- *Corresponding Author:
- Xinmin Wang
Department of Oncology, The First Affiliated Hospital of Shihezi University,
E-mail: wxmshzu@163.com
| This article was originally published in a special issue, “Recent Progression in Pharmacological and Health Sciences” |
| Indian J Pharm Sci 2024:86(2) Spl Issue “228-236” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A non-metastatic cell 1, also known as NM23 and NM23-H1, was the first gene identified among the 13 metastasis suppressor genes. While increasing research indicates the crucial role of non-metastatic cell 1 in the development of various tumors, its biological significance in urological tumors remains unclear. Therefore, we conducted a comprehensive analysis of the functional characteristics and prognostic value of the non-metastatic cell 1 gene in urological tumors. Our findings revealed high expression of non-metastatic cell 1 in five types of urological tumors, while only kidney chromophobe renal cell carcinoma exhibited downregulation. Survival analysis demonstrated a significant correlation between high non-metastatic cells 1 expression and poor prognosis in urological tumors. Furthermore, we investigated the mutation status and methylation levels of non-metastatic cells 1 in urological tumors, finding that amplification and mutation are the primary mutation types observed, with a substantial decrease in the promoter methylation level of non-metastatic cells 1 in urological tumor tissues. Immunoinfiltration analysis indicated a connection between aberrant non-metastatic cell 1 expression and immune cell infiltration, including B cells, clusters of differentiation 4 T cells, clusters of differentiation 8 T cells, neutrophils, macrophages, and dendritic cells. The functional enrichment results suggested that non-metastatic cells 1 may contribute to genomic instability by interfering with chromosome segregation, small nuclear ribonucleic acid binding, and spliceosome function, which could have significant implications for the mutations and immune evasion occurring during the development of urological tumors. In conclusion, our study comprehensively outlines the progress, prognosis, and immune significance of non-metastatic cell 1 in human urological tumors.
Keywords
Non-metastatic cell 1, pan-cancer, biological functions, ovarian cancer, bladder cancer
Cancer is a global health issue that significantly impacts human health and quality of life. With the increasing global aging population, the number of cancer-related deaths has risen considerably. Data has shown that cancer is the leading or second leading cause of death before the age of 70[1]. Among various tumor types, genitourinary tumors, such as Bladder Cancer (BLCA), prostate cancer, and kidney cancer, have a higher incidence in older adults and males, leading to a substantial societal burden[2]. Therefore, the identification of valuable tumor genes is crucial in elucidating the potential mechanisms underlying the occurrence and development of different genitourinary tumors.
Non-Metastatic Cell 1 (NME1) (also known as NM23 and NM23-H1) is the first gene identified among the 13 Metastasis Suppressor Genes (MSGs). It has been proven to be associated with the occurrence and development of various tumors. NME1 is located on chromosome 17q21 and encodes the NME1 protein, which consists of 166 amino acids and possesses metastasis-inhibitory functions[3,4]. NME1 messenger Ribonucleic Acid (mRNA) expression is reduced in highly metastatic cells. Previous studies have found a negative correlation between NME1 expression and metastatic potential in non-small cell lung cancer, Ovarian Cancer (OV), breast cancer, and liver cancer[5-8]. However, there is currently a lack of relevant research on the role of NME1 in genitourinary tumor occurrence and development.
This study utilized various bioinformatics methods to assess the expression of NME1 and investigate the potential molecular mechanisms of NME1 in the pathogenesis and clinical prognosis of human genitourinary tumors. The expression profiles of NME1 and the corresponding survival status were comprehensively analyzed using The Cancer Genome Atlas (TCGA) and UALCAN datasets. Additionally, gene enrichment analysis indicated the involvement of NME1 in the occurrence and development of genitourinary tumors. Furthermore, the potential significance of NME1 in anti-tumor immune response and gene alteration analysis was explored, providing insights for further functional experiments.
Materials and Methods
Gene expression analysis:
The TCGA database was downloaded from UCSC XENA (https://xenabrowser.net/datapages/) to obtain a standardized pan-cancer dataset. R version 4.0.3 was used for the normalization of the downloaded gene expression data, and the ggplot2 package was used for mRNA expression differential analysis. In addition, the Gene Expression Normal and Tumor Tissues 2 (GENT2) database (http://gent2.appex. kr/gent2/) was utilized to validate the differential expression. The expression of NME1 was analyzed in all TCGA genitourinary tumors in relation to patient pathological staging. Furthermore, quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) was performed to examine the expression of NME1 in clinical pathological tissues of BLCA. The tissue specimens were obtained from the First Affiliated Hospital of Shihezi University in Xinjiang. Total RNA was extracted from frozen lesion tissues collected during surgery using the RC101 (VAZYME, CN) reagent kit, followed by reverse transcription using the R312 (VAZYME, CN) reagent kit. The A260/A280 ratio was calculated to assess RNA quality and purity. Q711 (VAZYME, CN) Beta (β)- actin was used as the reference gene for qRT-PCR analysis under the recommended thermal cycling conditions. The Cycle Threshold (CT) values were recorded, and the 2-ΔΔCT method was employed to calculate the relative expression of NME1. Finally, the protein expression level of NME1 was further validated through immunohistochemically staining intensity in genitourinary tumors using the Human Protein Atlas (HPA).
