- *Corresponding Author:
- Pixiao WangDepartment of Hepatobiliary and Pancreatic Surgery, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province 430014, People’s Republic of China E-mail: pixiaowang_tch@163.com
| This article was originally published in a special issue, “Drug Discovery and Repositioning Studies in Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(4) Spl Issue “164-174” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Various types of cancers have been found to contain long non-coding RNAs. Hepatocellular carcinoma is closely associated with atypical expression of long non-coding RNAs. The cancer genome atlas databases were utilized to detect the expression of LINC00847 in different clinical staging and its clinical relevance in hepatocellular carcinoma. In this research, we found that LINC00847 was upregulated in different clinical stages in hepatocellular carcinoma compared with adjacent non-tumor tissues. Fluorescence in situ hybridization was conducted in addition to ribonucleic acid immunoprecipitation assays to indicate that LINC00847 has the ability of sponging microRNA-218-5p. Functional experiments demonstrated microRNA-218-5p repressed the capabilities of proliferation and invasion in hepatocellular carcinoma cells. Dual luciferase assays demonstrated microRNA-218-5p directly targeting to high mobility group box protein 1 messenger ribonucleic acid in its 3’-untranslated region which regulated high mobility group box protein 1/phosphatidylinositol 3-kinase/protein kinase B pathways to exert its antitumor effects. The discoveries of our research uncovered the vital role of LINC00847 could promote biological effect in hepatocellular carcinoma, through sponging microRNA-218-5p in post-transcriptional regulation of high mobility group box protein 1/phosphatidylinositol 3-kinase/protein kinase B axis.
Keywords
Hepatocellular carcinoma, LINC00847, high mobility group box protein 1, protein kinase B
Globally, Hepatocellular Carcinoma (HCC) is a prominent contributor to cancer-related fatalities, holding the position of the 6th most prevalent malignant neoplasm. In 2020, there were 906 000 new cases of liver cancer reported worldwide, resulting in 830 000 deaths[1]. Due to its characteristics such as its atypical symptoms at early stage, highly invasive characteristic, drug insensitivity, poor prognosis and other characteristics, HCC brings great challenge in early diagnosis and clinical treatment. Despite the progress in diagnostic and therapeutic innovations in recent times (such as surgical removal, liver transplant, chemo radiotherapy, targeted treatment, and immunotherapy), liver cancer still has a low overall survival rate, and the long-term outlook is frequently unfavorable. To achieve the best results for patients, it is crucial to acquire a thorough comprehension of the complex processes that control the beginning and advancement of liver cancer.
Non-coding RNAs, including Long noncoding RNAs (LncRNAs) and microRNAs (miRNAs), have been found as vital regulators in the pathogenesis of liver cancer[2,3]. Many diseases have extensively documented the presence of lncRNAs which are Ribonucleic Acid (RNA) molecules that are single-stranded and consist of >200 nucleotides[4]. Previous studies have investigated the involvement of LINC00847 in various types of cancer, including pancreatic cancer[5,6], non-small cell lung cancer[7], laryngeal squamous cell carcinoma[8], and thyroid cancer[9]. These research findings suggest that the role of LINC00847 may also be significant in other forms of cancer. In the context of Lung Adenocarcinoma (LUAD), LINC00847 exhibited associations with B cells, CD8 T cells, and dendritic cells. Additionally, it was found to reduce the expression of PD-1, thereby presenting a promising candidate for immunotherapy targeting[10]. Short RNA molecules called miRNAs, which do not have the ability to code, play a crucial part in regulating gene expression after transcription by attaching to the 3′ Untranslated Regions (3′ UTRs) of target messenger RNA (mRNAs). Multiple investigations have provided evidence indicating that liver cancer manifests abnormal expression patterns of miRNAs. These miRNAs participate in all kinds of biological processes, encompassing cell survival, death, invasion, and drug resistance[11-13]. High Mobility Group Box protein 1 (HMGB1), a multifunctional nonhistone protein, exerts a pivotal role in a multitude of cellular processes. These include Deoxyribonucleic Acid (DNA) organization, gene regulation, and the immune response, among others[14,15]. HMGB1 has been the subject of numerous research studies, which have shed light on its dual function in the development of tumors. It has been observed to have both promoting and suppressing effects on various types of cancers, including HCC. Additionally, its association with prognosis has been extensively investigated[16-19]. However, the exact mechanism by which LINC00847 functions as a sponge for miR-218-5p, and how miR-218-5p acts as a suppressor of HCC by targeting HMGB1, is still not fully understood.
In our study, the goal is to assess the impact of LINC00847 on HCC growth and progression, investigating the insights corresponding mechanisms. The outcomes reveal that LINC00847 effectively accelerates HCC cell growth and metastasis abilities by sponging to miR-218-5p. miR-218-5p can bind to HMGB1 3′ UTRs to regulate the Phosphatidylinositol 3-Kinase (PI3K)/Protein Kinase B (AKT) pathway. These promising findings suggest LINC00847 has the potential to be a targeted therapy for HCC.
Materials and Methods
Bioinformatics prediction involving access to The Cancer Genome Atlas (TCGA) database:
Previously established bioinformatics prediction methods were utilized in this study. The TCGA-Liver Hepatocellular Carcinoma (LIHC) data were acquired from the TCGA dataset (https://portal.gdc.com) and used for subsequent analysis. The survminer package is used for grouping, by the application of the survival package to conduct survival analysis on the resulting groups. In order to identify the possible targets of LINC00847 and miR-218-5p, we utilized various databases such as ENCORI (https://rnasysu.com/encori/agoClipRNA.php), and TargetScan (https://www.targetscan.org/mamm_31/).
Cell lines and culture:
MHCC97H and Hep3B, HCC cell lines, were acquired from CCTCC (Wuhan, China). They were cultured in 10 % Fetal Bovine Serum (FBS)- supplemented Dulbecco’s Modified Eagle Medium (DMEM) (HyClone, Utah, United States of America (USA)) at 37° with 5 % Carbon dioxide (CO2).
Plasmids and cell transfection:
RiboBio (Guangzhou, China) provided the siNC, siLINC00847, miR-218-5p mimics, miR-218-5p inhibitors, Negative Control (NC) mimics, and NC inhibitors. We obtained the psi-CHECKTM-2 dual- luciferase reporter vector from Wuhan AuGCT DNA-SYN Biotechnology Co., Ltd. All sequences were confirmed by DNA sequencing. Following the manufacturer’s guidelines, Lipofectamine 3000 (Invitrogen) was utilized to transfect the HCC cell lines with small interfering RNAs (siRNAs) and plasmids.
