- *Corresponding Author:
- Guohua Wang
Hunan Provincial People’s Hospital, The First Affiliated Hospital of Hunan Normal University, Changsha, Hunan 410005, People’s Republic of China
E-mail: w.guohua@outlook.com
| This article was originally published in a special issue,“Drug Discovery and Repositioning Studies in Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(4) Spl Issue “36-51” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To establish the information database of differentially expressed proteins in nucleus pulposus cells of the degeneration lumbar intervertebral disc. In this study, samples of nucleus pulposus cells from the degenerative lumbar intervertebral disc and normal lumbar intervertebral disc were collected. The tandem mass tag technique of proteomics was used to construct the map of differentially expressed proteome of nucleus pulposus cells between the lumbar intervertebral disc degeneration group and the control group. The differentially expressed proteins in nucleus pulposus cells in the degenerative lumbar intervertebral disc and the normal lumbar intervertebral disc are screened. Proteomics analysis of lumbar intervertebral disc degeneration provides some valuable differential protein markers for the diagnosis and treatment of intervertebral disc degeneration.
Keywords
Proteomics, lumbar intervertebral disc, annulus fibrosus, nucleus pulposus
Low Back Pain (LBP) is one of the most common medical diseases globally. It causes great suffering to many people around the world and also brings an enormous economic burden in many countries[1,2]. As early as 1934, some scholars found that lumbar Intervertebral Disc (IVD) factors may be the potential source of pain symptoms in LBP[3]. IVD is a soft tissue structure located between vertebral bodies of the vertebral column, which can form some intervertebral activities and distribute compression load evenly to adjacent vertebral bodies. IVD is composed of the Nucleus Pulposus (NP) in the center of the inner layer, the surrounding Annulus Fibrosus (AF), and the thin layer of cartilage Endplates (EP) between IVD and adjacent vertebral bodies[4,5]. Increasing evidence has shown that cells isolated from human IVD tissues, such as NP cells[6], AF cells[7], and EP cells[8], can exhibit most of the phenotypic markers similar to Mesenchymal Stromal Cells (MSCs). This supports the possibility that stem cells may be present in IVD. All of these cells maintain the potential for multiline differentiation, but they are diverse in protein phenotypic characteristics and biological capabilities. Studies have reported that NP-MSCs play a significant role in regulating apoptosis and dynamic balance and regeneration after IVD degeneration[9]. Adult IVD is usually vascular loss and neuron less, with very low cell density[10], which are perhaps one of the causes of lumbar disc degeneration. Freshly, it has been found that protein changes in IVD affect IVD Degeneration (IVDD)[11], which is not only a factor of LBP but also can cause sciatica and limited lumbar spine movement[12,13]. Some studies have also found that IVDD is associated with the Endplate Defect (ED) and facet joint orientation change to affect the development of LBP[14]. However, the precise mechanism of IVDD causing LBP has not been clarified[15], which is one of the research hotspots and focuses on the field of lumbar disc degenerative disease.
As early as 1994, Marc Wilkins and Keith Williams, two Australian researchers expounded the concept of the proteome as a whole set of proteins expressed by the genome for the first time at the Conference of Two-Dimensional Electrophoresis (2-DE) in Siena, followed by a paper published in the Journal of Electrophoresis in 1995[16]. Proteomics is a combination of proteins and genomics that includes all of the proteins present in a cell or even an organism. It allows accurate, quantitative and complete screening of most proteins expressed in genomes or most proteins in natural biological systems. Studies estimate that there are >100 000 proteins in the human proteome alone. This number illustrates the surprising challenge of proteomics analysis, which can be associated with a particular cell[17,18]. The study of IVD proteomics is a long and tortuous process, and it is difficult to evaluate how proteome changes in the nucleus pulpous cell line model, to find new biomarkers, and to understand the complexity of proteome and its impact on the balance between health and disease[19]. In the research field of IVDD, scholars devise that the inflammationrelated autocrine factor Chitinase-3-Like protein 1 (CHI3L1) showed high expression in degenerative NP cells by using high-throughput proteomics and isobaric Tags for Relative and Absolute Quantitation (iTRAQ). It was explained that the metabolic disorder of inflammation-oriented NP Extracellular Matrix (ECM) is one of the accounts of IVDD[20,21]. Research has reported the application of iTRAQ technology to the screening of differentially expressed proteins in embryonic, juvenile, and old bovine NP cells. Fetal collagen type XII and collagen type XIV, fibrinolectin in elderly NP cells, and collagen type XI enrichment in youth were found to help represent age-related IVDD[22].
To further explore human IVDD, this study collected different degrees of degenerative IVD clinically for proteomics analysis. In the pre-experiment, the quality control was performed with the emphasis on protein quantitative gumming, pre-mass spectrometry analysis, database comparison, labeling efficiency, and instrument status. In the formal experiment, peptide labeling, grading, mass spectrometry, database comparison, and bioinformatics analysis were carried out by the qualified samples in the preexperiment, and the experimental conclusion was finally obtained. This study aims to provide some valuable information for the diagnosis and treatment of IVDD through proteomics analysis of degenerative NP.
Materials and Methods
Collection of human lumbar NP tissue:
In this study, the NP specimens were collected from volunteers randomly enrolled in the spinal surgery department of the first affiliated hospital of Hunan Normal University (Hunan Provincial People’s Hospital), and IVD tissue was extracted during lumbar surgery. All enrolled patients voluntarily consented to sign the informed consent before surgery. The experiment was conducted in strict accordance with the requirements of the ethics committee.
Selection and implementation of clinical lumbar IVD case data:
The subjects of this study were patients who underwent lumbar surgery in the spinal surgery department of the First Affiliated Hospital of Hunan Normal University (Hunan Provincial People’s Hospital) from January 2018 to September 2020.
Inclusion criteria: Patients diagnosed with lumbar spinal stenosis, lumbar disc herniation, lumbar spondylolisthesis, or degenerative lumbar scoliosis requiring decompression and bone graft fusion were selected from the degenerative group and patients in the normal group who were diagnosed with idiopathic scoliosis or congenital spinal hemi vertebra deformity requiring intraoperative osteotomy and intervertebral bone graft were selected.
Exclusion criteria: Severe dysfunction of multiple organs, such as the heart, lung and brain; infective diseases of lumbar intervertebral space or severe systemic infection; lumbar tuberculosis infection; lumbar metastases of malignant tumors; lumbar metastases of multiple myeloma; pregnant and lactation women and patients suffering from systemic immune diseases.
Modified Pfirrmann grading of lumbar disc degeneration (Table 1) was utilized to classify the degree of degeneration of lumbar disc specimens by the same experienced Magnetic Resonance Imaging (MRI) diagnostic physician in combination with the causes and imaging data of the study subjects. The signal level of the NP and AF on MRI T2, the boundary between the posterior medial and lateral AF, and the height of the IVD was taken as the grading basis[23,24]. Then on 37 cases of information collected by the improved Pfirrmann grading of lumbar disc degeneration grade group, group A was suffering from lumbar degenerative diseases and modified Pfirrmann class IV or higher, in patients with lumbar spinal decompression and removal of the NP degeneration group. In group B; patients require intraoperative bone grafting due to idiopathic scoliosis or congenital hemi vertebra deformity (lumbar disc degeneration to grade I) were used as the normal control group.
| Grade | MRI-T2 signals of NP and AF | The boundary of AF medial to lateral IVD | The height of the IVD |
|---|---|---|---|
| I | Uniform high signal, consistent with Cerebrospinal Fluid (CSF) signal | Clear | Normal |
| II | High signal (>presacral fat, <CSF) with or without NP signal interruption | Clear | Normal |
| III | High signal<presacral fat | Clear | Normal |
| IV | Slight high signal (>AF) | Indistinct | Normal |
| V | Low signal=AF | Indistinct | Normal |
| VI | Low signal | Indistinct | Height is reduced by <30 % from normal |
| VII | Low signal | Indistinct | Height is reduced between 30 % and 60 % from normal |
| VIII | Low signal | Indistinct | Height is reduced by >60 % from normal |
Table 1: Modified Pfirrman Grading of Lumbar Disc Degeneration
NP and other IVD tissues were isolated by the same team of experienced spine surgeons during the decompression fusion procedure for NP removal. NP specimens were placed into 2 ml test tubes with sterile gloves, and 1 ml of sterile normal saline (0.9 % Sodium chloride (NaCl) isotonic solution) was added in each test tube to minimize the error caused by water content due to the preservation environment after NP removal. Then, the tube was sealed and immediately placed in a liquid nitrogen tank containing -196° in the operating room for cryogenic preservation, to reduce specimen transportation and sample deterioration caused by a long time during preservation. To decrease the experimental error brought by this situation.
