- *Corresponding Author:
- Surumi Beevisha
Department of Biotechnology, Inter University Centre for Genomics and Gene Technology, University of Kerala, Trivandrum, Kerala 695581, India
E-mail: surumib@keralauniversity.ac.in
| Date of Received | 14 September 2021 |
| Date of Revision | 04 July 2023 |
| Date of Acceptance | 01 September 2023 |
| Indian J Pharm Sci 2023;85(5):1190-1197 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Earlier, long non-coding ribonucleic acid molecules were considered as a part of transcriptional noise and ignored imprudently, but gradually got revealed as potential regulators in many biological processes and their roles in gene expression influencing almost every aspect associated with genes, including epigenetic, transcriptional, and post-transcriptional regulation. Apart from their involvement in normal physiology, long-non-coding ribonucleic acid expression functions are also related to adipose biology, indirectly leading to obesity. This review discusses the beneficial role and mechanisms of action of PU.1 antisense long-noncoding ribonucleic acids in normal adipogenesis and their implications for obesity. Extensive research and identification of prominent long-non-coding ribonucleic acids in adipose biology will not only grant insights into diseases associated with obesity but also give ensure therapeutic targets for it.
Keywords
Antisense long-non-coding ribonucleic acid, antisense oligonucleotides, long-non-coding ribonucleic acid, PU.1 antisense long-non-coding ribonucleic acid
In both developed and developing countries, obesity is becoming increasingly common, and by 2030 the proportion of overweight and obese individuals is predicted to hit 89 and 85 percent in men and women, respectively[1,2]. Even though excessive weight gain is associated with metabolic syndrome disorders, including hyperglycemia, dyslipidemia, high blood pressure, atherosclerosis[3,4], diabetes, and cardiovascular disease, hence the global obesity epidemic is a significant challenge to human health[5,6]. An unusual increase in the number and size of adipocytes causes obesity due to excessive fat accumulation in White Adipose Tissue (WAT)[7]. Apart from the widely accepted high-caloric intake, genetic and epigenetic factors play roles in obesity, as evident from different studies[8,9].
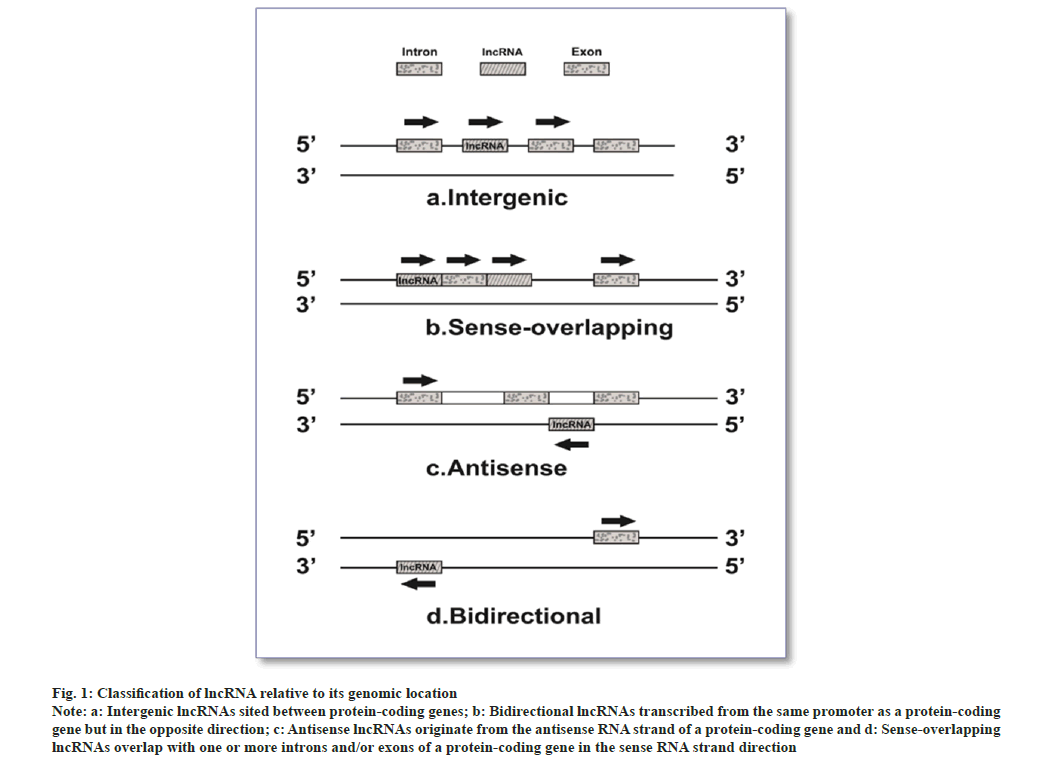
Non-coding Ribonucleic Acid (ncRNA) molecules are genomic sequence transcripts not intended to be translated[10]. Short ncRNA (sncRNAs equals 30 nt) and long ncRNA (lncRNA greater than 200 nt) are two ncRNAs categorized arbitrarily based on the length of RNA produced post-transcriptionally. With the discovery and functional characterization of lncRNAs, the family of regulatory Ribonucleic Acids (RNAs) has seen an explosion in the past decade. LncRNAs are a distinctive class of transcripts of more than 200 nucleotides, frequently polyadenylated and missing an active open reading frame[11-13]. LncRNAs categories are intergenic, antisense, divergent, intronic, and enhancer lncRNAs based on the relative location of the neighboring coding genes (fig. 1). The control of cellular functions by lncRNAs is by various mechanisms; they could act as scaffolds, decoys, or guides[12,14].