Survival analysis:
The expression of NME1 holds prognostic value for patients, obtained through the UCSC from the TCGA database for survival data. The TCGA tumor patients were divided into high-expression and lowexpression groups based on cut-off values. The survival analysis of the tumors was carried out using the survival package and survminer package, with the COX regression verifying the significance of the data.
Gene alteration analysis:
Mutation characteristics the alteration frequency, mutation types, mutation site information, and three-dimensional structures of candidate proteins in all TCGA tumors were collected using the cBioPortal data. The correlation between NME1 and Microsatellite Instability (MSI), Tumor Mutational Burden (TMB) in urological tumors was then calculated.
Methylation level:
The methylation data for NME1 was downloaded from the TCGA database, and the correlation between NME1 expression and gene promoter methylation was analyzed for six types of urological tumors.
Infiltration of immune cells:
The TIMER algorithm was used to analyze the correlation between NME1 expression and the level of immune infiltration, as well as the correlation with immune cells, in different urological tumors.
Gene enrichment:
The protein-protein interaction network was analyzed using the BioGRID website. The top 100 genes related to NME1 were obtained from TCGA urological tumor tissues and normal tissues using the GEPIA2.0 software. Pearson correlation analysis was then performed between NME1 and the selected genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were conducted to explore the potential biological functions and signaling pathways of NME1 in urological tumors. A p<0.05 was considered statistically significant.
Results and Discussion
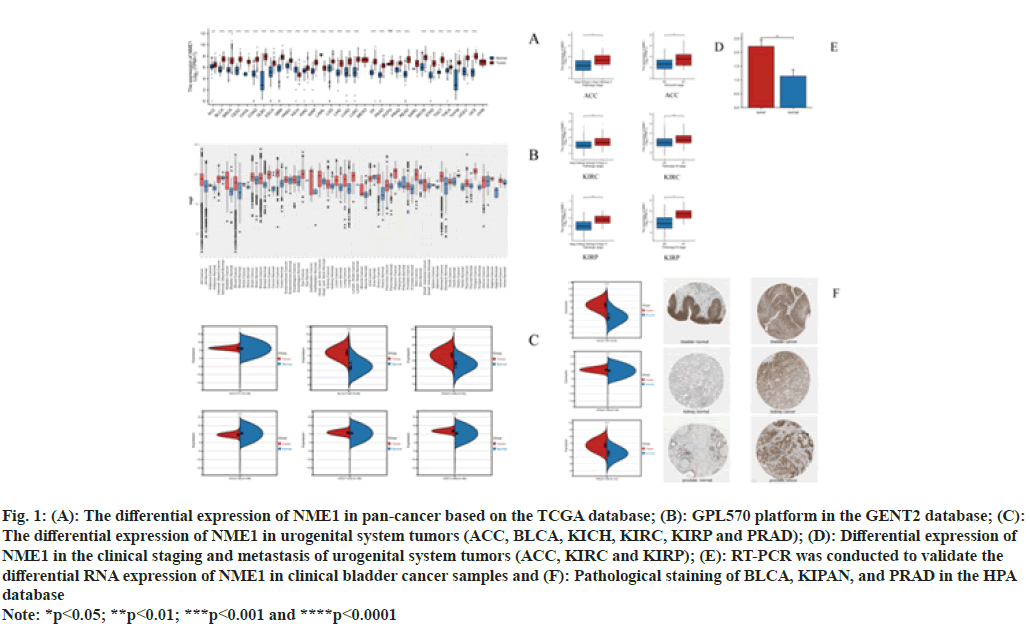
First, we investigated the expression levels of NME1 in pan-cancer using TCGA and GTEX databases. As shown in fig. 1A, except for Kidney Chromophobe Carcinoma (KICH) and Acute Myeloid Leukemia (LAML), NME1 expression in most tumors was significantly higher compared to adjacent normal tissues. To supplement the validation of expression differences due to the lack of data from some normal tissues, we used data from the GPL570 platform in the GENT2 database. As shown in fig. 1B, there was no significant difference in NME1 expression in Adrenal Cortex Carcinoma (ACC), BLCA, Blood Cancer (BC), bone cancer, Squamous Cell Carcinoma of the Head And Neck (HNSC), kidney Clear Cell Carcinoma (KIRC), lymph node cancer, OV, Laryngeal Cancer (LARY), Skin Cancer (SKCM), Small Intestinal Cancer (SIC), Splenic Cancer, Stomach Cancer (STAD), Oral Cavity Cancer (CESC), Vaginal Cancer (VA) and Vulvar Cancer (VUV) compared to adjacent normal tissues. Next, fig. 1C shows that NME1 expression in urinary system tumors such as ACC, BLCA, Prostate Adenocarcinoma (PRAD), KIRC, and Kidney Papillary Cell Carcinoma (KIRP) was upregulated, while it was downregulated in KICH. Furthermore, we explored the relationship between NME1 expression and pathological stage and lymphatic metastasis of urinary system tumor patients. We found significant effects of NME1 expression on the stage and metastasis of patients in ACC, KIRP, and KIRC (fig. 1D). Experimental evidence from RT-PCR showed significantly upregulated relative expression levels of NME1 in clinical BLCA specimens compared to adjacent tissues (fig. 1E). Finally, we further verified the immunohistochemically staining intensity of NME1 in urinary system tumors using the Human Proteome Atlas (HPA). The HPA results demonstrated strong or positive expression of NME1 primarily in BLCA, kidney cancer, and prostate cancer tissues (fig. 1F). In conclusion, we observed elevated expression of NME1 in these tumors.