Plan and implementation of pre-experiment:
Homogenate and Sonodynamic Therapy (SDT) pyrolysis extraction method: In this study, A1-4 was elected as the degenerated group and B1-4 as the normal control group. NP samples were chosen for precipitation and an appropriate amount of SDT lysate (4 % Sodium Dodecyl Sulfate (SDS), 100 mM Tris Hydrochloride (Tris HCl), pH 7.6) was added to the lysing matrix test tube. MP homogenizer was used for homogenization and crushing (24×2, 6.0 m/s, for 60 s, twice). After an ultrasound, they were subjected to a 15 min boiling water bath after centrifugation for 15 min. The supernatant was placed into a 0.22 micron centrifuge tube for filtration, and the filtrate was gathered. The quantitative method of protein is the selected Bicinchoninic Acid (BCA) assay method. Finally, they are packaged and stored in a low-temperature environment of -20°.
SDS-Polyacrylamide Gel Electrophoresis (SDSPAGE) method: Add 20 μg protein from each sample to 6X loading buffer. Then a boiling water bath was performed for 5 min. 12 % SDS-PAGE electrophoresis was executed for 40 min at 250 V constant pressure. Finally, it was stained with Coomassie Bright Blue.
Protein sample enzymatic hydrolysis (Filter- Aided Proteome Preparation (FASP)):
For the samples of group A and group B, 150 μg protein solution was respectively taken and put into Dithiothreitol (DTT) until the final concentration was adjusted to 100 mM. They were bathed in boiling water for 5 min and then cooled to room temperature. 200 μl UA buffer was added. Then centrifugation was carried out under a 30 kD ultrafiltration centrifuge tube for about 15 min, and the filtrate was abandoned. The above steps were repeated once.100 μl Iodoacetamide (IAA) buffer was added to it, and the vibration was done in 600 rpm for 1 min. Then, the centrifugation was conducted in room temperature for 30 min to avoid light and centrifuged again for 15 min. 100 μl UA buffer was added for centrifugation for 15 min and the previous step was repeated twice. After adding 100 μl 40 mm Ammonium Ion (NH4) Bicarbonate (HCO3) solution, centrifugation was carried out for 15 min. And this step was repeated twice. After centrifugation, 40 μl trypsin buffer was added to the solution, which was oscillated at 600 rpm for 1 min and stored in an incubator at 37° for 18 h. A pristine collection tube needs to be replaced and then centrifuged for 15 min. Then 20 μl 40 mm NH4HCO3 solution was added and centrifuged for 15 min to collect the obtained filtrate. Finally, the peptide was desalted using a C18 cartridge. After the peptide was lyophilized, 40 μl 0.1 % formic acid solution was added to resolution, and the peptide was quantified (Optical Density (OD) 280 nm). In all the above enzymatic hydrolysis steps, the centrifugation with a duration of 15 min was taken place at 12 500 g.
Mass spectrometric analysis and identification: All samples from both the degenerative group and the normal control group were separated by a system called the nanosecond flow rate easy NLC. 80 % acetonitrile aqueous solutions with 0.1 % formic acid were used as the buffer solution and buffer solution B with 0.1 % formic acid aqueous solution. The column was balanced with 100 % buffer solution A. At a flow rate of 300 nl/min, the separation of each sample is implemented by loading the sample into the column by an automatic sampler. The liquid phase gradient selected in this experiment was a 2 h gradient: The time was between 0 min and 5 min, and the buffer liquid was 6 %. The time varies from 5 min to 100 min, and the linear gradient range of buffer liquid A is between 6 %-28 %. The time was between 100 min and 110 min, and the linear gradient range of buffer liquid A was between 28 %-38 %. The time was between (110-115) min, and the linear gradient range of buffer liquid A was between 38 % and 100 %. The time ranges from (115-120) min, and buffer liquid a can be maintained at 100 %.
The Q executive plus mass spectrometer was selected for further mass spectrometry analysis of the NP isolated by chromatography. The conditions for mass spectrometry analysis are as follows; the mother ions and positive ions with a scanning range of 350-1800 mass charge ratio must be selected in the experiment. The resolution of primary and secondary mass spectra should be 70 000 and 35 000, respectively. The first and second level maximized IT should be set to 50 ms and 45 ms, respectively. The automatic gain control target value of the first stage mass spectrometry requires an accurate setting of 3E6 and the analysis time is 2 h.
Selection and data analysis of mass spectrometry database: The key step for qualitative analysis of mass spectral data consists of select the appropriate protein sequence database. The database utilized in the experiment was Uniprot Homo sapiens (H. sapiens) 20386 20180905 comprehensive protein database. The database is accessible to download at http://www.uniprot.org. Mass spectrometry raw file processing: In this experiment, the QexActive plus high-resolution mass spectrometer was used to fill out Tandem Mass Tag (TMT) quantitative proteomics analysis. QexActive belongs to the Orbitrap type mass spectrometer, which provides the data processing tool for this study, and also lays a broad path for the quantitative data of proteomics. The MS2 map constructed by QexActive effectively improves the quantitative ability, sensitivity, and ion transfer with low mass number. In this study, the database retrieval is accomplished by transferring the software to the Mascot Server 2.6. The process involves changing the initial map information. RAW format obtained by QexActive into. MGF format by proteome discoverer 2.1 software[25]. Next, an a DAT file was obtained on the MASCOT server and submitted to the software through Proteome Discoverer 2.1. The screening standard for the data is the False Discovery Rate (FDR), which is <0.01 because the FDR obtained after correcting the p-value of differential significance is an important indicator for differential expression screening. The qualitative results achieved in this way are reliable.
Sample evaluation and mass spectrometry: The NP evaluation measured according to the pre-experiment is carried out based on sample concentration, volume and total amount. The standard is divided into the following three points; the parallelism of each sample submitted for testing is proper. The parallelism within the sample group was superior, but the parallelism between the two groups was poor. The parallelism of the samples locally submitted for inspection in the group is not satisfactory, so these samples need to be submitted for inspection again.