Functional experiment:
In order to evaluate the viability of HCC cells in a laboratory setting, we utilized both the Cell Counting Kit-8 (CCK-8) assay and the colony formation test. Furthermore, we employed the transwell assay and wound-healing test to assess the migratory ability of HCC cells.
Around 100 000 cells were placed in a 24-well plate. Luciferase activity was observed after a 24 h period of transfection. Normalization of Renilla luciferase values was completed by calculating the ratio of Renilla-to-firefly luciferase activity, which was presented as the experimental result. This ratio is often used to determine the impact of miRNA on the target gene’s expression by measuring the inhibition or enhancement of luciferase activity.
RNA Immunoprecipitation (RIP):
In order to enhance Argonaute 2 (AGO2) binding RNA, we employed the Magna RIPTM RNA-binding protein immunoprecipitation kit (Millipore, Germany). Following the indicated transfection, cells were collected and lysed using a lysis buffer. Afterward, the cellular lysates were cultured with a RIP solution that included magnetic particles.
Consequently, AGO2 antibody (Abcam, UK) was used to conjugate these beads, while a separate set of beads was conjugated with anti-Immunoglobulin G (IgG) (Abcam) to serve as a NC. The samples underwent digestion using DNAse I and proteinase K. In the end, the enhanced RNA underwent quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) examination.
Fluorescence In Situ Hybridization (FISH):
In order to examine the localization of LINC00847 and miR-218-5p in HCC cells, our laboratory conducted a double FISH assay, building upon previous investigations[21]. RiboBio (Guangzhou, China) provided the Cy3-labeled LINC00847 and Fluorescein (FAM)-labeled miR-218-5p probes.
Western Blotting (WB):
A 180 kDa Prestained Protein Marker (MP102-01) from Vazyme in Nanjing, China, was used to load and compare the samples. Primary antibodies were as follows; primary antibodies against Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) (60004- 1-Ig for WB) were obtained from Proteintech (Wuhan, Hubei, China). Primary antibodies against phospho-mammalian Target of Rapamycin (mTOR) (FNab10006) and mTOR (FNab05417) were purchased from Fine test (Wuhan, Hubei, China). Primary antibodies against total AKT (#4685) and phospho (p)-AKT (ser473) (#4058), was purchased from Cell Signaling Technology (Shanghai, China).
Statistical analysis:
The mean and standard deviation were calculated for the analysis and visualization of the three separate experiments. For this purpose, the software GraphPad Prism-8 was employed. To determine the corresponding p value, a t-test was conducted. The levels of importance for (*p<0.05), (**p<0.01), and (***p< 0.001) are employed to signify the presence of a notable disparity.
Results and Discussion
The dysregulated expression of lncRNAs has consistently been observed in cases of HCC[22]. In a previous study, we utilized TCGA datasets to examine the upregulated expression of lncRNAs in HCC[22]. Through our analysis, we discovered a novel lncRNA known as LINC00847, which exhibited significantly elevated expression levels in HCC in different clinical stages compared to normal liver tissues (fig. 1A). Moreover, a significant correlation was observed between increased LINC00847 expression and decreased survival time and survival status, as well as Overall Survival (OS), Progression- Free Survival (PFS), Disease-Free Survival (DFS), and Disease-Specific Survival (DFS) (fig. 1B-fig. 1D). Also, we examined the correlation between high LINC00847 feature and the stoma score and other immune cells in LIHC (fig. 1E). We also evaluated LINC00847 level in PLC/PRF/5 cell lines and other HCC cell lines. The results showed that LINC00847 expression was higher in HCC cell lines (fig. 1F). Consequently, LINC00847 demonstrated a significant correlation with unfavorable prognosis in patients, indicating its potential as a valuable prognostic biomarker for HCC.