Fig. 1: (A): The differential expression of NME1 in pan-cancer based on the TCGA database; (B): GPL570 platform in the GENT2 database; (C):
The differential expression of NME1 in urogenital system tumors (ACC, BLCA, KICH, KIRC, KIRP and PRAD); (D): Differential expression of
NME1 in the clinical staging and metastasis of urogenital system tumors (ACC, KIRC and KIRP); (E): RT-PCR was conducted to validate the
differential RNA expression of NME1 in clinical bladder cancer samples and (F): Pathological staining of BLCA, KIPAN, and PRAD in the HPA
database
Note: *p<0.05; **p<0.01; ***p<0.001 and ****p<0.0001
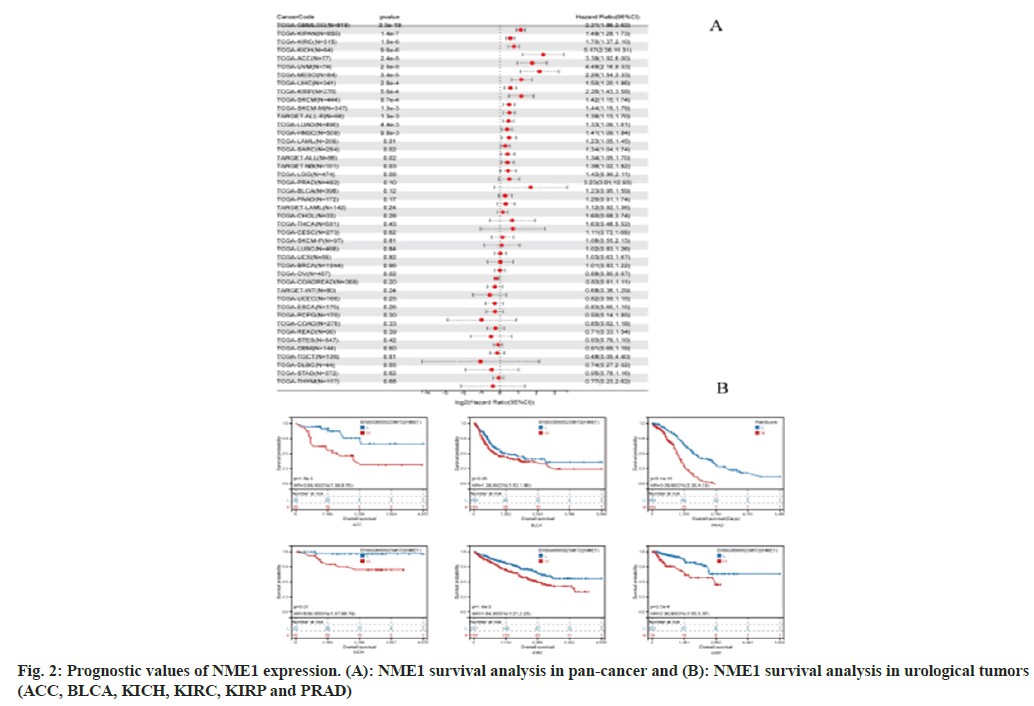
We used the R software’s survival package to analyze the relationship between gene expression and prognosis in each tumor. Statistical significance of the prognosis was obtained using the logrank test. Our findings revealed significant differences in prognosis between high expression and 18 tumor types, while low expression showed a significant difference in prognosis in one specific tumor, OV (fig. 2A). Additionally, we utilized the Kapan-Meier package to analyze survival data of urogenital system tumors, and the results indicated that high expression of NME1 was associated with a poorer prognosis in these tumors. These results suggest that NME1 may serve as a potential prognostic biomarker for various urogenital system tumors. Specifically, the expression profile and prognostic value of NME1 in ACC and PRAD patients indicate its potential role as an oncogene (fig. 2B).