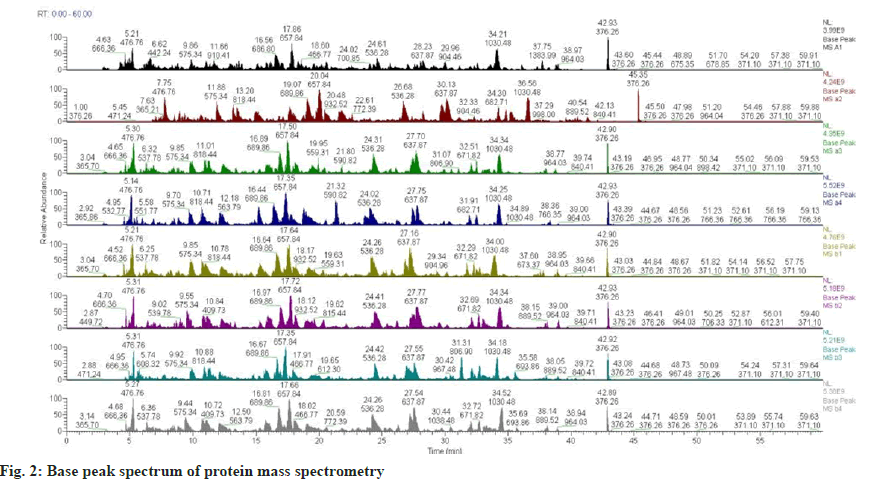
Base peak chromatogram: The chromatogram can compare the distribution of signal peaks between different samples and the similarity of mass spectrum signal intensity, and can reflect the composition complexity, chromatographic separation degree, and peptide signal intensity of the protein in the sample. It was further determined whether quality. The total amount and parallelism of the selected experimental samples could meet the requirements of the subsequent experiments.
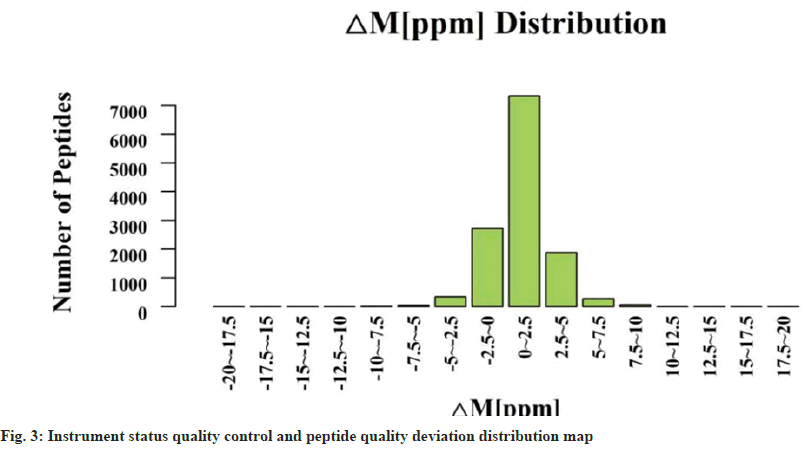
Quality control of sample marking efficiency and instrument status: To identify the accuracy of proteins, it is necessary not only to label the protein peptides detected in the NP samples submitted for testing but also to carry out quality control on the detection instrument-Liquid Chromatography tandem Mass Spectrometry (LC-MS/MS). When quality deviations of peptide segments ≥90 % are within the range of 8 ppm, it indicates that the instrument is in satisfactory condition and the data results are relatively reliable.
Sample marker and differential protein was detected in the formal experiment:
NP samples were labeled and divided into group A was degenerative group A1-4 and group B was normal control group B1-4. H. sapiens database name was chosen for the qualitative detection of proteins. In this study, the total number of peptides, the number of single peptides, the number of corresponding peptides, the number of proteins that could be screened, and the number of secondary mass spectra were detected respectively. In the process of quantitative protein detection of samples, the following two conditions must be met in screening to be used as differentially expressed proteins; the differential expression ratio was 1.2 times higher than that of up-regulation or down-regulation. In the independent sample t-test, p<0.05. Eventually, total and up-down-regulated differential protein numbers were counted in the experiment.
Protein cluster analysis:
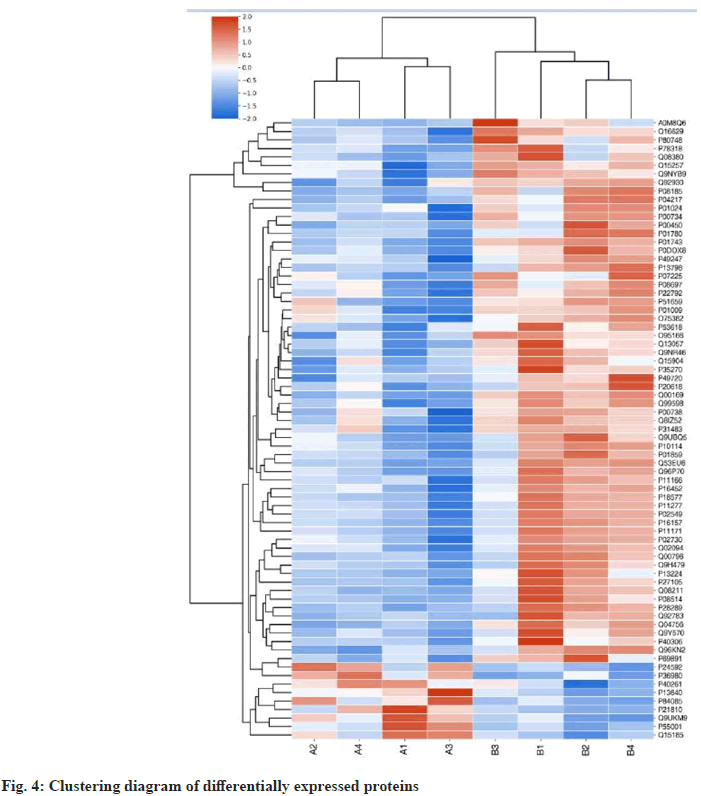
Cluster diagrams were constructed for all the differentially identified proteins in the degenerative group and normal samples. In the figure, the differential protein expression levels are illustrated in red, the down-regulation of differential protein expression levels is shown in green, and the degree of down-regulation is shown in the shade of color. The classification and grouping of differential protein data in the cluster diagram are built on similarity.
GO enrichment analysis:
GO annotation can be used to analyze the gene and protein functions of the samples from the aspects of cell components, biological processes and molecular functions[26]. In the experiment, this process can be divided into sequence alignment, item extraction, GO annotation and supplementary annotation[27]. In the subsequent enrichment analysis, Fisher’s exact test was used to further observe the protein enrichment level.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis:
NP proteins were annotated based on the KEGG database. The protein enrichment level of samples in each pathway was tested and counted. Signaling pathways with significant correlation were screened when p<0.05 was convinced. Enriched and differentially expressed proteins shared by GO annotation and KEGG pathways were identified.
Statistical analysis:
The data in this study were analyzed using GraphPad Prism 8.02 software. If the statistical data presented normal distribution and satisfied the homogeneity of variance, the measurement data would be represented by the mean±standard deviation. If the statistical data presented skewed distribution or variance heterogeneity, the median and quartile would be represented. After the logarithm was converted into a normal distribution and the homogeneity of variance was satisfied, statistical analysis would be completed. The purpose of this study consisted of the screen for differential protein between NP of degenerative IVD and normal IVD. When p≥0.05, the difference was not statistically significant, and the overall mean between the two groups was the same. When p<0.05, the difference was statistically significant, and the overall mean between the two groups was not exactly equal.