Fig. 1: Classification of lncRNA relative to its genomic location.
Note: a: Intergenic lncRNAs sited between protein-coding genes; b: Bidirectional lncRNAs transcribed from the same promoter as a protein-coding gene but in the opposite direction; c: Antisense lncRNAs originate from the antisense RNA strand of a protein-coding gene and d: Sense-overlapping lncRNAs overlap with one or more introns and/or exons of a protein-coding gene in the sense RNA strand direction.
LncRNAs control gene expression at both the transcriptional and post-transcriptional levels, resulting in several biological processes such as tumor initiation, growth, and metastasis in a variety of human diseases, including cancer[15-17] as well as in some obesity-related conditions.
LncRNAs in the adipogenesis process and its implication in obesity:
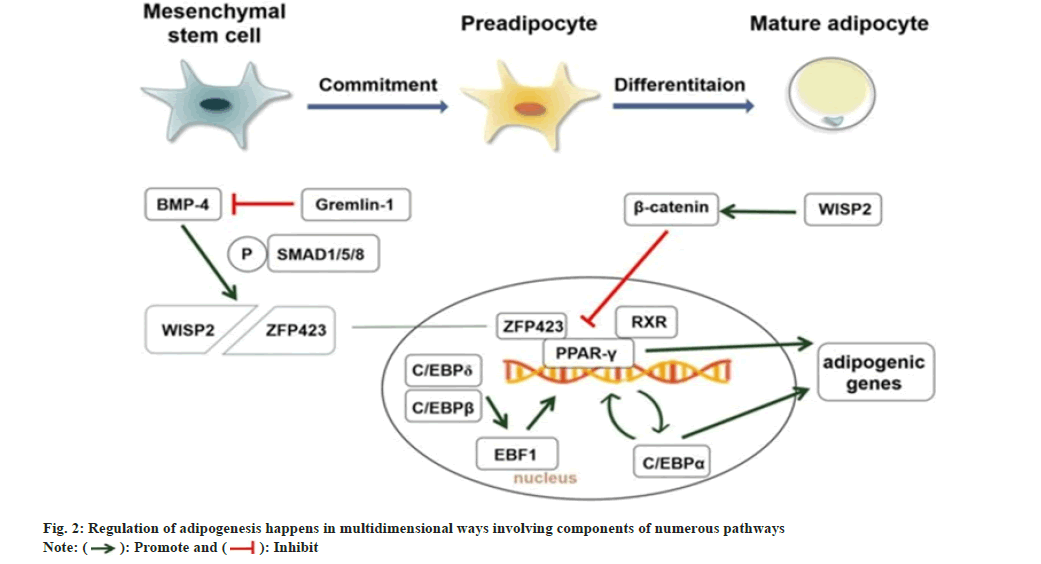
Adipocytes are peculiar cells destined to store excess energy as triglycerides and are involved in adipokine secretion that impairs systemic energy homeostasis[18]. Obesity typically happens when the volume or size of adipocytes increases[18]. WAT and Brown Adipose Tissue (BAT) are the two distinct kinds of mammalian adipose tissue. WAT is essential for storing and secretion of adipokines that affect energy homeostasis and metabolic processes, while BAT specializes in energy expenditure and thermogenesis[19,20]. Maintaining normal adiposity and optimizing lipid metabolism requires a proper balance of these processes. WAT consists of adipocytes generated by the differentiation of preadipocytes. Very high or low WAT leads to metabolic disorders such as hyperlipidemia, resistance to insulin, and type 2 diabetes[21]. For optimum health, maintenance of sufficient amounts of WAT is essential. The two primary types of white adipose tissue are Subcutaneous Adipose Tissue (SAT) below the skin and Visceral Adipose Tissue (VAT) found inside particular regions of the abdominal cavity[22]. Excessive fat accumulations relative to both SAT and VAT are responsible for the incidence of various metabolic diseases[23], especially fat accumulation in VAT, regarded as a high-risk factor for many metabolic disorders and cardiovascular diseases[24-26]. For a long time, studies on the differentiation of visceral adipocytes and their potential regulatory mechanisms have been at the forefront of obesity science. Adipose tissue also serves as an endocrine organ by secreting adipokines that impact the body's glucose and energy homeostasis[27]. A cascade of transcription factors, cofactors, and signaling intermediates from various pathways orchestrates the adipogenesis process[7]. Adipogenic differentiation is mainly monitored by the master regulator peroxisome proliferator for adipogenesis, together with other transcription factors and cofactors like CCAAT/enhancer binding proteins (C/EBPs), Kruppel-like factors (KLFs) or Wingless proteins (Wnt)[28]. The regulation of adipogenesis happens in multidimensional ways involving components of numerous pathways in a co-ordinated manner sequentially as seen in fig. 2.