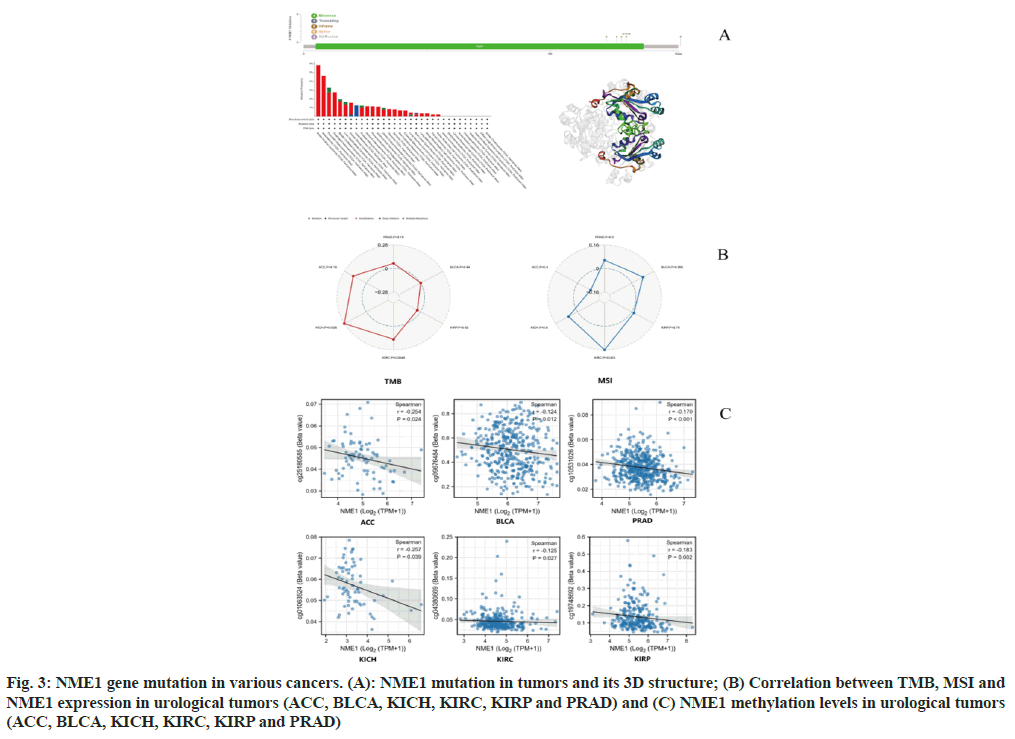
Pan-cancer analysis revealed high amplification of NME1 (>2 %) in breast infiltrating carcinoma, mesothelioma, pancreatic adenocarcinoma, and esophageal adenocarcinoma, with the highest occurrence rate of "deep deletion" in malignant melanoma at approximately 1.2 % (fig. 3A). As shown in fig. 3B, we found that amplification and mutation were the main mutation types of NME1, and they also demonstrated changes in the threedimensional structure of NME1. In addition, we analyzed the potential association between NME1 gene alterations and the survival prognosis of pancancer patients. However, we were unable to identify a significant impact of NME1 gene alterations on patient prognosis. These results require further validation with more clinical patient data.
We further investigated the relationship between NME1 expression levels and TMB and MSI. The sensitivity of tumor immune checkpoint inhibitors is significantly related to TMB and MSI. The results showed a significant positive correlation between NME1 expression and tumor mutation burden in ACC, KICH, and KIRC, with KICH having the highest correlation coefficient (core=0.28). NME1 expression was significantly positively associated with microsatellite instability in BLCA, KICH, KIRC, and PRAD, with KIRC having the largest correlation coefficient (core=0.16) (fig. 3B).
DNA methylation has been shown to affect transcriptional repression and is involved in the development of tumors[9]. Our data suggest that the methylation level of the NME1 promoter is significantly decreased in urinary system tumor tissues compared to normal tissues, indicating that the transcriptional expression of NME1 may be associated with changes in promoter methylation (fig. 3C).