Fig. 1: Bioinformatic analysis indicated LINC00847 is significantly higher in HCC and demonstrates an inverse association with patient survival. (A): Variations in LINC00847 expression; (B-D): Correlation between LINC00847 level and survival time and survival status in HCC; (E): qPCR experiment showed LINC00847 expression in different HCC cell lines and (F): The correlation between high LINC00847 feature and the stoma score and other immune cells in LIHC
Note:  High LINC00847 TPM and (D):
High LINC00847 TPM and (D):  Low groups
Low groups
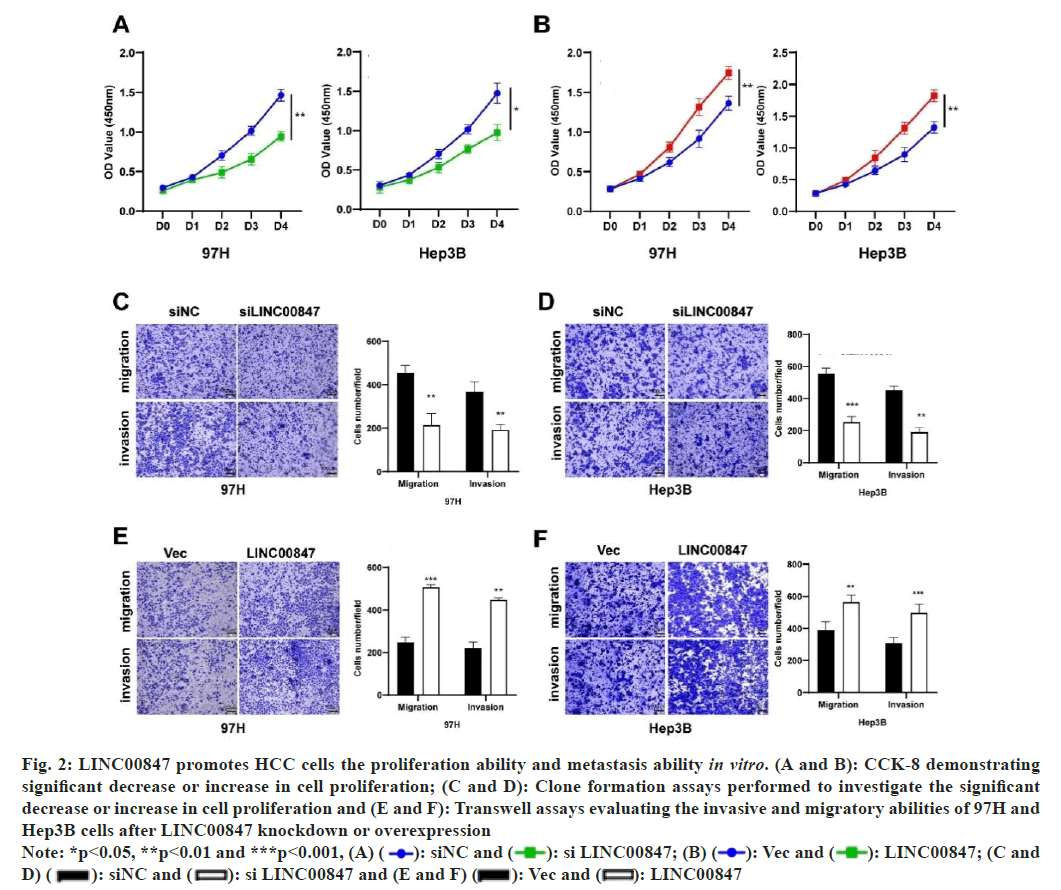
To appraise the biological function of LINC00847 in HCC, 97H and Hep3B were chosen for knockdown or overexpression according to the relative level of LINC00847 (fig. 1F). The pcDNA3.1-LINC00847 was generated and transfected into 97H or Hep3B cell lines to enhance its expression, while small interfering RNAs were used to suppress the expression of LINC00847. To assess the growth potential of LINC00847, the CCK-8 assay was employed. The experimental results suggested that the overexpression of LINC00847 promoted the proliferation, while downregulation of LINC00847 suppressed the proliferation ability (fig. 2A and fig. 2B). In general, the movement and infiltration of HCC cells are important biological processes. To investigate the impact of LINC00847 overexpression or knockdown on the migration and invasion of HCC cells, transwell assays were conducted as part of our research. In both 97H and Hep3B cell lines, it was observed that the reduction of LINC00847 resulted in a decline in cell migration and invasion. Conversely, the overexpression of LINC00847 was found to enhance cell migration and invasion (fig. 2A-fig. 2F).
Fig. 2: LINC00847 promotes HCC cells the proliferation ability and metastasis ability in vitro. (A and B): CCK-8 demonstrating significant decrease or increase in cell proliferation; (C and D): Clone formation assays performed to investigate the significant decrease or increase in cell proliferation and (E and F): Transwell assays evaluating the invasive and migratory abilities of 97H and Hep3B cells after LINC00847 knockdown or overexpression
Note: *p<0.05, **p<0.01 and ***p<0.001, (A)  LINC00847; (C and D)
LINC00847; (C and D)  LINC00847
LINC00847
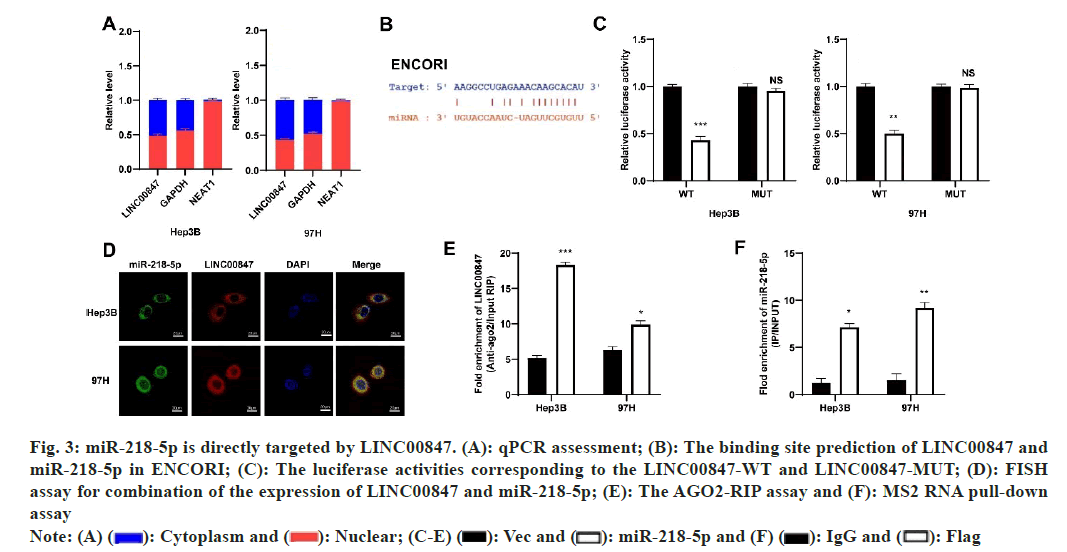
We investigated the distribution of LINC00847 expression in cells to determine its mechanisms in HCC and found that it was mostly found in the cytoplasm (fig. 3A). These findings indicated that LINC00847 potentially operated as a molecular sponge. Next, ENCORI (https://rnasysu.com/encori/agoClipRNA.php) was used to predict the possible target for LINC00847 (fig. 3B). To confirm the interaction between miR-218-5p and LINC00847, we utilized a dual luciferase reporter assay. Then specific binding site was confirmed in LINC00847. Co-transfecting miR-218-5p with LINC00847 WT or LINC00847 MUT resulted in a significant decrease in luciferase activity in LINC00847 WT when using miR-218-5p mimic, while there was no change in activity observed in LINC00847 MUT with miR- 218-5p mimic (fig. 3C). Furthermore, the HCC cells exhibited FISH assays, which revealed the coexistence of LINC00847 and miR-218-5p in the cytoplasm, enabling their potential interaction (fig. 3D). RIP assay also performed to show LINC00847 enriched more AGO2 pellet when miR-218-5p mimic came in existence (fig. 3E). MS2 RNA pull-down assay also further verified the recruiting function of miR-218-5p to LINC00847 (fig. 3F). These findings indicated that miR-218-5p was a target for LINC00847 in HCC cells.