The role of RNAs, especially ncRNAs, has recently been extensively studied for their contribution to the production and function of adipose tissue. The role of sncRNAs, including micro RNAs, in BAT and WAT biology is in many studies. While the role of lncRNAs in maintaining adipose tissue activities is in a few research studies only[29], after analyzing the differential expression of lncRNAs across primary WAT and BAT, preadipocytes, and cultured adipocytes, Sun et al.[29] characterized 175 lncRNAs regulated during adipogenesis. The group pointed out and analyzed 20 lncRNAs likely to be controlled by Peroxisome Proliferator-Activated Receptor gamma (PPARγ) and C/EBPs, the master regulators of adipogenesis. They also carried out a loss-of-function screen and demonstrated that 10 of them, including lncRAP-1 and lncRAP-2, function to modulate adipocyte differentiation[29]. Many other studies also showed definite roles of lncRNAs in adipogenesis and adipocyte biology networks. Utilizing RNA-seq analysis, Alvarez et al.[30] identified 1500 lncRNAs expressed in inguinal white, epididymal, and brown fat in mice. Exclusive expression of 127 lncRNAs was associated with BAT, most target the critical regulators of adipogenesis, including C/EBPα, C/EBPβ, and PPARγ. Table 1 describes the role of various lncRNAs involved during adipogenic differentiation. The exact role and function of lncRNAs in obesity and adipogenesis are still unknown, even if there is rapid research in this area[31-61].
| lncRNAs | Functions | References |
|---|---|---|
| SRA | Improves white adipogenesis | [31-33] |
| NEAT1 | Improves white adipogenesis | [34,35] |
| Lnc-RAP-n | Improves white adipogenesis | [29,36] |
| SlincRAD | Improves white adipogenesis | [37] |
| PU.1 AS | Improves white adipogenesis | [38,39] |
| ADINR | Improves white adipogenesis | [40] |
| Paral1 | Improves white adipogenesis | [41] |
| lnc-leptin | Improves white adipogenesis | [42] |
| HOTAIR | Improves white adipogenesis | [43] |
| ADNCR | Represses white adipogenesis | [44] |
| HoxA-AS3 | Improves white adipogenesis | [45] |
| lnc-U90926 | Represses white adipogenesis | [46] |
| MIR31HG | Improves white adipogenesis | [47] |
| Gm15290 | Improves white adipogenesis | [48] |
| TCONS_00041960 | Improves white adipogenesis | [49] |
| HoxA11-AS1 | Improves white adipogenesis | [50] |
| Adiponectin AS | Represses white adipogenesis | [51] |
| MEG3 | Represses white adipogenesis | [52] |
| H19 | Represses white adipogenesis | [53,54] |
| Blnc1 | Improves brown adipogenesis | [55,56,57] |
| lnc-BATE1; lnc-BATE | Improves brown adipogenesis | [30,58] |
| lnc-uc.417 | Represses brown adipogenesis | [59] |
| AK079912 | Improves brown adipogenesis | [60] |
| GM13133 | Improves brown adipogenesis | [61] |
Table 1: LncRNAs involved in regulating the Adipogenesis process.
Targeting RNA molecules as a promising therapeutic approach:
Identifying the potential targets modifiable therapeutically to deal with the broad clinical needs of patients with various ailments is critical. Most clinical drugs target proteins[62]; however, as they can also interact with proteins that aren't their targets, these frequently cause problems. RNA represents one class of targets, as proteins come from specific messenger RNAs (mRNAs); hence modulations in mRNAs or pre-mRNA levels could broaden the set of therapeutic targets. Nucleic acids are evolving therapeutics for unmet medical needs since they might cause fewer side effects than existing therapies[63]. The drawbacks of targeting proteins using conventional small-molecule or protein-based strategies (adapter proteins, transcription factors, etc.) can be easily targeted by modulating the mRNA levels and translation to the protein. Identifying the unique regulatory roles of ncRNAs and their roles in normal cellular physiology is expanding, as RNAs can directly promote pathology[64]. Current strategies to modulate the RNA functions in cells include the usage of small molecules targeting RNA, genome editing, gene therapy, delivery of exogenously expressed mRNAs genome editing, and synthetic Antisense Oligonucleotides (ASOs). ASOs are oligonucleotides artificially synthesized with a size range of 12-30 nucleotides that are designed to bind to RNA by Watson-Crick base pairing rules and can bind uniquely to only one target RNA and modulating its function by several different mechanisms[65]. Nowadays, antibacterial and anti-cancer therapies use drugs targeting nucleic acids. One primary approach to targeting lncRNAs for treatment is deregulating high lncRNA levels with ASOs, which block lncRNA activity, further leading to their degradation. Alternatively, the lncRNA function may be blocked by small molecules that cover the binding site of interrelating proteins or by antisense oligonucleotides that connect to the lncRNAs and restrain their protein binding capacity[66]. ASOs can target those lncRNAs that positively regulate white adipogenesis (up-regulated) instead of brown as a control method of obesity. Preliminary in vitro studies[67] showed that the ASO approach could be a critical tool for treating obesity. The practicability of ASO therapy for targeting lncRNAs is in some pre-clinical models; Antisense phosphorothioate oligonucleotides can target lncRNAs involved in Angelman syndrome and lung cancer in mice[68].