Fig. 3: NME1 gene mutation in various cancers. (A): NME1 mutation in tumors and its 3D structure; (B) Correlation between TMB, MSI and NME1 expression in urological tumors (ACC, BLCA, KICH, KIRC, KIRP and PRAD) and (C) NME1 methylation levels in urological tumors (ACC, BLCA, KICH, KIRC, KIRP and PRAD)
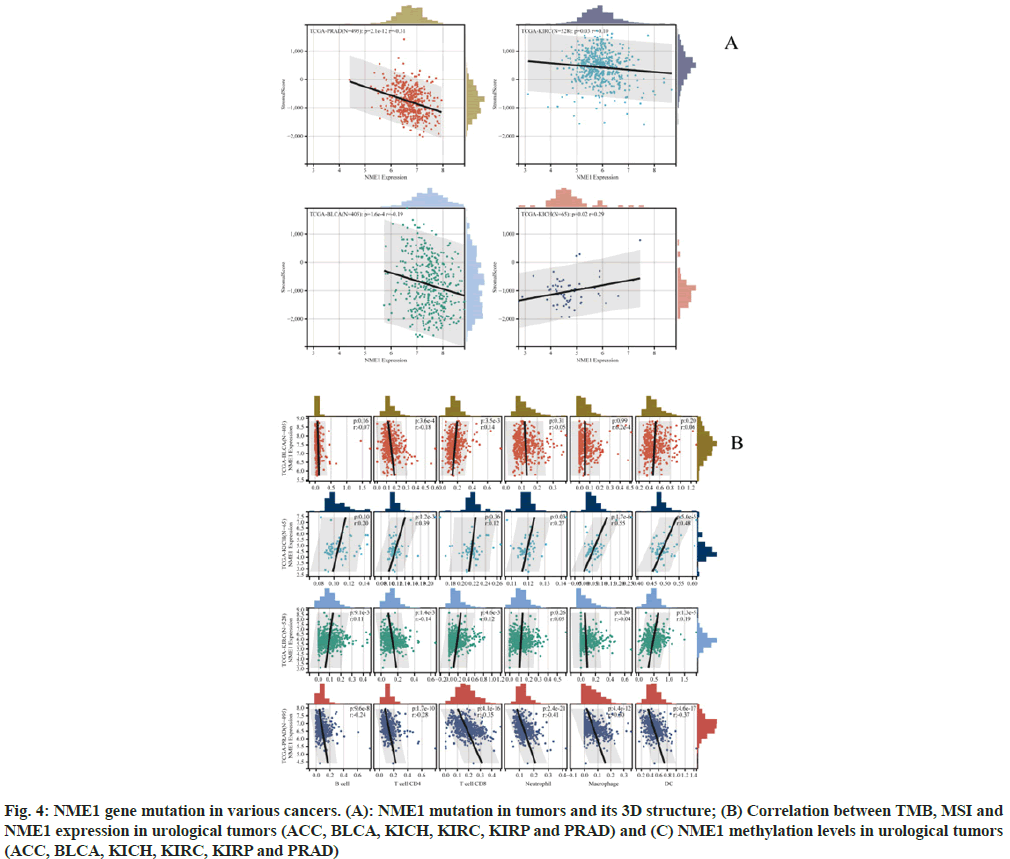
Recent studies have shown that immune infiltration is associated with the occurrence, progression, and metastasis of human cancers. Using the R software package ESTIMATE, stromal, immune, and ESTIMATE scores were calculated based on gene expression for each patient in each tumor. We obtained immune infiltration scores for six types of urinary system tumors and used the corr.test function from the R package psych to calculate the Pearson’s correlation coefficient between gene expression and immune infiltration scores in each tumor. Significant correlations between gene expression and immune infiltration were observed in four cancer types. Specifically, a significant positive correlation was found in KICH, while significant negative correlations were found in BLCA, KIRC, and PRAD (fig. 4A).
Using the Timer method from the R software package Immuno-Oncology Biological Research (IOBR), infiltration scores of B cells, T cells Clusters of Differentiation (CD) 4, T cells CD8, Neutrophils, Macrophages, and DCs were re-evaluated based on gene expression for each patient in each tumor. Eventually, we obtained immune cell infiltration levels for four types of urinary system tumors. Subsequently, the corr.test function from the R package psych was used to calculate the Pearson's correlation coefficient between gene expression and immune cell infiltration scores in each tumor to determine significant correlations. Ultimately, a significant positive correlation was observed between PRAD infiltration and NME1 expression. Additionally, a positive correlation was found between NME1 expression and KICH (fig. 4B).
Fig. 4: NME1 gene mutation in various cancers. (A): NME1 mutation in tumors and its 3D structure; (B) Correlation between TMB, MSI and NME1 expression in urological tumors (ACC, BLCA, KICH, KIRC, KIRP and PRAD) and (C) NME1 methylation levels in urological tumors (ACC, BLCA, KICH, KIRC, KIRP and PRAD)
We found that NME1 expression was significantly correlated with the infiltration values of B cells, T cells CD4, T cells CD8, Neutrophils, Macrophages, and DCs. These results suggest that NME1 might serve as a novel immune-related biomarker in tumor development.