Fig. 3: miR-218-5p is directly targeted by LINC00847. (A): qPCR assessment; (B): The binding site prediction of LINC00847 and miR-218-5p in ENCORI; (C): The luciferase activities corresponding to the LINC00847-WT and LINC00847-MUT; (D): FISH assay for combination of the expression of LINC00847 and miR-218-5p; (E): The AGO2-RIP assay and (F): MS2 RNA pull-down assay
Note: (A)  IgG and
IgG and  Flag
Flag
To assess the impact of miR-218-5p on HCC, functional tests were performed on cells that had either inhibited or overexpressed levels of miR- 218-5p. CCK8 assay and plate clone formation assay indicted miR-218-5p mimic and miR-218-5p inhibitor could repress and promoted the proliferation and growth capacity of two HCC cells, respectively (fig. 4A-fig. 4C). Trans-well assay was conducted to uncover the impact of miR-218-5p on the migration in HCC cells and the result uncovered that miR-218- 5p mimic restrained the HCC cells migration while miR-218-5p inhibitor had a contrary tendency in 97H and Hep3B cells (fig. 4D and fig. 4E). Moreover, patients with high miR-218 level predicted better overall survival.
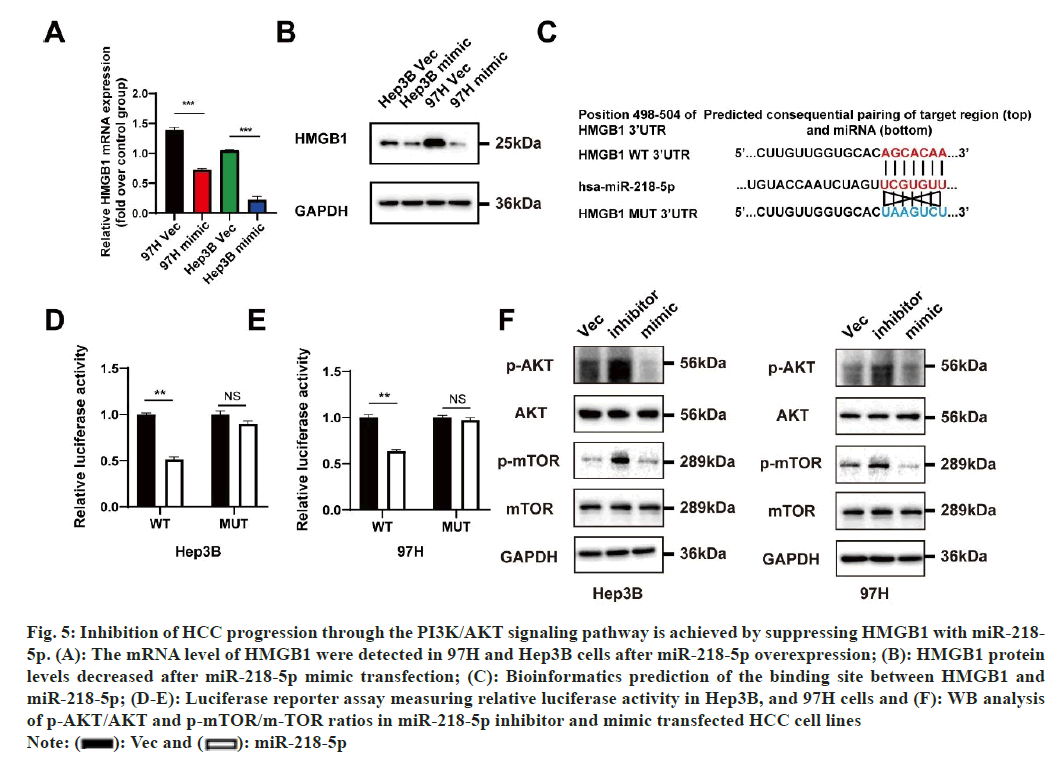
miRNAs are an innovative form of regulatory element that specifically targets the UTR of mRNAs in order to decrease translation effectiveness or promote mRNA breakdown. To further uncover the LINC00847 and miR-218-5p regulation mechanism, downstream targets HMGB1 of miR-218-5p was predicted. A comprehensive evaluation of HMGB1 expression in HCC was performed by extensively analyzing the TCGA database. The findings revealed a significant elevation in HMGB1 expression in HCC when compared to normal liver tissues, both in unpaired and paired groups. Furthermore, a positive relationship was found between the mRNA expression of HMGB1 and various factors, including T stage, pathologic stage, grade, and serum AFP level. Furthermore, a remarkable correlation was observed between increased HMGB1 mRNA expression and decreased OS, PFI, and a nearly significant association with DSS. We found miR- 218-5p could downregulated the mRNA and protein level of HMGB1 in MHCC97H and Hep3B cells (fig. 5A and fig. 5B). In addition, the psi-CHEECK-2 plasmid was constructed containing HMGB1-3’UTR region or HMGB1-3’UTR mutant region to further explore whether HMGB1 is a miR-218-5p target and the potential binding region in the HMGB1 3’- UTR were identified according to the TargetScan (https://www.targetscan.org/mamm_31/) databases information (fig. 5C). Following that, the cells were transfected with HMGB1-3’UTR binding sequences, which were either wild-type or mutated, along with either miR-218-5p mimic or a corresponding vector mimic. The findings suggested that the combination of HMGB1-wild-type and the miR-218-5p mimic resulted in a significant rise in luciferase activity compared to the control group. Conversely, the co-transfection of HMGB1-MUT and the miR-218- 5p mimic had no impact, implying the binding of miR-218-5p to HMGB1 mRNA (fig. 5D and fig. 5E). The PI3K/AKT/MTOR signaling pathway is regulated by miR-218-5p and HMGB1, which have been extensively supported by scientific literature in their role in tumor initiation and progression[19,23-25]. Our research findings demonstrate reduction in the phosphorylation levels of AKT and mTOR in MHCC97H and Hep3B cells upon the introduction of miR-218-5p mimic. In contrast, the application of a miR-218-5p suppressor led to an increase in the levels of phosphorylated AKT and mTOR proteins expression (fig. 5F). Moreover, the overexpression of HMGB1 notably enhanced the phosphorylation of AKT and mTOR in Hep3B cells, whereas the knockdown of HMGB1 substantially reduced their phosphorylation levels. Intriguingly, we also detected that the introduction of the miR-218-5p mimic partially counteracted the impact of HMGB1 overexpression on the phosphorylation of AKT and mTOR.