Natural Antisense (AS) transcripts (NATs) are RNA molecules transcribed from the opposite Deoxyribonucleic Acid (DNA) strand and partly overlap with sense mRNA. AS RNA is a rather uncommon term in a physiology environment until short interfering RNAs emerged as the tool of choice to knock down the expression of specific genes[69]. Recently, NAT levels have been dysregulated in various disease states[70]. Computational studies suggest that 15-25 % of mammalian genes overlap, giving rise to pairs of sense and antisense RNAs[71]. NATs, mostly categorized as AS lncRNAs play notable roles in the clarified regulation of animal genes in almost all stages of gene expression, from transcription initiation to translation to RNA degradation[72]. However, we know little about their exact functions and molecular mechanisms in many biological processes, especially in animal adipogenesis. The mammalian genome contains large spans of AS lncRNAs and recent studies have indicated that some of these AS lncRNAs might be functional[73]. The biological role of antisense lncRNAs, despite their low expression, could still be rationalized because there are two copies of DNA for any given gene in a cell; consequently, just two antisense lncRNA molecules are sufficient to interact with the two gene copies and elicit regulatory effects[74] AS lncRNAs at numerous gene loci silences sense transcription by affecting histone acetylation and methylation states and regulating mRNA dynamics at a post-transcriptional level[75]. Many studies indicated that AS lncRNA decreases mRNA levels, such as, AS lncRNAs of tie?1[76], Fibroblast Growth Factor?2 (FGF?2)[77], and Multiple peptide resistance factor (MprF)[78]. The regulatory mechanism of AS lncRNA remains unclear, although there is evidence for the regulation by similar mechanisms as for protein-coding genes. AS lncRNAs play a positive or negative role in translation[79], transcription[80-82] and stabilization of mRNA[83].
PU.1 AS lncRNA in adipogenesis:
The PU.1 gene was initially recognized as a proviral integration site for the Spleen Focus-Forming Virus (SFFV) in erythroleukaemias[84]. The transcription factor Spi1/PU.1 (SFFV proviral integration oncogene/PU box binding protein) is a hematopoietic ETS family member that is influential in the immune system generation[85]. PU.1 is a critical transcription factor in biological processes, for it played important roles not only in the hematopoiesis and immune system development[85] but also in cell cycle exit[86,87] and epigenetic silencing[88]. PU.1 functions solely in a cell-intrinsic manner to monitor the development of granulocytes, macrophages, and B and T lymphocytes[89]. PU.1 deficiency generally arrests lymphopoiesis and myelopoiesis in mice fatally, very recently human congenital PU.1 disorder has been recognized in six agammaglobulinemia patients with varying SPI1 mutations but shared insufficient levels of PU.1 and absence of B cells with consequently, zero antibodies and the condition got reversed on CRISPR editing of SPI1 in cord blood in vitro[90].
Overexpression of PU.1 downregulated essential adipogenic genes C/EBPβ and PPARγ in the C/EBPα/β-PPARγ terminal pathway of adipogenic differentiation[91]; however, the underlying mechanism that PU.1 suppressed the expression of C/EBPb and PPARc remains elusive. Two master regulators-C/EBPβ/α and PPARγ regulate adipogenesis which in turn could be strongly inhibited by PU box-binding protein (PU.1). Proven to be expressed in the adipose tissue of humans and other animals, PU.1 could suppress the C/EBPβ/α-PPARγ pathway, significantly and negatively influence adipogenesis[91].