Results and Discussion
The results of protein quantification and mass spectrometry are described below as shown in Table 2. In sample evaluation, the protein of group A (degenerative group) and group B (normal control group) in the pre-experiment all reached the detection standard. The total amount of protein can be identified twice or more. In mass spectrometric evaluation, the parallelism between the two groups was general. Partial parallelism in the degenerative group was weak. Intra-group parallelism was favorable in the normal control group.
| Sample name and number | 101 | 104 | 105 | 106 | 102 | 103 | 107 | 108 |
|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | B1 | B2 | B3 | B4 | |
| Concentration (µg/µl) | 6.2 | 10.0 | 6.1 | 11.8 | 8.9 | 8.1 | 9.9 | 9.5 |
| Volume (µl) | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
| Gross (µg) | 1240 | 2000 | 1220 | 2360 | 1780 | 1620 | 1980 | 1900 |
| Sample evaluation | I | I | I | I | I | I | I | I |
| Database | Uniprot H. Sapiens _20386_20180905 | |||||||
| Proteome | 364 | 808 | 433 | 803 | 483 | 410 | 513 | 412 |
| (Independent peptide ≥2) proteome | 262 | 517 | 298 | 501 | 340 | 270 | 348 | 279 |
| Mass spectrum evaluation | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
Table 2: Protein Quantification and Mass Spectrometry
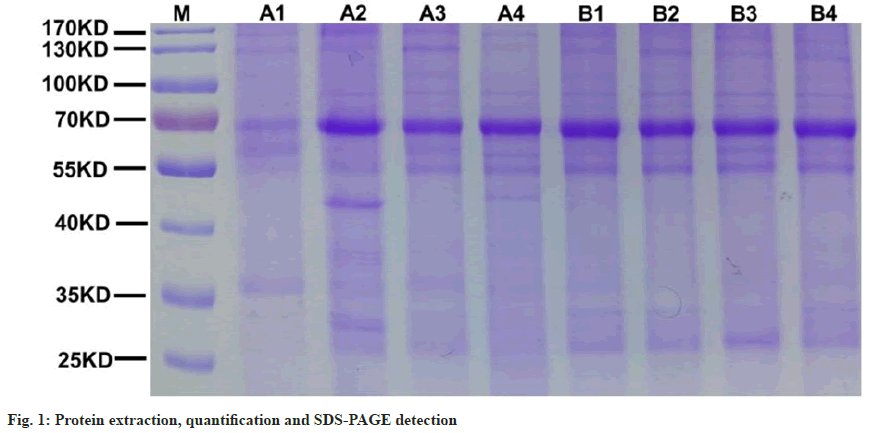
In the pre-experiment, the results of protein SDSPAGE and mass spectrometry analysis of the base peak spectrum are summarized as follows. The consequences of mass spectrometry showed that there were no abnormalities in the way of chromatographymass spectrometry and enzymatic hydrolysis of protein. The number of proteins in the samples of the degenerative group was significantly different. The parallelism of the degenerative group was general. SDS-PAGE and protein quantification showed that the total amount of NP was sufficient and the quality was high. However, the parallel difference between A1 (101) sample and other samples is great. To sum up to ensure the quality of protein data identified in the follow-up. In the follow-up experiment, A2 (the first sample send). A3 (the first sample sends) and A4 (the first sample sends) were used in group A. Reselect the sample of A1 for the second time and submit it for inspection. In the control group, B1, B2, B3 and B4 samples were utilized for subsequent experiments. Protein-SDS-PAGE (fig. 1) method found that 8 NP samples (group A1-4, group B1-4) could meet the detection standard, and the feasible detection was greater than or equal to two times.
The results of base peak spectrum analysis by mass spectrometry (fig. 2) showed that the intragroup parallelism of the NP samples (A1-4 of the degenerated group and B1-4 of the control group) in the pre-mass spectrometry library was superb. The parallelism between the two groups was worse as shown in Table 3.
| Sample number | A1 | A2 | A3 | A4 | B1 | B2 | B3 | B4 |
|---|---|---|---|---|---|---|---|---|
| Proteome | 364 | 808 | 433 | 803 | 383 | 410 | 513 | 412 |
| Peptides | 262 | 517 | 298 | 501 | 340 | 270 | 348 | 279 |
Table 3: Data of Parallel Lines were Analyzed by Mass Spectrometry
A1-4 and B1-4 were selected for quality control of marking efficiency. The marking efficiency of this project is 99.5 % as shown in Table 4. The mark is complete. The protein quantity and differential analysis list showed significant protein expression and difference. The quality deviations of the ≥99.6 % peptides in this experiment were all within 8 ppm, which indicated that the instrument was in good condition and the results of experimental data were reliable (fig. 3). Subsequent experiments can be performed.
| Number of peptides | Labeled peptides | Mark efficiency |
|---|---|---|
| 2528 | 2516 | 99.5 % |
Table 4: Number of Protein-Peptide Markers and Efficiency Values
The database selected in the experiment was H. sapiens. The total number of proteins is 1885. The number of peptides was 12 678. The number of the single peptide was 11 626. The number of peptides corresponding to the mass spectrum was 51 303. The number of grade 2 mass spectrogram was 284 386 (Table 5).
| Database | Proteins | Peptides | Single peptide | Peptides corresponding to the mass spectrum | Grade 2 mass spectrogram |
|---|---|---|---|---|---|
| H. sapiens | 1885 | 12 678 | 11 626 | 51 303 | 284 386 |
Table 5: Protein Characterization Results
In the experiment, there were 9 differential proteins upregulated (Table 6). Increase diversity protein genes are presented below; Metallothionein 1G (MT1G), Insulin-Like Growth Factor-Binding Protein 6 (IGFBP6), ADP-Ribosylation Factor 5 (ARF5), Microfibrillar-Associated Protein 2 (MFAP2), Complement Factor H-Related Protein 2 (CFHR2), RNA-Binding Protein Raly (RALY), Biglycan (BGN), Nicotinamide N-Methyltransferase (NNM), Prostaglandin E Synthase 3 (PTGES3). There were 64 down-regulated differential proteins (Table 7). The genes for the down-regulated differential proteins are listed below; Alpha-2- antiplasmin SERPINF2, Tropomodulin-1 (TMOD1), Corticosteroid-Binding Globulin (SERPINA6), Complement C3 (C3), Ribose-5-Phosphate Isomerase (RPIA), Proteasome Subunit Beta Type (PSMB)- 1, Acylamino-Acid-Releasing Enzyme (APEH), Phosphatidylinositol Transfer Protein Alpha Isoform (PITPNA), Serine/Threonine-Protein Phosphatase 2A Activator (PTPA), Sorbitol Dehydrogenase (SORD), SERPINA1, Gamma-Aminobutyric Acid Receptor-Associated Protein (GABARAP), Beta- Ala-His Dipeptidase (CNDP1), Ceruloplasmin (CP), Ras-related Protein Rab-8B (RAB8B), Peroxisomal Multifunctional Enzyme Type 2 (HSD17B4), Prothrombin (F2), Tripartite Motif- Containing Protein 3 (TRIM3), Chondroitin Sulfate Synthase 2 (CHPF), Proteasome Subunit Beta Type- 3 (PSMB3), Protein Phosphatase Methylesterase 1 (PPME1), Ras-Related Protein Rap-2A (RAP2A), Proteasome Subunit Beta Type-10 (PSMB10), Abl Interactor 2 (ABI2), V-Type Proton ATPase Subunit S1 (ATP6AP1), Hepatocyte Growth Factor Activator (HGFAC), Nucleolysin TIA-1 Isoform p40 (TIA1), ATP-Dependent RNA Helicase A (DHX9), Stomatin (STOM), Immunoglobulin Heavy Variable 1 (IGHV1)-46, Translin-Associated Protein X (TSNAX), Vitamin K-Dependent Protein S (PROS1), Fructosamine-3-Kinase (FN3K), Eukaryotic Translation Initiation Factor 3 Subunit K (EIF3K), Alpha-1B-Glycoprotein (A1BG), Coatomer Subunit Beta (COPB1), Haptoglobin (HP), Serine/Arginine- Rich Splicing Factor 7 (SRSF7), Immunoglobulin Heavy Constant Gamma 2 (IGHG2), spectrin beta chain, Erythrocytic (SPTB), Bifunctional Coenzyme A Synthase (COASY), Ankyrin-1 (ANK1), Signal transducing Adapter Molecule (STAM), Spectrin Alpha Chain, Erythrocytic 1 (SPTA1), Erythrocyte Membrane Protein Band 4.1 (EPB41), IGHV7, Erythrocyte Membrane Protein Band 4.2 (EPB42), Immunoglobulin-Binding Protein 1 (IGBP1), Solute Carrier Family 4 Member 1 (SLC4A1) Galectin- 3-Binding Protein (LGALS3BP), Endophilin-B2 (SH3GLB2) Integrin Alpha-IIB (ITGA2B), blood group Rh (CE) Polypeptide (RHCE), Solute Carrier Family 2, Facilitated Glucose Transporter Member 1 (SLC2A1), Glycerol-3-Phosphate Acyltransferase 3 (GPAT3), Importin-9 (IPO9), Sepiapterin Reductase (SPR), Immunoglobulin Lambda Variable 3-21 (IGLV3-21), Ammonium Transporter Rh Type A (RHAG), Carboxypeptidase N Subunit 2 (CPN2), Platelet Glycoprotein IB Beta Chain (GP1BB), Hemoglobin Subunit Gamma-1 (HBG1), Immunoglobulin Lambda Constant 7 (IGLC7). There were 73 differential proteins detected by screening. NP of the degenerative group and the control group were qualitatively identified and obtained 1885 proteins. After quantitative statistics, 73 differentially expressed proteins were found. A cluster plot was drawn with 73 differentially expressed proteins. In fig. 4, the up-down degree of the differentially expressed protein amount is indicated by the color shade. The amount of up-regulated differentially expressed proteins is depicted in red. The amount of down-regulated differentially expressed proteins is depicted in green. The clustering results in the fig. 4 show that there is a high similarity among A1, A2, A3, and A4 in the degenerative group. There was a high similarity among B1, B2, B3 and B4 in the control group. It is ascertained that the target differential protein obtained by screening is reasonable for the experiment. There was a low similarity between the degenerative group and the control group. This indicates that each sample is less affected by biological treatment during the experiment.