Fig. 5: Inhibition of HCC progression through the PI3K/AKT signaling pathway is achieved by suppressing HMGB1 with miR-218- 5p. (A): The mRNA level of HMGB1 were detected in 97H and Hep3B cells after miR-218-5p overexpression; (B): HMGB1 protein levels decreased after miR-218-5p mimic transfection; (C): Bioinformatics prediction of the binding site between HMGB1 and miR-218-5p; (D-E): Luciferase reporter assay measuring relative luciferase activity in Hep3B, and 97H cells and (F): WB analysis of p-AKT/AKT and p-mTOR/m-TOR ratios in miR-218-5p inhibitor and mimic transfected HCC cell lines
Note:  miR-218-5p
miR-218-5p
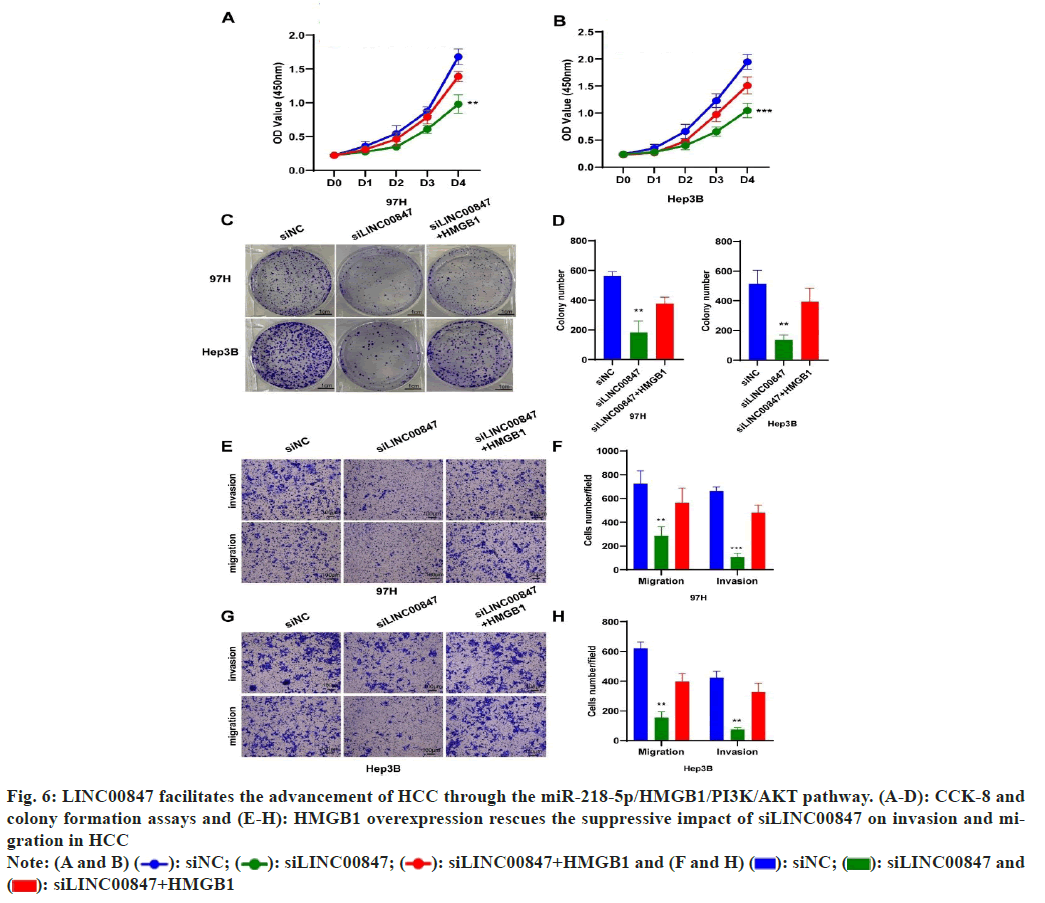
In order to provide additional evidence of the impact of HMGB1 on the advancement of HCC, we enhanced the HMGB1 expression in HCC cells by suppressing LINC00847. Our results demonstrated that the enhanced expression of HMGB1 effectively counteracted the suppressive effects caused by LINC00847 silencing, resulting in restored growth and migratory abilities of HCC cells (fig. 6A-fig. 6D). The transwell assay also presented the similar results in HCC cells (fig. 6E-fig. 6H). Moreover, we investigated the correlation between LINC00847 and HMGB1 according to spearman’s correlation analysis in TCGA database, which showed LINC00847 was positively correlated with HMGB1 (p=8.15e-10).
Fig. 6: LINC00847 facilitates the advancement of HCC through the miR-218-5p/HMGB1/PI3K/AKT pathway. (A-D): CCK-8 and colony formation assays and (E-H): HMGB1 overexpression rescues the suppressive impact of siLINC00847 on invasion and migration in HCC
Note: (A and B)  siLINC00847+HMGB1 and (F and H)
siLINC00847+HMGB1 and (F and H)  siLINC00847 and
siLINC00847 and  siLINC00847+HMGB1
siLINC00847+HMGB1
Several studies have shown that lncRNAs have a notable impact on the progression of different types of cancers[26]. LINC00847 regulated miR-643-STK31 axis in pancreatic cancer[5,6]. LINC00847 promoter region was bond with E2F1 which promoted its expression. miR-147a/IFITM1 was a identified as a target downstream in non-small cell lung cancer[7]. LINC00847 specifically engaged with miR-181a-5p in laryngeal squamous cell carcinoma, resulting in the reduction of miR-181a-5p expression. Following this, ZEB2, a gene that is targeted by miR-181a-5p, underwent upregulation due to an indirect positive influence from LINC00847[8]. LINC00847 interaction with miR-146b-5p influenced the advancement of thyroid cancer, exhibiting an inverse correlation with miR-146b-5p levels[9]. In the context of LUAD, a correlation was observed between LINC00847 and the infiltration of immune cells including B cells, CD8 T cells, and dendritic cells. The PD-L1 gene expression, which is associated with Immune Checkpoint Blockade (ICB) treatments, was downregulated by LINC00847. This suggests that LINC00847 could potentially be utilized as a new target for cancer immunotherapy[10]. The development and progression of hepatocellular carcinoma have been highlighted in published studies, emphasizing the importance of HMGB1 and miR-218-5p[27-29]. HMGB1, a nuclear protein primarily participated in pathological and physiological processes, is normally localized within the cell nucleus[14,15]. HMGB1 is widely expressed in various cell types and tissues, serving functions in nuclear structure stability, regulation of DNA- binding proteins, and transcriptional control of genes. Abnormal expression or release of HMGB1 in disease states can lead to various pathological effects. The involvement of HMGB1 in inflammatory diseases is significant, particularly through its interaction with Toll-like Receptors (TLRs) or the Receptor for Advanced Glycosylation End products (RAGE)[15,30]. Upon its binding to TLRs or RAGE, HMGB1 initiates an immune response by releasing of pro-inflammatory cytokines and chemical mediators. Notably, HMGB1 overexpression has been discovered in many cancers, including liver cancer, and it can be actively secreted by cancer cells[17]. Moreover, high HMGB1 expression often predicted a poorer prognosis, as evidenced by a systematic literature review of multiple cancer types[31]. Additionally, HMGB1 has demonstrated its involvement in the initiation and progression of tumours, showcasing