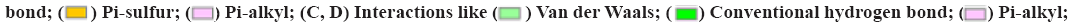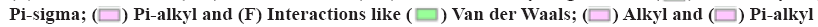- *Corresponding Author:
- L. Zhao
Department of Gastroenterology, The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Luzhou, Longmatan 646000, China
E-mail: zhaolong1982@swmu.edu.cn
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
|
Indian J Pharm Sci 2022:84(5) Spl Issue “291-303” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Banxia Xiexin decoction, a traditional herbal mixture, has been widely used for chronic atrophic gastritis in clinical practice in China. However, research on the bioactive components and underlying mechanisms of Banxia Xiexin decoction in chronic atrophic gastritis is still scarce. Components and corresponding targets of Banxia Xiexin decoction were collected from the traditional Chinese medicine systems pharmacology database and analysis platform. The chemical structures of each component were obtained from PubChem. Meanwhile, targets of chronic atrophic gastritis were collected from DrugBank and Online Mendelian Inheritance in Man. Then, the herb-compound-target-disease network and the protein-protein interaction network were constructed based on the data obtained above. Cytoscape network centrality analysis was utilized to filter hub genes and VarElect was used to analyze the relationship between genes and diseases. At last, Metascape was employed for systematic analysis of the potential targets of herbals against chronic atrophic gastritis and AutoDock was applied for molecular docking to verify the results. A total of 214 components were screened as active components and there were 21 shared targets between Banxia Xiexin decoction and chronic atrophic gastritis. Two function modules of Banxia Xiexin decoction were found in the protein-protein interaction network. Further systematic analysis of shared genes and function modules explained the potential mechanism of Banxia Xiexin decoction in the treatment of chronic atrophic gastritis; molecular docking has verified the interactions. Banxia Xiexin decoction could be employed for chronic atrophic gastritis through mechanisms including complex interactions between related components and targets, as predicted by network pharmacology and molecular docking. This work confirmed that Banxia Xiexin decoction could apply for the treatment of chronic atrophic gastritis and promoted the explanation of Banxia Xiexin decoction for chronic atrophic gastritis in the molecular mechanisms.
Keywords
Banxia Xiexin decoction, network pharmacology, chronic atrophic gastritis, herb-compound- target-disease network
Chronic Atrophic Gastritis (CAG) is a complex syndrome, defined as gastritis with the atrophy of gastric mucosa, gastric parietal cells and gastric glands, with or without metaplasia. It is mainly caused by H. pylori infection or autoimmunity reaction[1]. Although most CAG cases are benign courses, some of them can also lead to malignant complications such as life-threatening severe anemia and cancer[2].
In clinical practice, mucosal-protective agents and Proton Pump Inhibitors (PPIs) are the most commonly prescribed medications for treating CAG[3]. However, the current therapy strategy still lacks effective means and drugs for treating CAG. At the same time, Traditional Chinese Medicine (TCM) has a clinical- based development history of over 2000 y and is safe and effective for the treatment of various diseases[4]. Although there are no records of a disease named CAG in TCM literature, according to the clinical symptoms and pathogenesis characteristics of CAG, it can be classified as “distention and fullness”, “epigastric pain” and “stomachache” in TCM[5].
Banxia Xiexin Decoction (BXD) is first recorded in “Treatise on Febrile and Miscellaneous Diseases” by Zhang Zhongjing in the Eastern Han Dynasty, consisting of seven Chinese herbs: Banxia, Huanglian, Huangqin, Renshen, Ganjiang, Gancao and Dazao (Table 1), which is widely used for gastrointestinal diseases and witness great curative effect. The clinical effect of BXD in patients with CAG has been confirmed, but research on its pharmacological mechanism and the exact target remains vacant.
| Herbs | Latin name |
|---|---|
| Banxia | Arum ternatum Thunb |
| Huanglian | Coptidis rhizoma |
| Huangqin | Scutellariae radix |
| Renshen | Panax ginseng C. A. Mey. |
| Ganjiang | Zingiberis Rhizoma |
| Dazao | Fructus Jujubae |
| Gancao | Licorice |
Table 1: The Latin Name of each Herb
Therefore, further related molecular and pathway research on BXD is still a giant challenge for us.