| Protein# | Description | Gene | A/B# | p |
|---|---|---|---|---|
| P13640 | Metallothionein-1G | MT1G | 2.27694 | 0.04436 |
| P24592 | Insulin-like growth factor-binding protein 6 | IGFBP6 | 1.55233 | 0.01729 |
| P84085 | ADP-ribosylation factor 5 | ARF5 | 1.45248 | 0.02756 |
| P55001 | Microfibrillar-associated protein 2 | MFAP2 | 1.37954 | 0.04765 |
| P36980 | Complement factor H-related protein 2 | CFHR2 | 1.36436 | 0.01114 |
| Q9UKM9 | RNA-binding protein Raly | RALY | 1.33987 | 0.01959 |
| P21810 | Biglycan | BGN | 1.26565 | 0.03014 |
| P40261 | Nicotinamide N-methyltransferase | NNMT | 1.23283 | 0.04419 |
| Q15185 | Prostaglandin E synthase 3 | PTGES3 | 1.22503 | 0.03074 |
Notes: (#): Protein codes from UniProt database and (#A/B): Represents the multiple of the difference in protein expression between the two groups
Table 6: Nine Differentially Expressed Proteins were Upregulated
| Protein# | Description | Gene | A/B# | p |
|---|---|---|---|---|
| P08697 | Alpha-2-antiplasmin | SERPINF2 | 0.82315 | 0.03334 |
| P28289 | Tropomodulin-1 | TMOD1 | 0.82255 | 0.01541 |
| P08185 | Corticosteroid-binding globulin | SERPINA6 | 0.81919 | 0.01164 |
| P01024 | Complement C3 | C3 | 0.8186 | 0.03566 |
| P49247 | Ribose-5-phosphate isomerase | RPIA | 0.81736 | 0.04521 |
| P20618 | Proteasome subunit beta type-1 | PSMB1 | 0.81511 | 0.04846 |
| P13798 | Acylamino-acid-releasing enzyme | APEH | 0.81283 | 0.01706 |
| Q00169 | Phosphatidylinositol transfer protein alpha isoform | PITPNA | 0.80955 | 0.00032 |
| Q15257 | Serine/threonine-protein phosphatase 2A activator | PTPA | 0.80646 | 0.02266 |
| Q00796 | Sorbitol dehydrogenase | SORD | 0.8046 | 0.01967 |
| P01009 | Alpha-1-antitrypsin | SERPINA1 | 0.80239 | 0.04011 |
| O95166 | Gamma-aminobutyric acid receptor-associated protein | GABARAP | 0.79856 | 0.00797 |
| Q96KN2 | Beta-Ala-His dipeptidase | CNDP1 | 0.79811 | 0.01081 |
| P00450 | Ceruloplasmin | CP | 0.79757 | 0.01886 |
| Q92930 | Ras-related protein Rab-8B | RAB8B | 0.79757 | 0.01265 |
| P51659 | Peroxisomal multifunctional enzyme type 2 | HSD17B4 | 0.79735 | 0.03309 |
| P00734 | Prothrombin | F2 | 0.79654 | 0.01127 |
| O75382 | Tripartite motif-containing protein 3 | TRIM3 | 0.7935 | 0.04045 |
| Q8IZ52 | Chondroitin sulfate synthase 2 | CHPF | 0.79171 | 0.02185 |
| P49720 | Proteasome subunit beta type-3 | PSMB3 | 0.78829 | 0.02356 |
| Q9Y570 | Protein phosphatase methylesterase 1 | PPME1 | 0.78607 | 0.0307 |
| P10114 | Ras-related protein Rap-2a | RAP2A | 0.78509 | 0.02444 |
| P40306 | Proteasome subunit beta type-10 | PSMB10 | 0.78328 | 0.03993 |
| Q9NYB9 | Abl interactor 2 | ABI2 | 0.77541 | 0.0142 |
| Q15904 | V-type proton ATPase subunit S1 | ATP6AP1 | 0.77288 | 0.04239 |
| Q04756 | Hepatocyte growth factor activator | HGFAC | 0.77209 | 0.00124 |
| P31483 | Nucleolysin TIA-1 isoform p40 | TIA1 | 0.76913 | 0.04844 |
| Q08211 | ATP-dependent RNA helicase A | DHX9 | 0.766 | 0.01064 |
| P27105 | Stomatin | STOM | 0.76362 | 0.0077 |
| P01743 | Immunoglobulin heavy variable 1-46 | IGHV1-46 | 0.7625 | 0.00076 |
| Q99598 | Translin-associated protein X | TSNAX | 0.75786 | 0.00944 |
| P07225 | Vitamin K-dependent protein S | PROS1 | 0.7573 | 0.04194 |
| Q9H479 | Fructosamine-3-kinase | FN3K | 0.7567 | 0.03911 |
| Q9UBQ5 | Eukaryotic translation initiation factor 3 subunit K | EIF3K | 0.74422 | 0.02692 |
| P04217 | Alpha-1B-glycoprotein | A1BG | 0.74292 | 0.00549 |
| P53618 | Coatomer subunit beta | COPB1 | 0.74276 | 0.02699 |
| P00738 | Haptoglobin | HP | 0.73951 | 0.03446 |
| Q16629 | Serine/arginine-rich splicing factor 7 | SRSF7 | 0.73929 | 0.00862 |
| P01859 | Immunoglobulin heavy constant gamma 2 | IGHG2 | 0.73595 | 0.02967 |
| P11277 | Spectrin beta chain, erythrocytic | SPTB | 0.73289 | 0.0205 |
| Q13057 | Bifunctional coenzyme A synthase | COASY | 0.73176 | 0.02061 |
| P16157 | Ankyrin-1 | ANK1 | 0.72749 | 0.01102 |
| Q92783 | Signal transducing adapter molecule 1 | STAM | 0.72504 | 0.04575 |
| P02549 | Spectrin alpha chain, erythrocytic 1 | SPTA1 | 0.72451 | 0.02174 |
| P11171 | Erythrocyte Membrane Protein Band 4.1 | EPB41 | 0.71991 | 0.01606 |
| P0DOX8 | Immunoglobulin lambda-1 light chain | 0.71248 | 0.01315 | |
| P01780 | Immunoglobulin heavy variable 3-7 | IGHV3-7 | 0.7105 | 0.04093 |
| P16452 | Erythrocyte membrane protein band 4.2 | EPB42 | 0.70977 | 0.02184 |