its potential impact on cancer development. Research has indicated that HMGB1 facilitates the advancement of tumours in prostate cancer by attaching to TNFR1 and stimulating the NF-κB signalling pathway[32]. In hepatocellular carcinoma, VCP interaction with HMGB1 has been shown to facilitate cancer progression by activating the PI3K/AKT/mTOR pathway[19]. Additionally, the suppression of circ-CSPP1 impeded the advancement of HCC by means of miR-493-5p-induced repression of HMGB1[33]. In the same way, the inhibition of HMGB1 by miR-495-3p impedes the proliferation and migration of colorectal cancer cells[34]. miR-218- 5p has been documented to lower cellular migration, invasion, and proliferation in cervical cancer, and its decreased expression is linked with a poorer prognosis[35]. MAGI2-AS3 has been found to act as a competing endogenous RNA in nasopharyngeal carcinoma, inhibiting the binding of miR-218-5p to its target genes. This ceRNA activity of MAGI2- AS3 increases the expression of GDPD5, which in turn influences tumor progression and contributes to cisplatin resistance in this cancer type[36]. Furthermore, miR-218-5p has been shown to target CD44, PPME1, IKBKB, EGFR, TRIM9, MTF2, and HS3ST3B1[37-42]. Nonetheless, the particular function and mechanism of miR-218-5p, as well as its correlation with HMGB1, in HCC have been given minimal investigation thus far.
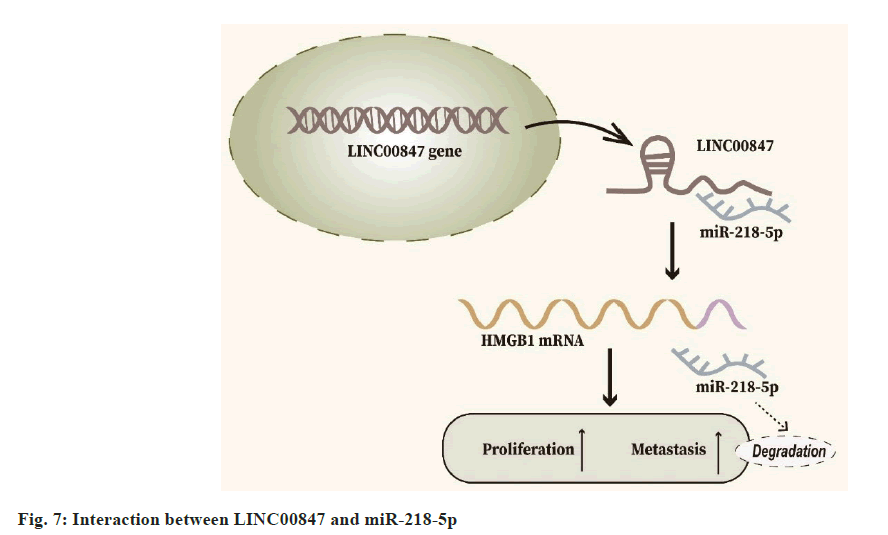
In our investigation, we uncovered a previously uncharted molecular pathway involving miR- 218-5p and LINC00847 in the context of HCC pathogenesis. Through meticulous experimentation, including luciferase reporter assessments, RIP, and RT-qPCR analyses, our data established a direct interaction between LINC00847 and miR-218-5p. This interaction appears to play a pivotal role in driving HCC progression. Moreover, our research pinpointed HMGB1 as a crucial downstream effector of miR-218-5p. Confirmatory luciferase reporter assays elucidated this connection by demonstrating miR-218-5p affinity for binding to the HMGB1 messenger RNA 3’UTR. To further elucidate the functional implications of these molecular interplays, we employed CCK8 and Trans-well assays. These experiments reinforced our hypothesis, illustrating that elevating HMGB1 levels can mitigate the inhibitory effects on HCC cell proliferation and migration caused by silencing LINC00847, thereby accentuating the vital role of LINC00847/miR-218- 5p/HMGB1 axis in HCC progression (fig. 7).
Our findings suggest that LINC00847 facilitates HCC progression through the regulation of the miR-218- 5p/HMGB1/AKT axis. Additionally, the LINC00847/ miR-218-5p/HMGB1/AKT axis further implies the potential therapeutic targeting of LINC00847 in HCC, thereby emphasizing its significance as a valuable therapeutic target.
In summary, our findings suggest that LINC00847 facilitates HCC progression through the regulation of the miR-218-5p/HMGB1/AKT axis. Additionally, the LINC00847/miR-218-5p/HMGB1/AKT axis further implies the potential therapeutic targeting of LINC00847 in HCC, thereby emphasizing its significance as a valuable therapeutic target.
Author’s contribution:
Ling Zhu and Pixiao Wang conceived the idea of the manuscript, designed experiments and wrote the manuscript. Fuliang Jiang conducted the experiments, analyzed data, and drafted the manuscript. Xiaolin Zheng analyzed data and drafted the manuscript. All authors have read and approved the final manuscript. Fuliang Jiang and Xiaolin Zheng contributed equally to the study and are co-first authors. Pixiao Wang and Ling Zhu have contributed equally to the article and are the corresponding authors.
Acknowledgment:
This work was supported by grant from the Natural Science Foundation of Hubei Province of China (Grant No: 2021CFB207).
Conflict of interests:
The authors declared no conflict of interests.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Khan S, Zhang DY, Zhang JY, Hayat MK, Ren J, Nasir S, et al. The key role of microRNAs in initiation and progression of hepatocellular carcinoma. Front Oncol 2022;12:950374.
[Crossref] [Google Scholar] [PubMed]
- Oura K, Morishita A, Masaki T. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma-a review. Int J Mol Sci 2020;21(21):8362.
