.
[Crossref] [Google Scholar] [PubMed]
- Hossain MM, Zhao G, Woo MS, Wang JH, Jin JP. Deletion of calponin 2 in mouse fibroblasts increases myosin II-dependent cell traction force. Biochemistry 2016;55(43):6046-55.
[Crossref] [Google Scholar] [PubMed]
- Huang QQ, Hossain MM, Sun W, Xing L, Pope RM, Jin JP. Deletion of calponin 2 in macrophages attenuates the severity of inflammatory arthritis in mice. Am J Physiol Cell Physiol 2016;311(4):C673-85.
[Crossref] [Google Scholar] [PubMed]
- Hu J, Xie W, Shang L, Yang X, Li Q, Xu M, et al. Knockdown of calponin 2 suppressed cell growth in gastric cancer cells. Tumor Biol 2017;39(7):1010428317706455.
[Crossref] [Google Scholar] [PubMed]
- Qiu Z, Chu Y, Xu B, Wang Q, Jiang M, Li X, et al. Increased expression of calponin 2 is a positive prognostic factor in pancreatic ductal adenocarcinoma. Oncotarget 2017;8(34):56428-42.
[Crossref] [Google Scholar] [PubMed]
- Ji T, Ma F, Huo L, Guo X, Chen B, Zhou Q. Calponin‑h2 is upregulated in the tissues and plasma of patients with breast cancer. Mol Med Rep 2015;12(2):2886-92.
[Crossref] [Google Scholar] [PubMed]
- Hossain MM, Wang X, Bergan RC, Jin JP. Diminished expression of h2-calponin in prostate cancer cells promotes cell proliferation, migration and the dependence of cell adhesion on substrate stiffness. FEBS Open Bio 2014;4:627-36.
[Crossref] [Google Scholar] [PubMed]
- Kang X, Wang F, Lan X, Li X, Zheng S, Lv Z, et al. Lentivirus-mediated shRNA targeting CNN2 inhibits hepatocarcinoma in vitro and in vivo. Int J Med Sci 2018;15(1):69-76.
[Crossref] [Google Scholar] [PubMed]
- Deng F, Zhou K, Li Q, Liu D, Li M, Wang H, et al. iTRAQ-based quantitative proteomic analysis of esophageal squamous cell carcinoma. Tumor Biol 2016;37:1909-18.
[Crossref] [Google Scholar] [PubMed]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA methylation in the mammalian genome. Cell 2016;167(1):233-47.
[Crossref] [Google Scholar] [PubMed]
- Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med 2018;24(10):1611-24.
[Crossref] [Google Scholar] [PubMed]
- Li J, Li Y, Li W, Luo H, Xi Y, Dong S, et al. Guide Positioning Sequencing identifies aberrant DNA methylation patterns that alter cell identity and tumor-immune surveillance networks. Genome Res 2019;29(2):270-80.
[Crossref] [Google Scholar] [PubMed]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017;543(7643):72-7.
[Crossref] [Google Scholar] [PubMed]
- Izquierdo AG, Crujeiras AB. Obesity-related epigenetic changes after bariatric surgery. Front Endocrinol 2019;10:232.
[Crossref] [Google Scholar] [PubMed]
- Yao T, Cao R, Xiao W, Pan F, Li X. An optical study of drug resistance detection in endometrial cancer cells by dynamic and quantitative phase imaging. J Biophotonics 2019;12(7):e201800443.
[Crossref] [Google Scholar] [PubMed]
- Liu C, Luo JW, Zhang KK, Lin LX, Liang T, Luo ZP, et al. Tendon-derived stem cell differentiation in the degenerative tendon microenvironment. Stem Cells Int 2018;2018:2613821.
[Crossref] [Google Scholar] [PubMed]
- Zhu D, Su Y, Zheng Y, Fu B, Tang L, Qin YX. Zinc regulates vascular endothelial cell activity through zinc-sensing receptor ZnR/GPR39. Am J Physiol Cell Physiol 2018;314(4):C404-14.
[Crossref] [Google Scholar] [PubMed]
- Zhang B, Shetti D, Fan C, Wei K. miR-29b-3p promotes progression of MDA-MB-231 triple-negative breast cancer cells through downregulating TRAF3. Biol Res 2019;52:38.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Gu J, Xu F, Zhu Q, Ge D, Lu C. Novel methylation-driven genes identified as prognostic indicators for lung squamous cell carcinoma. Am J Transl Res 2019;11:1997-2012.
[Google Scholar] [PubMed]
- Malinova TS, Huveneers S. Sensing of cytoskeletal forces by asymmetric adherens junctions. Trends Cell Biol 2018;28(4):328-41.
[Crossref] [Google Scholar] [PubMed]
- Tsukita S, Tanaka H, Tamura A. The claudins: From tight junctions to biological systems. Trends Biochem Sci 2019;44(2):141-52.
[Crossref] [Google Scholar] [PubMed]
- Piotrowska Ż, Niezgoda M, Młynarczyk G, Acewicz M, Kasacka I. Comparative assessment of the WNT/β-catenin pathway, CacyBP/SIP, and the immunoproteasome subunit LMP7 in various histological types of renal cell carcinoma. Front Oncol 2020;10:566637.
[Crossref] [Google Scholar] [PubMed]
- Han D, Sun J, Fan D, Zhang C, Du S, Zhang W. Simvastatin ameliorates oxygen glucose deprivation/reoxygenation-induced pulmonary endothelial barrier dysfunction by restoring cell-cell junctions and actin cytoskeleton dynamics via the PI3K/Akt signaling pathway. Am J Transl Res 2020;12(9):5586-96.