| P78318 | Immunoglobulin-binding protein 1 | IGBP1 | 0.70249 | 0.04211 |
| P02730 | Solute Carrier Family 4 Member 1 | SLC4A1 | 0.69527 | 0.02264 |
| Q08380 | Galectin-3-binding protein | LGALS3BP | 0.69026 | 0.02743 |
| Q9NR46 | Endophilin-B2 | SH3GLB2 | 0.68159 | 0.00738 |
| P08514 | Integrin alpha-IIb | ITGA2B | 0.67015 | 0.0195 |
| P18577 | Blood group Rh(CE) polypeptide | RHCE | 0.6488 | 0.00649 |
| P11166 | Solute carrier family 2, facilitated glucose transporter member 1 | SLC2A1 | 0.64779 | 0.02963 |
| Q53EU6 | Glycerol-3-phosphate acyltransferase 3 | GPAT3 | 0.62946 | 0.0058 |
| Q96P70 | Importin-9 | IPO9 | 0.62866 | 0.00946 |
| P35270 | Sepiapterin reductase | SPR | 0.60881 | 0.03237 |
| P80748 | Immunoglobulin lambda variable 3-21 | IGLV3-21 | 0.57947 | 0.03628 |
| Q02094 | Ammonium transporter Rh type A | RHAG | 0.5542 | 0.0223 |
| P22792 | Carboxypeptidase N subunit 2 | CPN2 | 0.54848 | 0.01883 |
| P13224 | Platelet glycoprotein Ib beta chain | GP1BB | 0.53973 | 0.01445 |
| P69891 | Hemoglobin subunit gamma-1 | HBG1 | 0.51966 | 0.01723 |
| A0M8Q6 | Immunoglobulin lambda constant 7 | IGLC7 | 0.3893 | 0.04266 |
Notes: (#): Protein codes from UniProt database and (#A/B): Represents the multiple of the difference in protein expression between the two groups
Table 7: Sixty-Four Differentially Expressed Proteins were Down-Regulated
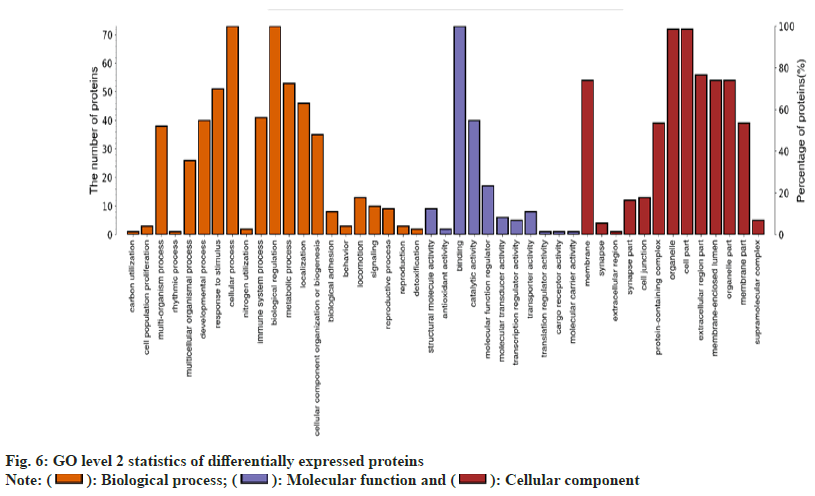
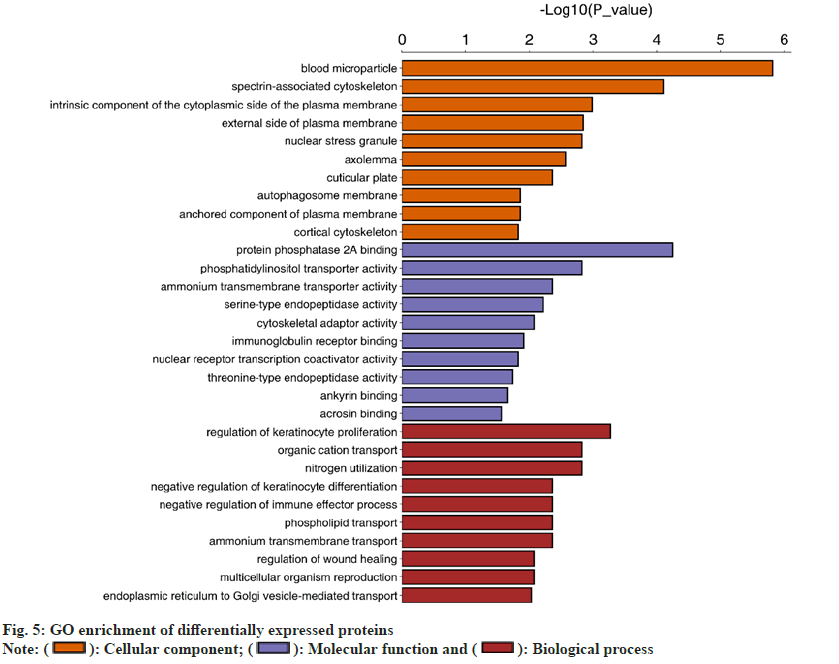
In terms of the biological processes involved, there is one protein involved in carbon utilization. There are three proteins involved in cell population proliferation. The protein of the multi-biological process is 38. The number of proteins engaged in response to stimuli is 52. 73 proteins participated in cellular processes. One protein is associated with nitrogen utilization. One protein is related to rhythm. The number of proteins implicated in the immune system is 41. There are 73 proteins involved in biological regulation. There are 53 proteins involved in the metabolic process. 45 proteins are involved in localization. There are 35 proteins involved in the organization or biogenesis of cell components. There are six proteins involved in bioadhesion. There are three proteins involved in behavior. There are twentyfive proteins involved in multicellular processes. The number of proteins involved in exercise is twelve. There are 10 proteins involved in signaling. There are 9 proteins involved in the reproductive process. There are three proteins implicated in reproduction. The number of proteins during development is forty. There are two proteins related to detoxification (fig. 5).
In terms of molecular function, there are nine proteins involved in the structural molecular activity. Two proteins are linked with antioxidant activity. There are 73 proteins involved in the binding activity. There are 38 proteins involved in catalytic activity. 14 proteins are involved in molecular function regulation. Four proteins participate in molecular transduction activity. Three proteins are related to transcriptional regulation. Five proteins are implicated in transporter activity. One protein is active in translation regulation. One protein is involved in transporter receptor activity. One protein is associated with the activity of the molecular carrier. In terms of cell composition, there are 53 proteins in the membrane. Three proteins make up a synapse. The extracellular domain consists of one protein. 11 proteins make up the synaptic part. 12 proteins make up the connections between cells. 38 proteins make up the protein-containing complex. There are 73 proteins that make up the organelle. There are 73 proteins that compose part of the cell. The extracellular region consists of 54 proteins. There are 52 proteins in the membrane encapsulation cavity. 52 proteins make up the organelle part. The membrane consists of 38 proteins. The supramolecular complex consists of four proteins.
In terms of biological processes, 73 differentially expressed proteins are associated with biological regulation. In terms of molecular function, 73 differential proteins play a part in the binding activity. In terms of cellular components, 73 differential proteins are involved in organelle and cellular components.