[Crossref] [Google Scholar] [PubMed]
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018;172(3):393-407.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Wei J, Wang C, Meng Z, Luo D, Zhao X, et al. MicroRNA-543 controls pancreatic cancer development by LINC00847-microRNA-543-STK31 axis. J Gastrointest Oncol 2022;13(6):3263.
[Crossref] [Google Scholar] [PubMed]
- Office E. Erratum to microRNA-543 controls pancreatic cancer development by LINC00847-microRNA-543-STK31 axis. J Gastrointest Oncol 2023;14(3):1657-8
[Crossref] [Google Scholar] [PubMed]
- Li H, Chen YK, Wan Q, Shi AQ, Wang M, He P, et al. Long non-coding RNA LINC00847 induced by E2F1 accelerates non-small cell lung cancer progression through targeting miR-147a/IFITM1 axis. Front Med 2021;8:663558.
[Crossref] [Google Scholar] [PubMed]
- Li W, Hu X, Huang X. Long intergenic non-protein coding RNA 847 promotes laryngeal squamous cell carcinoma progression through the microRNA-181a-5p/zinc finger E-box binding homeobox 2 axis. Bioengineered 2022;13(4):9987-10000.
[Crossref] [Google Scholar] [PubMed]
- Hei G, Yu Y, Wu Y, Huang J. Aberrantly expressed lncRNA LINC00847 may serve as a promising prognostic factor for thyroid cancer. Horm Metabol Res 2023;55(11):794-800.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Zhang L. Integrative analysis revealed LINC00847 as a potential target of tumor immunotherapy. Appl Biochem Biotechnol 2023;195(10):6345-58.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019;234(5):5451-65.
[Crossref] [Google Scholar] [PubMed]
- Han Q, Wang M, Dong X, Wei F, Luo Y, Sun X. Non-coding RNAs in hepatocellular carcinoma: Insights into regulatory mechanisms, clinical significance, and therapeutic potential. Front Immunol 2022;13:985815.
[Crossref] [Google Scholar] [PubMed]
- Hajizadeh M, Hajizadeh F, Ghaffarei S, Doustvandi MA, Hajizadeh K, Yaghoubi SM, et al. MicroRNAs and their vital role in Apoptosis in Hepatocellular carcinoma: miRNA-based diagnostic and treatment methods. Gene 2023;888:147803.
[Crossref] [Google Scholar] [PubMed]
- Chen R, Kang R, Tang D. The mechanism of HMGB1 secretion and release. Exp Mol Med 2022;54(2):91-102.
[Crossref] [Google Scholar] [PubMed]
- Xue J, Suarez JS, Minaai M, Li S, Gaudino G, Pass HI, et al. HMGB1 as a therapeutic target in disease. J Cell Physiol 2021;236(5):3406-19.
[Crossref] [Google Scholar] [PubMed]
- Dong H, Zhang L, Liu S. Targeting HMGB1: An available therapeutic strategy for breast cancer therapy. Int J Biol Sci 2022;18(8):3421-4.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Liu Q, Wu E, Ma Z, Tuo B, Terai S, et al. The role of HMGB1 in digestive cancer. Biomed Pharmacother 2023;167:115575.
[Crossref] [Google Scholar] [PubMed]
- Cheng KJ, Alshawsh MA, Mejia Mohamed EH, Thavagnanam S, Sinniah A, Ibrahim ZA. HMGB1: An overview of its versatile roles in the pathogenesis of colorectal cancer. Cell Oncol 2020;43:177-93.
[Crossref] [Google Scholar] [PubMed]
- Pu Z, Duda DG, Zhu Y, Pei S, Wang X, Huang Y, et al. VCP interaction with HMGB1 promotes hepatocellular carcinoma progression by activating the PI3K/AKT/mTOR pathway. J Transl Med 2022;20(1):212.
[Crossref] [Google Scholar] [PubMed]
- Dong S, Wang W, Liao Z, Fan Y, Wang Q, Zhang L. MYC-activated LINC00607 promotes hepatocellular carcinoma progression by regulating the miR-584-3p/ROCK1 axis. J Gene Med 2023;25(4):e3477.
[Crossref] [Google Scholar] [PubMed]
- Han M, Liu F, Li X, Zhang H, Pan Y, Liu Y, et al. LINC01608 activated by YY1 facilitate hepatocellular carcinoma progression by modulating the EGFR/ERK axis. Liver Int 2023;43(2):471-89.
[Crossref] [Google Scholar] [PubMed]
- Liao Z, Zhang H, Su C, Liu F, Liu Y, Song J, et al. Long noncoding RNA SNHG14 promotes hepatocellular carcinoma progression by regulating miR-876-5p/SSR2 axis. J Exp Clin Cancer Res 2021;40:1-2.
[Crossref] [Google Scholar] [PubMed]
- Du X, Zhang X, Dong J, Zou N, Guo D, Yao W, et al. Irradiation-induced exosomal HMGB1 to confer radioresistance via the PI3K/AKT/FOXO3A signaling pathway in ESCC. J Transl Med 2022;20(1):507.
[Crossref] [Google Scholar] [PubMed]
- Chen SY, Hsu YH, Wang SY, Chen YY, Hong CJ, Yen GC. Lucidone inhibits autophagy and MDR1 via HMGB1/RAGE/PI3K/Akt signaling pathway in pancreatic cancer cells. Phytother Res 2022;36(4):1664-77.
[Crossref] [Google Scholar] [PubMed]
- Liu QZ, Yu HR, Wang LP, Zhou MJ, Chen Z, Zhou DH, et al. Up-regulation of PUM1 by miR-218-5p promotes colorectal tumor-initiating cell properties and tumorigenesis by regulating the PI3K/AKT axis. J Gastrointest Oncol 2023;14(1):233.
[Crossref] [Google Scholar] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19(3):143-57.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Xiang L, Li H, Chen P, Feng Y, Zhang J, et al. The role of HMGB1 signaling pathway in the development and progression of hepatocellular carcinoma: A review. Int J Mol Sci 2015;16(9):22527-40.
[Crossref] [Google Scholar] [PubMed]
- Feng W, Chen J, Huang W, Wang G, Chen X, Duan L, et al. HMGB1-mediated elevation of KLF7 facilitates hepatocellular carcinoma progression and metastasis through upregulating TLR4 and PTK2. Theranostics 2023;13(12):4042.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Xie Z, Tang G, Wei S, Chen G. Knockdown of terminal differentiation induced ncRNA (TINCR) suppresses proliferation and invasion in hepatocellular carcinoma by targeting the miR-218-5p/DEAD-box helicase 5 (DDX5) axis. J Cell Physiol 2020;235(10):6990-7002.