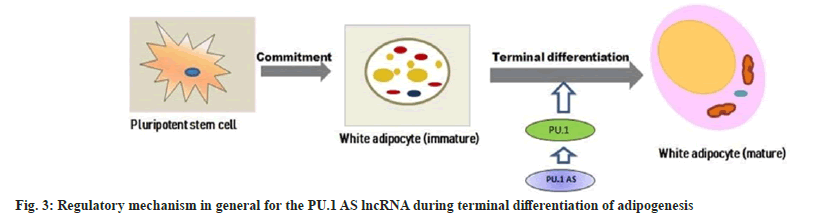
Recent studies validated that a novel mechanism of gene regulation found in mouse PU.1 locus, as the locus gave rise to both mRNA and NATs, also AS lncRNAs, which originated from an intronic promoter, and PU.1 gene level regulated through coordinated expression of its mRNA and AS lncRNAs[92]. Antisense RNAs could regulate the expression of their respective gene-altering processes in which they are involved. Previous studies indicated that PU.1 AS lncRNA promoted adipogenesis in 3T3-L1 adipocytes by preventing PU.1 mRNA translation[93,94]. PU.1 AS-PU1 RNA duplex results in inhibition of adipogenesis and modulation of sense gene expression by altering its protein expression and a decrease in expression of PPARγ, fatty acid synthase, and adiponectin in the mouse. The porcine PU.1 locus transcribed both PU.1 mRNA and PU.1 AS lncRNA, which regulates adipogenesis[93]. Antisense lncRNA overlaps PU.1 mRNA and negatively affects PU.1 protein expression via blocking translation without down-regulating mRNA levels. The regulatory mechanism in general for the PU.1 AS lncRNA during terminal differentiation of adipogenesis is shown in fig. 3. Knockdown of PU.1 AS lncRNA in zebrafish or mice up-regulated levels of PU.1 mRNA, causing expression changes of downstream genes[93]. These findings suggest that the same AS lncRNA showed distinct regulatory mechanisms, which is crucial because of its size and position in different species. Moreover, lncRNAs are so long and complicated that a slight disparity in the sequence may lead to a tremendous change in the secondary structure, distinctly altering their functions and mechanisms.
Conclusion
Therapeutic interventions in obesity depend on knowledge of molecular mechanisms that could help prevent adipogenesis. This review presents a recent approach to targeting adipogenesis utilizing antisense non-coding transcripts, indirectly, obesity. The data revealed the definite mechanism of PU.1 inhibiting adipogenesis and provided insight into the adipogenesis regulatory networks. Shortly, we will witness comprehensive and functional aspects of lncRNAs during all stages of metabolism. It is interesting to consider a combination of multiple schemes aiming at molecular mechanisms targeting adipogenesis could emerge for beneficial personalized treatment of obesity and its related complications.
Acknowledgments:
Taking account of the abundant literature on adipogenesis, we apologize if some relevant papers were not cited here due to space limitations; we thank Prof. (Dr) P. R. Sudhakaran, Prof. (Dr) G. Mohanadasan Nair and Prof. (Dr) A. Gangaprasad for stimulating discussions.
Conflicts of interest:
The authors express that no conflict of interest exists.
Funding:
Department of Higher Education, Govt. of Kerala, India.
References
- You W, Henneberg M. Relaxed natural selection contributes to global obesity increase more in males than in females due to more environmental modifications in female body mass. PloS one 2018;13(7):e0199594.
[Crossref] [Google Scholar] [PubMed]
- Rtveladze K, Marsh T, Barquera S, Romero LM, Levy D, Melendez G, et al. Obesity prevalence in Mexico: Impact on health and economic burdenv. Public Health Nutr 2014;17(1):233-9.
[Crossref] [Google Scholar] [PubMed]
- Sharma BR, Kim DW, Rhyu DY. Korean Chungtaejeon tea extract attenuates weight gain in C57BL/6J-Lep ob/ob mice and regulates adipogenesis and lipolysis in 3T3-L1 adipocytes. J Integr Med 2017;15(1):56-63.
[Crossref] [Google Scholar] [PubMedv]
- Wang WJ, Zhang T. Integration of traditional Chinese medicine and Western medicine in the era of precision medicine. J Integr Med 2017;15(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Wang YC, McPherson K, Marsh T, Gortmaker SL, Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 2011;378(9793):815-25.
[Crossref] [Google Scholar] [PubMed]
- Rutter H, Bes-Rastrollo M, de Henauw S, Lahti-Koski M, Lehtinen-Jacks S, Mullerova D, et al. Balancing upstream and downstream measures to tackle the obesity epidemic: A position statement from the European association for the study of obesity. Obes Facts 2017;10(1):61-3.
[Crossref] [Google Scholar] [PubMed]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006;444(7121):847-53.
[Crossref] [Google Scholar] [PubMed]
- Bell CG. The epigenomic analysis of human obesity. Obesity 2017;25(9):1471-81.
[Crossref] [Google Scholar] [PubMed]
- Tam V, Turcotte M, Meyre D. Established and emerging strategies to crack the genetic code of obesity. Obes Rev 2019;20(2):212-40.
[Crossref] [Google Scholar] [PubMed]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 2009;23(13):1494-504.
[Crossref] [Google Scholar] [PubMed]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012;22(9):1775-89.