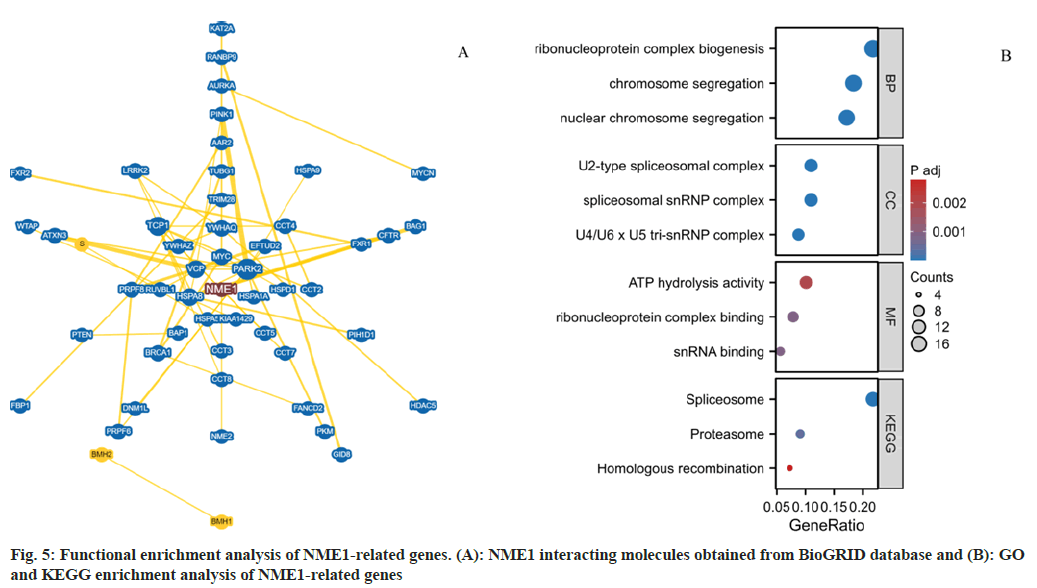
Next, we used functional enrichment analysis to evaluate the potential molecular mechanisms of NME1 in the occurrence and development of urogenital tumors. As shown in the figure, we obtained 53 molecules that interact with NME1 through the BioGRID web (fig. 5A). In addition, we obtained the top 100 genes related to urogenital tumors and NME1 from GEPIA2.0, and performed enrichment analysis on them. GO and KEGG enrichment analysis suggested that NME1 might affect genome instability by influencing chromosome segregation, small nuclear RNA (snRNA) binding, and spliceosome function, leading to abnormal cell cycle, lack of cell differentiation, and excessive cell proliferation. On the other hand, the dysfunction of cell cycle checkpoints would disrupt the balance between oncogenes and tumor suppressor genes, activate cell proliferation-related pathways, and promote excessive cell division, ultimately promoting tumor occurrence and progression (fig. 5B).
Tumor metastasis is the leading cause of death in cancer patients. In a series of tumor cohort studies, NME1 has been associated with poor prognosis, survival, and lymph node infiltration in various cancer patients[10,11]. However, no relevant relationship between NME1 and genitourinary tumors has been established. In our study, we conducted a comprehensive analysis of NME1, a metastasis-suppressive gene, in 51 different tumors. The expression of NME1 was significantly upregulated in tumor tissues such as BRCA, BLCA, CESC, KIRC, KIRP, PRAD, ACC, and UCEC, but showed low expression in tumor tissues such as KICH, LAML, and oral cancer. Overexpression of NME1 in 18 tumor types, including KIRC, KICH, ACC, and UVM, was associated with poor prognosis. Similarly, high expression of NME1 was correlated with poor prognosis in genitourinary tumors. These results suggest that NME1 may serve as a potential prognostic marker in genitourinary tumors. NME1 may function as an oncogene in ACC and PRAD tumors.
NME1 has a dual regulatory role in the process of tumor occurrence and development. In the early stages of tumor formation, NME1 is overexpressed in primary tumors compared to adjacent nontumor tissues, but its expression is subsequently downregulated during metastasis development[12]. NME1 has multiple effects in regulating metastasis cascade steps, including cell migration[13], growth and differentiation[14,15], signal transduction, transcription regulation[16-18], and apoptosis. Additionally, NME1 has other molecular activities, such as histidine-dependent protein kinase (histidine phosphorylation transferase) activity[19,20], abnormal nucleoside triphosphates activity[21], and binding to lipid bilayers[22]. Therefore, the NME1 gene family exhibits multifunctionality, and its impact on metastatic progression may be contradictory in various cancers. Previous studies have found that the expression of NME1 is negatively correlated with metastatic potential in melanoma and epithelial tumors such as breast, liver, colon, and cervical cancer[3,23-26]. However, in hematological malignancies such as ovarian and prostate cancer, the opposite relationship is observed, where upregulation of NM23-H1 is associated with poor prognosis[27,28]. Research on neuroblastoma has also reported a positive correlation between NM23-H1 expression and tumor progression[29]. Furthermore, the S120G missense mutation has been found in invasive cases, which appears to be a tumor-specific mutation in this tumor type[30]. However, few studies have identified the functional connection between NME1 and genitourinary tumor development. Although NME1 has been shown to be upregulated in melanoma[25], its detailed role and potential mechanisms in genitourinary tumors remain unclear and require further research. Our study revealed that amplification and mutations are the main mutation types of NME1, which can be observed in melanoma, infiltrating breast cancer, mesothelioma, pancreatic adenocarcinoma, and esophageal adenocarcinoma. Additionally, the promoter methylation level of NME1 was significantly decreased in genitourinary tumor tissues. The expression of NME1 was significantly positively correlated with tumor mutation burden in ACC, KICH, and KIRC, and with microsatellite instability in BLCA, KICH, KIRC, and PRAD. Single-cell sequencing and gene enrichment analysis indicated that genes associated with NME1 may regulate various cancer biological functions, such as chromosome segregation and snRNA binding.