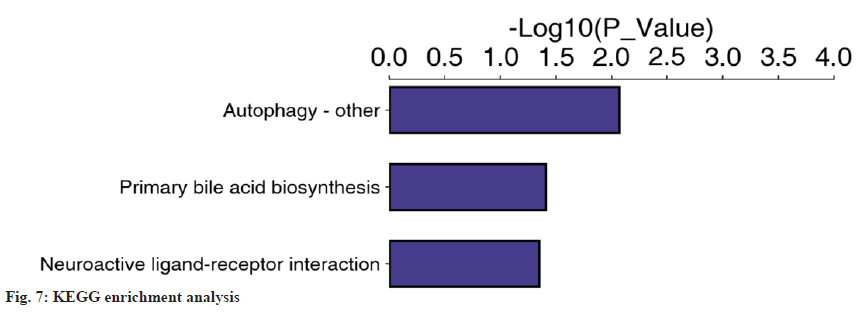
The differentially expressed proteins were annotated based on GO enrichment and KEGG enrichment. Fisher’s exact test was performed. The GO enrichment results indicated that 48 of the 73 differential proteins were significantly enriched in 30 GO terms belonging to biological processes (fig. 6). Then, KEGG enrichment was adopted. When the corrected p<0.05, the difference was statistically significant. Three pathways (fig. 7) were found; autophagy-other, primary bile acid biosynthesis and neuroactive ligand-receptor interaction. In the course of this experiment, we focused on the common differentially expressed proteins in GO and KEGG enrichment, of which there were five types as shown in Table 8.
| Proteins | Definition | GO-ID# | KO# | Map name# |
|---|---|---|---|---|
| P01024 | CO3_HUMAN complement C3 | 0006957 | 04080 | Neuroactive ligand-receptor interaction |
| P00734 | THRB_human prothrombin | 0016021 | 04080 | Neuroactive ligand-receptor interaction |
| O95166 | GBRAP_HUMAN gamma-aminobutyricacidreceptor-associated | 0005930 | 04136 | Autophagy-other |
| P78318 | IGBP1_HUMAN immunoglobulin-binding 1 | 0051721 | 04136 | Autophagy-other |
| P51659 | DHB4_HUMAN peroxisomal multifunctional enzyme type 2 | 0016508 | 00120 | Primary bile acid biosynthesis |
Notes: (#GO-ID): The ID of the annotated Go term; (#KO): The corresponding protein ID of the target protein in the KEGG database and (#Map Name): Name of the pathway in which the target protein is involved
Table 8: Common Differentially Expressed Proteins in Go and Kegg Enrichment
Studies have proved that Degenerative Lumbar Disc Disease (DDD) (such as lumbar disc herniation, lumbar canal stenosis, lumbar spondylolisthesis, degenerative lumbar scoliosis, etc.,) is the potential cause of lumbar back pain. DDD is an NP-mediated biological change caused by IVD under the comprehensive action of multiple factors[28]. The loss of proteoglycan and apoptosis of IVD cells may affect the degenerative process of the lumbar disc. DDD process involves the destruction of IVD structural integrity and changes in cell number and composition[29].
IVDD is characterized by progressive NP cell dehydration and annular rupture. In IVDD, interactions mediated by ECM-Integrins (ITG) can affect cell proliferation, cell signaling, cell survival and protein production. NP may express changes in NP-ECM interactions with age[30]. Previous studies have shown that intervertebral macrophage infiltration leads to inflammatory pain response, leading to blocked controlled CCR3-CCL1 signal[31]. Fibronectin fragments produced during NP degeneration are known to upregulate matrix metalloproteinases such as MMP-3 and MMP-13 through the Extracellular Signal-Regulated Kinase (ERK) pathway, leading to NP degeneration[32]. Axon growth is stimulated in degenerative NP but inhibited in normal NP. This suggests that NP plays a part in neuroregulation during degeneration[33].
Proteomics methods are in constant development. Relative and absolute quantitative protein results can be obtained by using tandem mass spectrometry with high accuracy and sensitivity, without the need for gel. The TMT and iTRAQ are part of the major advances in the rise of proteomics[34,35]. In the field of spine surgery, Sun et al.[36] used proteomics techniques to screen 16 serum differential proteins between patients with congenital scoliosis and patients without scoliosis and constructed a database of differentially expressed proteins. Xie et al.[37] used proteomics to analyze the serum samples between the group with lumbar disc herniation and the group without lumbar disc degeneration by 2D electrophoresis and mass spectrometry and found 6 different proteins. This study is a direct examination of lumbar disc NP. It is somewhat innovative in comparison with previous proteomic studies of serum samples.
This study analyzed the protein profiles of human IVDs and their quantitative changes during disc degeneration. The study has mapped a total of 1885 proteins present in NP, including 73 that were prevalent in both the degenerative and normal control groups. In quantitative differential proteomics analysis of normal and degenerative IVDs, we found differential regulation of 9 up-regulated and 64 down-regulated proteins, respectively, in NP tissues. GO enrichment and level 2 were used to calculate the differentially expressed proteins. The experimental results indicated that all the differential proteins participated in the biological regulation of biological processes. In terms of molecular function, all the differential proteins were involved in the binding activity. In terms of cellular components, all differential proteins were involved in organelle and cellular components. According to Fisher’s exact test, enrichment results of biological proteins indicated that 48 of the 73 different proteins in NP cells were significantly enriched in 30 GO terms belonging to biological processes. 3 pathways were identified by KEGG enrichment; autophagy-other, primary bile acid biosynthesis and neuroactive ligand-receptor interaction. 5 common differentially expressed proteins were identified in the three screened pathways. Besides, various signaling pathways, cellular interactions, inhibition and activation of IVD degeneration may be linked.
A total of 73 differentially expressed proteins were screened for NP in human lumbar IVDs using proteomics analysis. 9 of the up-regulated differentially expressed proteins were associated with the induction of lumbar disc degeneration. 64 down-regulated differentially expressed proteins were involved in the maintenance of normal lumbar disc degeneration. Species pathways of differentially expressed proteins were screened and differentially expressed proteins that were enriched together in the pathways were found. A database of NP differentially expressed proteins in degenerative lumbar IVDs was established. In this study, our preliminary proteomics analysis of human IVD provided useful information on the differential regulation of various proteins in IVDD. This may link the function and expression of proteins in IVDD in novel ways.
Acknowledgements:
The authors acknowledge the Scientific Research Project of Hunan Provincial Department of Education (Project No: 21C0003).
Conflict of interests:
The authors declared no conflict of interests.
References
- Cheung JP, Luk KD. The relevance of high-intensity zones in degenerative disc disease. Int Orthop 2019;43:861-7.
[Crossref] [Google Scholar] [PubMed]
- Gansau J, McDonnell EE, Buckley CT. Development and characterization of antacid microcapsules to buffer the acidic intervertebral disc microenvironment. J Biomed Mater Res A 2024.
[Crossref] [Google Scholar] [PubMed]
- Mixter WJ, Barr JS. Rupture of the intervertebral disc with involvement of the spinal canal. N Engl J Med 1934;211(5):210-5.
- Wu H, Zeng X, Yu J, Shang Y, Tu M, Cheang LH, et al. Comparison of nucleus pulposus stem/progenitor cells isolated from degenerated intervertebral discs with umbilical cord derived mesenchymal stem cells. Exp Cell Res 2017;361(2):324-32.
[Crossref] [Google Scholar] [PubMed]
- Lama P, Claireaux H, Flower L, Harding IJ, Dolan T, Le Maitre CL, et al. Physical disruption of intervertebral disc promotes cell clustering and a degenerative phenotype. Cell Death Discov 2019;5(1):154.
[Crossref] [Google Scholar] [PubMed]
- Zhao YD, Huang YC, Lin JL, Li WS. Intervertebral disc progenitors: Lessons learned from single-cell RNA sequencing and the role in intervertebral disc regeneration. Bioengineering 2023;10(6):713.