[Crossref] [Google Scholar] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66.
[Crossref] [Google Scholar] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013;152(6):1298-307.
[Crossref] [Google Scholar] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet 2014;15(1):7-21.
[Crossref] [Google Scholar] [PubMed]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non?coding RNAs: Regulators of disease. J Pathol 2010;220(2):126-39.
[Crossref] [Google Scholar] [PubMed]
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29(4):452-63.
[Crossref] [Google Scholar] [PubMed]
- Mathieu EL, Belhocine M, Dao LT, Puthier D, Spicuglia S. Functions of lncRNA in development and diseases. Med Sci 2014;30(8-9):790-6.
[Crossref] [Google Scholar] [PubMed]
- Lu S, Guan Q, Liu Y, Wang H, Xu W, Li X, et al. Role of extrathyroidal TSHR expression in adipocyte differentiation and its association with obesity. Lipids Health Dis 2012;11(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell 2014;156(1):20-44.
[Crossref] [Google Scholar] [PubMed]
- Merlin J, Sato M, Nowell C, Pakzad M, Fahey R, Gao J, et al. The PPARγ agonist rosiglitazone promotes the induction of brite adipocytes, increasing β-adrenoceptor-mediated mitochondrial function and glucose uptake. Cell Signal 2018;42:54-66.
[Crossref] [Google Scholar] [PubMed]
- Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Ruth TY, Mangelsdorf DJ, et al. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell 2012;148(3):556-67.
[Crossref] [Google Scholar] [PubMed]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 2011;121(1):96-105.
[Crossref] [Google Scholar] [PubMed]
- Arner P. Human fat cell lipolysis: Biochemistry, regulation and clinical role. Best Pract Res Clin Endocrino Metab 2005;19(4):471-82.
[Crossref] [Google Scholar] [PubMed]
- Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, et al. Abdominal visceral and subcutaneous adipose tissue compartments: Association with metabolic risk factors in the Framingham heart study. Circulation 2007;116(1):39-48.
[Crossref] [Google Scholar] [PubMed]
- Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, et al. General and abdominal adiposity and risk of death in Europe. N Engl J Med 2008;359(20):2105-20.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Rimm EB, Stampfer MJ, Willett WC, Hu FB. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am J Clin Nutr 2005;81(3):555-63.
[Crossref] [Google Scholar] [PubMed]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006;444(7121):847-53.
[Crossref] [Google Scholar] [PubMed]
- Tontonoz P, Graves RA, Budavari AI, Erdjument-Bromage H, Lui M, Hu E, et al. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR7 and RXRa. Nucleic Acids Res 1994;22(25):5628-34.
[Crossref] [Google Scholar] [PubMed]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci 2013;110(9):3387-92.
[Crossref] [Google Scholar] [PubMed]
- Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab 2015;21(5):764-76.
[Crossref] [Google Scholar] [PubMed]
- Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PloS one 2010;5(12):e14199.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Sheng L, Miao H, Saunders TL, MacDougald OA, Koenig RJ, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem 2014;289(19):13000-9.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Xu R, Gerin I, Cawthorn WP, MacDougald OA, Chen XW, et al. SRA regulates adipogenesis by modulating p38/JNK phosphorylation and stimulating insulin receptor gene expression and downstream signaling. PloS one 2014;9(4):e95416.
[Crossref] [Google Scholar] [PubMed]
- Cooper DR, Carter G, Li P, Patel R, Watson JE, Patel NA. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1 cells. Genes 2014;5(4):1050-63.
[Crossref] [Google Scholar] [PubMed]
- Gernapudi R, Wolfson B, Zhang Y, Yao Y, Yang P, Asahara H, et al. MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol Cell Biol 2015;36(1):30-8.
[Crossref] [Google Scholar] [PubMed]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 2014;21(2):198-206.
[Crossref] [Google Scholar] [PubMed]
- Yi F, Yang F, Liu X, Chen H, Ji T, Jiang L, et al. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol 2013;10(6):990-1001.
[Crossref] [Google Scholar] [PubMed]
- Pang WJ, Lin LG, Xiong Y, Wei N, Wang Y, Shen QW, et al. Knockdown of PU. 1 AS lncRNA inhibits adipogenesis through enhancing PU. 1 mRNA translation. J Cell Biochem 2013;114(11):2500-12.
[Crossref] [Google Scholar] [PubMed]
- Wei N, Wang Y, Xu RX, Wang GQ, Xiong Y, Yu TY, et al. PU. 1 antisense lnc RNA against its m RNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet 2015;46(2):133-40.
[Crossref] [Google Scholar] [PubMed]
- Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Rep 2015;5(5):856-65.
[Crossref] [Google Scholar] [PubMed]
- Firmin FF, Oger F, Gheeraert C, Dubois-Chevalier J, Vercoutter-Edouart AS, Alzaid F, et al. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci Rep 2017;7(1):14087.