Infiltrating immune cells play a crucial role in regulating cancer cell recognition and tumor growth[31,32]. B cells are best known for their ability to produce antibodies, such as Immunoglobulin (Ig) M, IgG, IgE, and IgA[33]. Exhaustion of effector B and T cells can help tumor cells evade immune surveillance, thereby reducing the overall survival of cancer patients[34]. In this study, we found a close correlation between NME1 expression and immune cell infiltration, including B cells, T cells (CD4 and CD8), neutrophils, macrophages, and dendritic cells. These results suggest that NME1 may be an effective immune therapy target, providing new hope for clinical treatment of tumor patients. The relationship between NME1 expression in cancer patients and immune checkpoint is a topic that needs to be explored further in more preclinical and clinical trials.
In summary, utilizing comprehensive bioinformatics analysis techniques, NME1 exhibits abnormal expression in various tumor tissues and may serve as a novel potential prognostic and immunerelated biomarker in genitourinary tumors. It plays a significant potential role in the occurrence and development of tumors. We explored the expression levels of NME1, clinical prognosis, methylation values, gene mutations, and immune modulation in genitourinary tumors. The results indicate that the role of NME1 in tumors involves a complex network regulation process that is influenced by multiple factors. Further experimental validation is required to elucidate its specific biological behavior and mechanisms.
Conflict of interests:
The authors declared no conflict of interests.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Dy GW, Gore JL, Forouzanfar MH, Naghavi M, Fitzmaurice C. Global burden of urologic cancers, 1990–2013. Eur Urol 2017;71(3):437-46.
[Crossref] [Google Scholar] [PubMed]
- Shi X, Jin H, Peng M, Li B, She M, Zhu T, et al. Association between NME1 polymorphisms and cancer susceptibility: A meta-analysis based on 1644 cases and 2038 controls. Pathol Res Pract 2018;214(4):467-74.
[Crossref] [Google Scholar] [PubMed]
- Han W, Shi CT, Cao FY, Cao F, Chen MB, Lu RZ, et al. Prognostic value of NME1 (NM23-H1) in patients with digestive system neoplasms: A systematic review and meta-analysis. PLoS One 2016;11(8):e0160547.
[Crossref] [Google Scholar] [PubMed]
- Goncharuk VN, del-Rosario A, Kren L, Anwar S, Sheehan CE, Carlson JA, et al. Co-downregulation of PTEN, KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in non-small cell lung cancer. Ann Diagn Pathol 2004;8(1):6-16.
[Crossref] [Google Scholar] [PubMed]
- Gao QL, Ma D, Meng L, Wang SX, Wang CY, Lu YP, et al. Association between Nm23-H1 gene expression and metastasis of ovarian carcinoma. Ai Zheng 2004;23(6):650-4.
[Google Scholar] [PubMed]
- Chagpar A, Magliocco A, Kerviche A, Tan L, Walley B, deCoteau JF. The replication error phenotype is associated with the development of distant metastases in hormonally treated patients with breast carcinoma. Cancer 2004;100(5):913-9.
[Crossref] [Google Scholar] [PubMed]
- Guo HB, Liu F, Zhao JH, Chen HL. Down-regulation of N-acetylglucosaminyltransferase V by tumorigenesis-or metastasis-suppressor gene and its relation to metastatic potential of human hepatocarcinoma cells. J Cell Biochem 2000;79(3):370-85.
[Crossref] [Google Scholar] [PubMed]
- Nestler EJ, Luscher C. The molecular basis of drug addiction: Linking epigenetic to synaptic and circuit mechanisms. Neuron 2019;102(1):48-59.
[Crossref] [Google Scholar] [PubMed]
- Hammerbacher J, Snyder A. Informatics for cancer immunotherapy. Ann Oncol 2017;28(12):56-73.
[Crossref] [Google Scholar] [PubMed]
- Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr 2000;32:301-8.
[Crossref] [Google Scholar] [PubMed]
- Mátyási B, Farkas Z, Kopper L, Sebestyén A, Boissan M, Mehta A, et al. The function of NM23-H1/NME1 and its homologs in major processes linked to metastasis. Pathol Oncol Res 2020;26(1):49-61.
[Crossref] [Google Scholar] [PubMed]
- Marino N, Marshall JC, Steeg PS. Protein–protein interactions: A mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch Pharmacol 2011;384(4-5):351-62.
[Crossref] [Google Scholar] [PubMed]
- Lee MY, Jeong WJ, Oh JW, Choi KY. NM23H2 inhibits EGF-and Ras-induced proliferation of NIH3T3 cells by blocking the ERK pathway. Cancer Lett 2009;275(2):221-6.
[Crossref] [Google Scholar] [PubMed]
- Mochizuki T, Bilitou A, Waters CT, Hussain K, Zollo M, Ohnuma SI. Xenopus NM23-X4 regulates retinal gliogenesis through interaction with p27Xic1. Neural Dev 2009;4(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 1993;261(5120):478-80.