[Google Scholar] [PubMed]
- Jia Y, Qin T, Zhang X, Liu S, Liu Z, Zhang C, et al. Effect of bevacizumab on the tight junction proteins of vascular endothelial cells. Am J Transl Res 2019;11(9):5546-59.
[Google Scholar] [PubMed]
- Tekpli X, Lien T, Røssevold AH, Nebdal D, Borgen E, Ohnstad HO, et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Commun 2019;10(1):5499.
[Crossref] [Google Scholar] [PubMed]
- Aiello I, Fedele MM, Román F, Marpegan L, Caldart C, Chiesa JJ, et al. Circadian disruption promotes tumor-immune microenvironment remodeling favoring tumor cell proliferation. Sci Adv 2020;6(42):eaaz4530.
[Crossref] [Google Scholar] [PubMed]
- Alvero AB, Hanlon D, Pitruzzello M, Filler R, Robinson E, Sobolev O, et al. Transimmunization restores immune surveillance and prevents recurrence in a syngeneic mouse model of ovarian cancer. Oncoimmunol 2020;9(1):1758869.
[Crossref] [Google Scholar] [PubMed]
- Castella M, Caballero-Baños M, Ortiz-Maldonado V, González-Navarro EA, Suñé G, Antoñana-Vidósola A, et al. Point-of-care CAR T-cell production (ARI-0001) using a closed semi-automatic bioreactor: Experience from an academic phase I clinical trial. Front Immunol 2020;11:482.
[Crossref] [Google Scholar] [PubMed]
- Sheng H, Huang Y, Xiao Y, Zhu Z, Shen M, Zhou P, et al. ATR inhibitor AZD6738 enhances the antitumor activity of radiotherapy and immune checkpoint inhibitors by potentiating the tumor immune microenvironment in hepatocellular carcinoma. J Immunother Cancer 2020;8(1):e000340.
[Crossref] [Google Scholar] [PubMed]
- Singh K, Martinell M, Luo Z, Espes D, Stålhammar J, Sandler S, et al. Cellular immunological changes in patients with LADA are a mixture of those seen in patients with type 1 and type 2 diabetes. Clin Exp Immunol 2019;197(1):64-73.
[Crossref] [Google Scholar] [PubMed]
- Moreno‐Nieves UY, Mundy DC, Shin JH, Tam K, Sunwoo JB. The aryl hydrocarbon receptor modulates the function of human CD56bright NK cells. Eur J Immunol 2018;48(5):771-6.
[Crossref] [Google Scholar] [PubMed]
- Gong W, Xiao W, Qian L, Gong C, Hu M, Pan X, et al. Immobilized MHC class I chain-related protein A synergizes with IL-15 and soluble 4-1BB ligand to expand NK cells with high cytotoxicity ex vivo. Cell Mol Immunol 2010;7(6):477-84.
[Crossref] [Google Scholar] [PubMed]
- Cao Y, Wang X, Jin T, Tian Y, Dai C, Widarma C, et al. Immune checkpoint molecules in natural killer cells as potential targets for cancer immunotherapy. Signal Transduct Target Ther 2020;5(1):250.
[Crossref] [Google Scholar] [PubMed]
- Gunesch JT, Angelo LS, Mahapatra S, Deering RP, Kowalko JE, Sleiman P, et al. Genome-wide analyses and functional profiling of human NK cell lines. Mol Immunol 2019;115:64-75.
[Crossref] [Google Scholar] [PubMed]
- Li N, Xie C, Lu NH. Transforming growth factor-β: an important mediator in Helicobacter pylori-associated pathogenesis. Front Cell Infect Microbiol 2015;5:77.
[Crossref] [Google Scholar] [PubMed]
- Dunne MR, Ryan C, Nolan B, Tosetto M, Geraghty R, Winter DC, et al. Enrichment of inflammatory IL-17 and TNF-α secreting CD4+ T cells within colorectal tumors despite the presence of elevated CD39+ T regulatory cells and increased expression of the immune checkpoint molecule, PD-1. Front Oncol 2016;6:50.
[Crossref] [Google Scholar] [PubMed]
- Sakkal S, Miller S, Apostolopoulos V, Nurgali K. Eosinophils in cancer: Favourable or unfavourable? Curr Med Chem 2016;23(7):650-66.
[Crossref] [Google Scholar] [PubMed]
- Pinto A, Rega A, Crother TR, Sorrentino R. Plasmacytoid dendritic cells and their therapeutic activity in cancer. Oncoimmunology 2012;1(5):726-34.
[Crossref] [Google Scholar] [PubMed]
- Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 2015;15(8):471-85.
[Crossref] [Google Scholar] [PubMed]
- Yu J, Qin B, Moyer AM, Nowsheen S, Tu X, Dong H, et al. Regulation of sister chromatid cohesion by nuclear PD-L1. Cell Res 2020;30(7):590-601.
[Crossref] [Google Scholar] [PubMed]
- Li X, Song W, Shao C, Shi Y, Han W. Emerging predictors of the response to the blockade of immune checkpoints in cancer therapy. Cell Mol Immunol 2019;16(1):28-39.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Dang F, Ren J, Wei W. Biochemical aspects of PD-L1 regulation in cancer immunotherapy. Trends Biochem Sci 2018;43(12):1014-32.
[Crossref] [Google Scholar] [PubMed]