[Crossref] [Google Scholar] [PubMed]
- Lo KA, Huang S, Walet AC, Zhang ZC, Leow MK, Liu M, et al. Adipocyte long-noncoding RNA transcriptome analysis of obese mice identified Lnc-leptin, which regulates leptin. Diabetes 2018;67(6):1045-56.
[Crossref] [Google Scholar] [PubMed]
- Divoux A, Karastergiou K, Xie H, Guo W, Perera RJ, Fried SK, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity 2014;22(8):1781-5.
[Crossref] [Google Scholar] [PubMed]
- Li M, Sun X, Cai H, Sun Y, Plath M, Li C, et al. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim Biophys Acta 2016;1859(7):871-82.
[Crossref] [Google Scholar] [PubMed]
- Zhu XX, Yan YW, Chen D, Ai CZ, Lu X, Xu SS, et al. Correction: Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2018;9(25):17978.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Liu Y, Lu S, Yin L, Zong C, Cui S, et al. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int J Obes 2017;41(2):299-308.
[Crossref] [Google Scholar] [PubMed]
- Huang Y, Jin C, Zheng Y, Li X, Zhang S, Zhang Y, et al. Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci Rep 2017;7(1):8080.
[Crossref] [Google Scholar] [PubMed]
- Liu W, Ma C, Yang B, Yin C, Zhang B, Xiao Y. LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun 2017;493(3):1168-75.
[Crossref] [Google Scholar] [PubMed]
- Shang G, Wang Y, Xu Y, Zhang S, Sun X, Guan H, et al. Long non?coding RNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis of rat bone marrow mesenchymal stem cell by targeting miR?204?5p and miR?125a?3p. J Cell Physiol 2018;233(8):6041-51.
[Crossref] [Google Scholar] [PubMed]
- Nuermaimaiti N, Liu J, Liang X, Jiao Y, Zhang D, Liu L, et al. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem Biophys Res Commun 2018;495(2):1878-84.
[Crossref] [Google Scholar] [PubMed]
- Cai R, Sun Y, Qimuge N, Wang G, Wang Y, Chu G, et al. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim Biophys Acta Mol Cell Biol Lipids 2018;1863(4):420-32.
[Crossref] [Google Scholar] [PubMed]
- Li Z, Jin C, Chen S, Zheng Y, Huang Y, Jia L, et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem 2017;433:51-60.
[Crossref] [Google Scholar] [PubMed]
- Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep 2016;6(1):28897.
- Wang Y, Liu W, Liu Y, Cui J, Zhao Z, Cao H, et al. Long noncoding RNA H19 mediates LCoR to impact the osteogenic and adipogenic differentiation of mBMSCs in mice through sponging miR?188. J Cell Physiol 2018;233(9):7435-46.
[Crossref] [Google Scholar] [PubMed]
- Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 2014;55(3):372-82.
[Crossref] [Google Scholar] [PubMed]
- Mi L, Zhao XY, Li S, Yang G, Lin JD. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol Metab 2017;6(1):101-10.
[Crossref] [Google Scholar] [PubMed]
- Li S, Mi L, Yu L, Yu Q, Liu T, Wang GX, et al. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci 2017;114(34):E7111-20.
[Crossref] [Google Scholar] [PubMed]
- Bai Z, Chai XR, Yoon MJ, Kim HJ, Lo KA, Zhang ZC, et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulatorsk. PLoS Biol 2017;15(8):e2002176.
[Crossref] [Google Scholar] [PubMed]
- Cui X, You L, Li Y, Zhu L, Zhang F, Xie K, et al. A transcribed ultraconserved noncoding RNA, uc. 417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J 2016;30(12):4301-12.
[Crossref] [Google Scholar] [PubMed]
- Xiong Y, Yue F, Jia Z, Gao Y, Jin W, Hu K, et al. A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim Biophys Acta 2018;1863(4):409-19.
[Crossref] [Google Scholar] [PubMed]
- You L, Zhou Y, Cui X, Wang X, Sun Y, Gao Y, et al. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. J Cell Physiol 2018;233(1):313-24.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006;34(suppl_1):D668-72.
[Crossref] [Google Scholar] [PubMed]
- Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med 2018;24(3):257-77.
[Crossref] [Google Scholar] [PubMed]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009;136(4):777-93.
[Crossref] [Google Scholar] [PubMed]
- Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med 2019;70:307-21.
[Crossref] [Google Scholar] [PubMed]
- Sánchez Y, Huarte M. Long non-coding RNAs: Challenges for diagnosis and therapies. Nucleic Acid Ther 2013;23(1):15-20.
[Crossref] [Google Scholar] [PubMed]
- Gao H, Kerr A, Jiao H, Hon CC, Rydén M, Dahlman I, et al. Long non-coding RNAs associated with metabolic traits in human white adipose tissue. EBioMedicine 2018;30:248-60.