[Crossref] [Google Scholar] [PubMed]
- Yang H, Wang H, Andersson U. Targeting inflammation driven by HMGB1. Front Immunol 2020 Mar;11:484.
[Crossref] [Google Scholar] [PubMed]
- Wu T, Zhang W, Yang G, Li H, Chen Q, Song R. HMGB1 overexpression as a prognostic factor for survival in cancer: A meta-analysis and systematic review. Oncotarget 2016;7(31): 50417-27.
[Crossref] [Google Scholar] [PubMed]
- Jung AR, Kim GE, Kim MY, Ha US, Hong SH, Lee JY, et al. HMGB1 promotes tumor progression and invasion through HMGB1/TNFR1/NF-κB axis in castration-resistant prostate cancer. Am J Cancer Res 2021;11(5):2215-27.
[Google Scholar] [PubMed]
- Yang G, Xu Q, Wan Y, Zhang L, Wang L, Meng F. Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation. Cell Signal 2021;86:110065.
[Crossref] [Google Scholar] [PubMed]
- Zhang JL, Zheng HF, Li K, Zhu YP. miR-495-3p depresses cell proliferation and migration by downregulating HMGB1 in colorectal cancer. World J Surg Oncol 2022;20(1):101.
[Crossref] [Google Scholar] [PubMed]
- Cruz-De la Rosa MI, Jiménez-Wences H, Alarcón-Millán J, Romero-López MJ, Castañón-Sánchez CA, Salmerón-Bárcenas EG, et al. miR-218-5p/RUNX2 axis positively regulates proliferation and is associated with poor prognosis in cervical cancer. Int J Mol Sci 2022;23(13):6993.
[Crossref] [Google Scholar] [PubMed]
- Cao C, Zhou S, Hu J. Long noncoding RNA MAGI2-AS3/miR-218-5p/GDPD5/SEC61A1 axis drives cellular proliferation and migration and confers cisplatin resistance in nasopharyngeal carcinoma. Int Forum Allergy Rhinol 2020;10(8)-1012-23.
[Crossref] [Google Scholar] [PubMed]
- Young MJ, Chen YC, Wang SA, Chang HP, Yang WB, Lee CC, et al. Estradiol-mediated inhibition of Sp1 decreases miR-3194-5p expression to enhance CD44 expression during lung cancer progression. J Biomed Sci 2022;29(1):3.
[Crossref] [Google Scholar] [PubMed]
- Feng N, Jiao Z, Zhang Y, Yang B. Hsa_circ_0050102 regulates the pancreatic cancer development viamiR-218-5p/PPME1 axis. J Clin Lab Anal 2022;36(3):e24247.
[Crossref] [Google Scholar] [PubMed]
- Pan F, Zhang J, Tang B, Jing L, Qiu B, Zha Z. The novel circ_0028171/miR-218-5p/IKBKB axis promotes osteosarcoma cancer progression. Cancer Cell Int 2020;20:1-4.
[Crossref] [Google Scholar] [PubMed]
- Islam R, Zhao L, Zhang X, Liu LZ. MiR-218-5p/EGFR signaling in arsenic-induced carcinogenesis. Cancers 2023;15(4):1204.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Wang B, Wang K. Targeting TRIM9 by miR-218-5p restricts cell proliferation and epithelial-mesenchymal transition in non-small cell lung cancer. Ann Clin Lab Sci 2023;53(1):106-15.
[Google Scholar] [PubMed]
- Li Y, Shi B, Dong F, Zhu X, Liu B, Liu Y. LncRNA KCNQ1OT1 facilitates the progression of bladder cancer by targeting miR-218-5p/HS3ST3B1. Cancer Gene Ther 2021;28(3):212-20.
[Crossref] [Google Scholar] [PubMed]
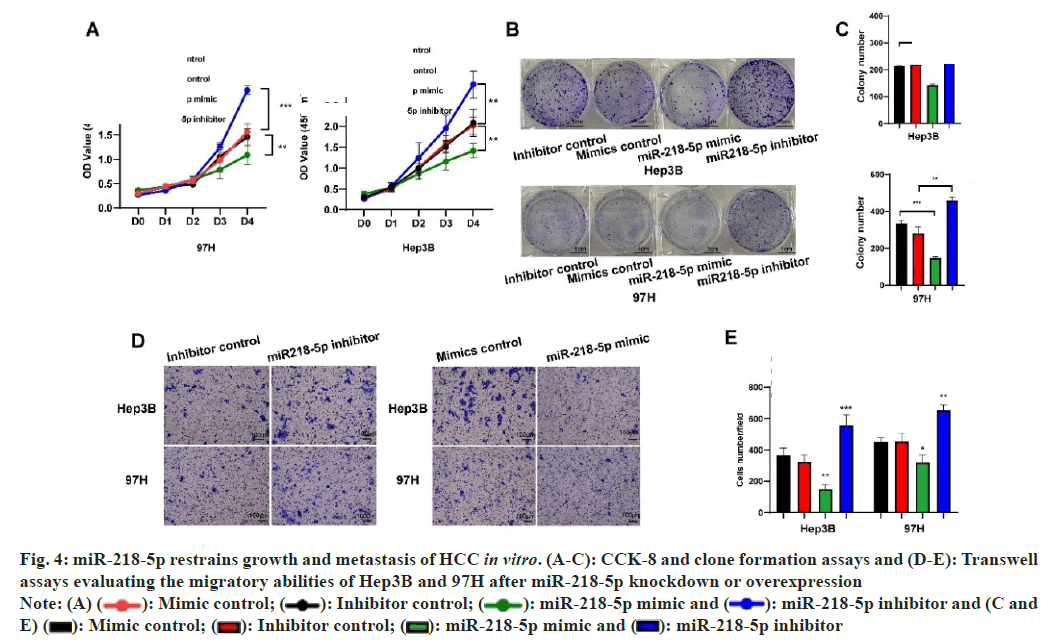
 miR-218-5p inhibitor and (C and E)
miR-218-5p inhibitor and (C and E)  miR-218-5p inhibitor
miR-218-5p inhibitor