[Crossref] [Google Scholar] [PubMed]
- Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, et al. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucl Acids Res 2009;37(1):172-83.
[Crossref] [Google Scholar] [PubMed]
- Khan I, Steeg PS. The relationship of NM23 (NME) metastasis suppressor histidine phosphorylation to its nucleoside diphosphate kinase, histidine protein kinase and motility suppression activities. Oncotarget 2018;9(12):10185.
[Crossref] [Google Scholar] [PubMed]
- Adam K, Hunter T. Histidine kinases and the missing phosphoproteome from prokaryotes to eukaryotes. Lab Investig 2018;98(2):233-47.
[Crossref] [Google Scholar] [PubMed]
- Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM. Metastasis suppressor function of NM23-H1 requires its 3′-5′ exonuclease activity. Int J Cancer 2011;128(1):40-50.
[Crossref] [Google Scholar] [PubMed]
- Tokarska-Schlattner M, Boissan M, Munier A, Borot C, Mailleau C, Speer O, et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J Biol Chem 2008;283(38):26198-207.
[Crossref] [Google Scholar] [PubMed]
- Leonard MK, Puts GS, Pamidimukkala N, Adhikary G, Xu Y, Kwok E, et al. Comprehensive molecular profiling of UV-induced metastatic melanoma in Nme1/Nme2-deficient mice reveals novel markers of survival in human patients. Oncogene 2021;40(45):6329-42.
[Crossref] [Google Scholar] [PubMed]
- Lodillinsky C, Fuhrmann L, Irondelle M, Pylypenko O, Li XY, Bonsang-Kitzis H, et al. Metastasis-suppressor NME1 controls the invasive switch of breast cancer by regulating MT1-MMP surface clearance. Oncogene 2021;40(23):4019-32.
[Crossref] [Google Scholar] [PubMed]
- Yang J, Li DZ, Pang Y, Zhou T, Sun J, Cheng XY, et al. MicroRNA-139-5p negatively regulates NME1 expression in hepatocellular carcinoma cells. Adv Clin Exp Med 2022;31(6):655-70.
[Crossref] [Google Scholar] [PubMed]
- Eliason S, Hong L, Sweat Y, Chalkley C, Cao H, Liu Q, et al. Extracellular vesicle expansion of PMIS-miR-210 expression inhibits colorectal tumour growth via apoptosis and an XIST/NME1 regulatory mechanism. Clin Transl Med 2022;12(9):e1037.
[Crossref] [Google Scholar] [PubMed]
- Li MQ, Shao J, Meng YH, Mei J, Wang Y, Li H, et al. NME1 suppression promotes growth, adhesion and implantation of endometrial stromal cells via Akt and MAPK/Erk1/2 signal pathways in the endometriotic milieu. Hum Reprod 2013;28(10):2822-31.
[Crossref] [Google Scholar] [PubMed]
- Fang J, Guo X, Zheng B, Han W, Chen X, Zhu J, et al. Correlation between NM23 protein overexpression and prognostic value and clinicopathologic features of ovarian cancer: A meta-analysis. Arch Gynecol Obstetr 2018;297:449-58.
[Crossref] [Google Scholar] [PubMed]
- Andolfo I, de Martino D, Liguori L, Petrosino G, Troncone G, Tata N, et al. Correlation of NM23-H1 cytoplasmic expression with metastatic stage in human prostate cancer tissue. Naunyn-Schmiedebergs Arch Pharmacol 2011;384:489-98.
[Crossref] [Google Scholar] [PubMed]
- Chang CL, Zhu XX, Thoraval DH, Ungar D, Rawwas J, Hora N, et al. Nm23-H1 mutation in neuroblastoma. Nature 1994;370(6488):335-6.
- Tan CY, Chang CL. NDPKA is not just a metastasis suppressor-Be aware of its metastasis-promoting role in neuroblastoma. Lab Investig 2018;98(2):219-27.
[Crossref] [Google Scholar] [PubMed]
- Shihab I, Khalil BA, Elemam NM, Hachim IY, Hachim MY, Hamoudi RA, et al. Understanding the role of innate immune cells and identifying genes in breast cancer microenvironment. Cancers 2020;12(8):2226.
[Crossref] [Google Scholar] [PubMed]
- Talty R, Olino K. Metabolism of innate immune cells in cancer. Cancers 2021;13(4):904.
[Crossref] [Google Scholar] [PubMed]
- Kim SS, Sumner WA, Miyauchi S, Cohen EE, Califano JA, Sharabi AB. Role of B cells in responses to checkpoint blockade immunotherapy and overall survival of cancer patients. Clin Cancer Res 2021;27(22):6075-82.
[Crossref] [Google Scholar] [PubMed]
- Chakraborty S, Khamaru P, Bhattacharyya A. Regulation of immune cell metabolism in health and disease: Special focus on T and B cell subsets. Cell Biol Int 2022;46(11):1729-46.
[Crossref] [Google Scholar] [PubMed]