[Crossref] [Google Scholar] [PubMed]
- Cai F, Wu XT, Xie XH, Wang F, Hong X, Zhuang SY, et al. Evaluation of intervertebral disc regeneration with implantation of Bone Marrow Mesenchymal Stem Cells (BMSCs) using quantitative T2 mapping: A study in rabbits. Int Orthop 2015;39:149-59.
[Crossref] [Google Scholar] [PubMed]
- Tang S, Gantt C, Salazar Puerta A, Bodine L, Khan S, Higuita-Castro N, et al. Nonviral overexpression of Scleraxis or Mohawk drives reprogramming of degenerate human annulus fibrosus cells from a diseased to a healthy phenotype. JOR Spine 2023;6(3):e1270.
- Fontes RB, Baptista JS, Rabbani SR, Traynelis VC, Liberti EA. Normal aging in human lumbar discs: An ultrastructural comparison. PloS One 2019;14(6):e0218121.
[Crossref] [Google Scholar] [PubMed]
- Mulholland RC. The myth of lumbar instability: The importance of abnormal loading as a cause of low back pain. Eur Spine J 2008;17(5):619-25.
[Crossref] [Google Scholar] [PubMed]
- Karunanayake AL, Pathmeswaran A, Wijayaratne LS. Chronic low back pain and its association with lumbar vertebrae and intervertebral disc changes in adults. A case control study. Int J Rheum Dis 2018;21(3):602-10.
[Crossref] [Google Scholar] [PubMed]
- Lv B, Yuan J, Ding H, Wan B, Jiang Q, Luo Y, et al. Relationship between endplate defects, modic change, disc degeneration and facet joint degeneration in patients with low back pain. BioMed Res Int 2019;2019(1):9369853.
- Ishigami S, Tarui S, Goto T, Ousaka D, Baba K, Kasahara S, et al. Transcoronary infusion of cardiac progenitor cells in hypoplastic left heart syndrome: 3-year results of the TICAP Trial. Circulation 2015;132(3):A11909.
[Crossref] [Google Scholar] [PubMed]
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, et al. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 1996;13(1):19-50.
[Crossref] [Google Scholar] [PubMed]
- Mwape K, Mubanga C, Chilyabanyama ON, Chibesa K, Chisenga CC, Silwamba S, et al. Application of a novel proteomic microarray reveals high exposure to diarrhoeagenic Escherichia coli among children in Zambia participating in a phase I clinical trial. Microorganisms 2024;12(3):420.
[Crossref] [Google Scholar] [PubMed]
- Mongia A, Zohora FT, Burget NG, Zhou Y, Saunders DC, Wang YJ, et al. AnnoSpat annotates cell types and quantifies cellular arrangements from spatial proteomics. Nat Commun 2024;15(1):3744.
[Crossref] [Google Scholar] [PubMed]
- Patiño-Lopez G, Encarnación-Guevara S. Clinical proteomics in Mexico: Where do we stand? Bol Med Hosp Infant Mex 2017;74(3):173-4.
[Crossref] [Google Scholar] [PubMed]
- Wang R, Xu C, Zhong H, Hu B, Wei L, Liu N, et al. Inflammatory-sensitive CHI3L1 protects nucleus pulposus via AKT3 signaling during intervertebral disc degeneration. FASEB J 2020;34(3):3554-69.
[Crossref] [Google Scholar] [PubMed]
- Rajasekaran S, Tangavel C, Ks SV, Soundararajan DC, Nayagam SM, Matchado MS, et al. Inflammaging determines health and disease in lumbar discs-evidence from differing proteomic signatures of healthy, aging and degenerating discs. Spine J 2020;20(1):48-59.
[Crossref] [Google Scholar] [PubMed]
- Caldeira J, Santa C, Osorio H, Molinos M, Manadas B, Gonçalves R, et al. Matrisome profiling during intervertebral disc development and ageing. Sci Rep 2017;7(1):11629.
[Crossref] [Google Scholar] [PubMed]
- Griffith JF, Wang YX, Antonio GE, Choi KC, Yu A, Ahuja AT, et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine 2007;32(24):E708-12.
[Crossref] [Google Scholar] [PubMed]
- Yu LP, Qian WW, Yin GY, Ren YX, Hu ZY. MRI assessment of lumbar intervertebral disc degeneration with lumbar degenerative disease using the Pfirrmann grading systems. PloS one 2012;7(12):e48074.
[Crossref] [Google Scholar] [PubMed]
- Stadlmann J, Hoi DM, Taubenschmid J, Mechtler K, Penninger JM. Analysis of PNGase F-resistant N-Glycopeptides using SugarQb for proteome discoverer 2.1 reveals cryptic substrate specificities. Proteomics 2018;18(13):1700436.
[Crossref] [Google Scholar] [PubMed]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25(1):25-9.
[Crossref] [Google Scholar] [PubMed]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucl Acids Res 2008;36(10):3420-35.
[Crossref] [Google Scholar] [PubMed]
- Yang G, Chen L, Gao Z, Wang Y. Implication of microglia activation and CSF1/CSF1R pathway in lumbar disc degeneration-related back pain. Mol Pain 2018;14:1744806918811238.
[Crossref] [Google Scholar] [PubMed]
- Kos N, Gradisnik L, Velnar T. A brief review of the degenerative intervertebral disc disease. Med Arch 2019;73(6):421.
- Babu NS, Krishnan S, Swamy CV, Subbaiah GP, Reddy AV, Idris MM. Quantitative proteomic analysis of normal and degenerated human intervertebral disc. Spine J 2016;16(8):989-1000.
[Crossref] [Google Scholar] [PubMed]
- Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat Rev Rheumatol 2014;10(1):44-56.
[Crossref] [Google Scholar] [PubMed]
- Ding L, Guo D, Homandberg GA. The cartilage chondrolytic mechanism of fibronectin fragments involves MAP kinases: Comparison of three fragments and native fibronectin. Osteoarthr Cartil 2008;16(10):1253-62.
[Crossref] [Google Scholar] [PubMed]
- Kim JW, Jeon N, Shin DE, Lee SY, Kim M, Han DH, et al. Regeneration in spinal disease: Therapeutic role of hypoxia-inducible factor-1 alpha in regeneration of degenerative intervertebral disc. Int J Mol Sci 2021;22(10):5281.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Elias JE. Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol Biol 2017;1550:185-98.
[Crossref] [Google Scholar] [PubMed]
- Moulder R, Bhosale SD, Goodlett DR, Lahesmaa R. Analysis of the plasma proteome using iTRAQ and TMT-based isobaric labeling. Mass Spectrom Rev 2018;37(5):583-606.
[Crossref] [Google Scholar] [PubMed]
- Jiang H, Qin H, Yang Q, Huang L, Liang X, Wang C, et al. Effective delivery of miR-150-5p with nucleus pulposus cell-specific nanoparticles attenuates intervertebral disc degeneration. J Nanobiotechnol 2024;22(1):292.
- Wang D, Zhang L, He D, Zhang Y, Zhao L, Miao Z, et al. A natural hydrogel complex improves intervertebral disc degeneration by correcting fatty acid metabolism and inhibiting nucleus pulposus cell pyroptosis. Mater Today Bio 2024;26:101081.
- Sun W. Comparative proteomic study of congenital scoliosis. Peking Union Med Coll; 2011.
- Xie FG, Cai DZ, Rong LM. Serum proteomics in patients with lumbar disc herniation. J Sun Yat-Sen Univ 2009;3:313-7.
 ): Cellular component; (
): Cellular component; ( ): Molecular function and (
): Molecular function and ( ): Biological process
): Biological process