[Crossref] [Google Scholar] [PubMed]
- Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov 2017;16(3):167-79.
[Crossref] [Google Scholar] [PubMed]
- Werner A, Berdal A. Natural antisense transcripts: sound or silence. Physiol Genomics 2005;23(2):125-31.
[Crossref] [Google Scholar] [PubMed]
- Faghihi MA, Kocerha J, Modarresi F, Engström PG, Chalk AM, Brothers SP, et al. RNAi screen indicates widespread biological function for human natural antisense transcripts. PloS One 2010;5(10):e13177.
[Crossref] [Google Scholar] [PubMed]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science 2005;309(5740):1564-6.
[Crossref] [Google Scholar] [PubMed]
- Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013;14(12):880-93.
[Crossref] [Google Scholar] [PubMed]
- Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010;115(1):133-9.
[Crossref] [Google Scholar] [PubMed]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009;10(9):637-43.
[Crossref] [Google Scholar] [PubMed]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 2007;131(4):706-17.
[Crossref] [Google Scholar] [PubMed]
- Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010;115(1):133-9.
[Crossref] [Google Scholar] [PubMed]
- MacFarlane LA, Murphy PR. Regulation of FGF?2 by an endogenous antisense RNA: Effects on cell adhesion and cell?cycle progression. Mol Carcinog 2010;49(12):1031-44.
- Rubio A, Conrad M, Haselbeck RJ, GC K, Brown-Driver V, Finn J, et al. Regulation of mprF by antisense RNA restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob Agents Chemother 2011;55(1):364-7.
[Crossref] [Google Scholar] [PubMed]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012;491(7424):454-7.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Ji Y, Li M, Wang M, Yi X, Yin C, et al. Integrated analysis of long noncoding RNA and mRNA expression profile in children with obesity by microarray analysis. Sci Rep 2018;8(1):8750.
- Lu J, Wu X, Hong M, Tobias P, Han J. A potential suppressive effect of natural antisense IL-1β RNA on lipopolysaccharide-induced IL-1β expression. J Immunol 2013;190(12):6570-8.
[Crossref] [Google Scholar] [PubMed]
- Li Q, Su Z, Xu X, Liu G, Song X, Wang R, et al. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci 2012;109(35):14110-5.
[Crossref] [Google Scholar] [PubMed]
- Kimura T, Jiang S, Nishizawa M, Yoshigai E, Hashimoto I, Nishikawa M, et al. Stabilization of human interferon-α1 mRNA by its antisense RNA. Cell Mol Life Sci 2013;70:1451-67.
[Crossref] [Google Scholar] [PubMed]
- Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 1988;331(6153):277-80.
[Crossref] [Google Scholar] [PubMed]
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood 1997;90(2):489-519.
[Crossref] [Google Scholar] [PubMed]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU. 1 and the cell cycle controls myeloid differentiation. Science 2013;341(6146):670-3.
[Crossref] [Google Scholar] [PubMed]
- Ziliotto R, Gruca MR, Podder S, Noel G, Ogle CK, Hess DA, et al. PU. 1 promotes cell cycle exit in the murine myeloid lineage associated with downregulation of E2F1. Exp Hematol 2014;42(3):204-17.
[Crossref] [Google Scholar] [PubMed]
- Ridinger-Saison M, Evanno E, Gallais I, Rimmele P, Selimoglu-Buet D, Sapharikas E, et al. Epigenetic silencing of Bim transcription by Spi-1/PU. 1 promotes apoptosis resistance in leukaemia. Cell Death Differ 2013;20(9):1268-78.
[Crossref] [Google Scholar] [PubMed]
- Dahl R, Simon MC. The importance of PU. 1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis 2003;31(2):229-33.
[Crossref] [Google Scholar] [PubMed]
- Le Coz C, Nguyen DN, Su C, Nolan BE, Albrecht AV, Xhani S, et al. Constrained chromatin accessibility in PU. 1-mutated agammaglobulinemia patients. J Exp Med 2021;218(7):e20201750.
[Crossref] [Google Scholar] [PubMed]
- Wang F, Tong Q. Transcription factor PU. 1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol 2008;295(1):C213-20.
[Crossref] [Google Scholar] [PubMed]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, et al. PU. 1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 2008;22(15):2085-92.
[Crossref] [Google Scholar] [PubMed]
- Pang WJ, Lin LG, Xiong Y, Wei N, Wang Y, Shen QW, et al. Knockdown of PU. 1 AS lncRNA inhibits adipogenesis through enhancing PU. 1 mRNA translation. J Cell Biochem 2013;114(11):2500-12.
[Crossref] [Google Scholar] [PubMed]
- Liu F, He J, Wang H, Zhu D, Bi Y. Adipose morphology: A critical factor in regulation of human metabolic diseases and adipose tissue dysfunction. Obes Surg 2020;30:5086-100.
[Crossref] [Google Scholar] [PubMed]