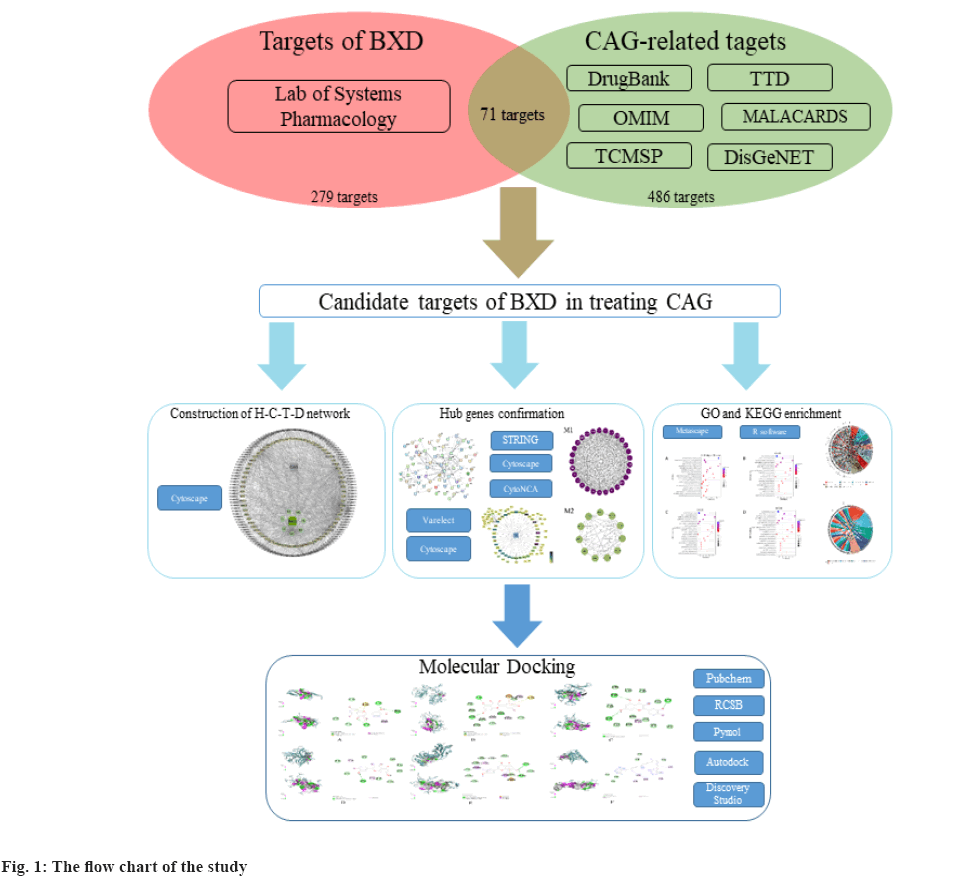
In recent years, the research methods of network pharmacology have been gradually systematized. Based on system biology and polypharmacology, network pharmacology provided a novel network mode of “multiple targets, multiple effects, complex diseases”, which contributes to the revelation of complex network connections among drugs, genes, targets and diseases[6-8]. Here, we adopted network pharmacology to identify the potential therapeutic targets and relevant pathways of BXD in treating CAG and try to provide reliable proof for further molecular mechanisms research of BXD against CAG (fig. 1).
Materials and Methods
Screening active ingredients:
The ingredients of the seven herbals consisted BXD were searched and collected from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://lsp.nwu.edu. cn/tcmsp.php). The active ingredients of each herb were screened by the criteria of Oral Bioavailability (OB)≥30 % and Drug-Likeness (DL)≥0.18[9,10].
Target prediction and verification:
Although the ingredients screened by OB and DL have a higher possibility of being absorbed by the human body, we need to explore the potential ability of these ingredients in regulating the corresponding target to judge the effectiveness of these ingredients[11]. Thus, we collected the corresponding targets from TCMSP and only the targets with clinical trial verification records in the DrugBank database were adopted[12].
Taking “chronic atrophic gastritis” or “atrophic gastritis” as keywords, we obtained the therapy- related targets of CAG from the following databases: Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org), Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/), DrugBank database (https://www.drugbank.ca) and GeneCards database (https://www.genecards. org). Then, duplicate records were deleted and the results were combined to form the cluster of potential therapeutic targets of CAG. Furthermore, the intersection targets of the BXD and CAG targets section were considered as the therapeutic targets of BXD against CAG.
Protein-Protein Interaction (PPI) and hub-gene analysis:
To reveal the complex relationships between the therapy targets and CAG, we performed PPI network analysis. PPI makes the interactions of proteins clear and helps to explain the function of possible protein complexes or functional modules[13]. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a web database providing online PPI analysis[13]. After uploading the common targets to STRING, we constructed the PPI network. Then, the result was imported to Cytoscape (version 3.8.0) for further analysis[14]. Cytoscape Network Centrality Analysis (CytoNCA) plugin in Cytoscape was applied to analyze the centrality of certain targets and evaluate protein interaction networks[15]. The VarElect online tool can analyze direct and indirect relationships between genes and diseases[16]. In this study, the relationships of potential targets of BXD against CAG were analyzed with VarElect, the results helped to determine which targets will be included in the next molecular docking.
Biology functional analysis:
Since Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) can contribute to the interpretation of system-level data and enable discoveries[17], in this study, Metascape was utilized for GO and KEGG analysis to further explore the complex mechanism of BXD in the treatment of CAG. Metascape is a web-based platform providing gene annotation, functional enrichment and interactome analysis services; monthly database update could keep our analysis results up to date[17]. In this work, GO and KEGG terms with p<0.03 were considered as significant enrichment analyses.
Molecular docking:
Molecular docking was used to assess interactions between components and hub targets. All hub targets were included in the molecular progress. The structures of these targets were collected from Protein Data Bank (PDB)[18]. We used AutoDock and PyMOL for molecular docking and binding energy assessment[19,20]. Finally, we used Discovery Studio to visualize the results of molecular docking.
Results and Discussion
Screening of active components was shown here. We obtained 212 candidates active ingredients of BXD from the TCMSP database with the screening conditions of OB≥30 % and DL≥0.18. Among them, 13 candidate ingredients were from Banxia, 14 from Huanglian, 36 from Huangqin, 22 from Renshen, 5 from Ganjiang, 93 from Gancao and 29 from Dazao. After deduplication, a total of 279 ingredients of BXD were collected for the further screening.
Target prediction and verification was discussed here. Meanwhile, taking “chronic atrophic gastritis” or “atrophic gastritis” as keywords, a total of 486 validated therapeutic targets for CAG were obtained from OMIM, TTD, DrugBank and GeneCards databases. Then, we combined the targets collected from the different databases and deduplicated the duplicate records to construct the cluster of therapy targets of CAG.
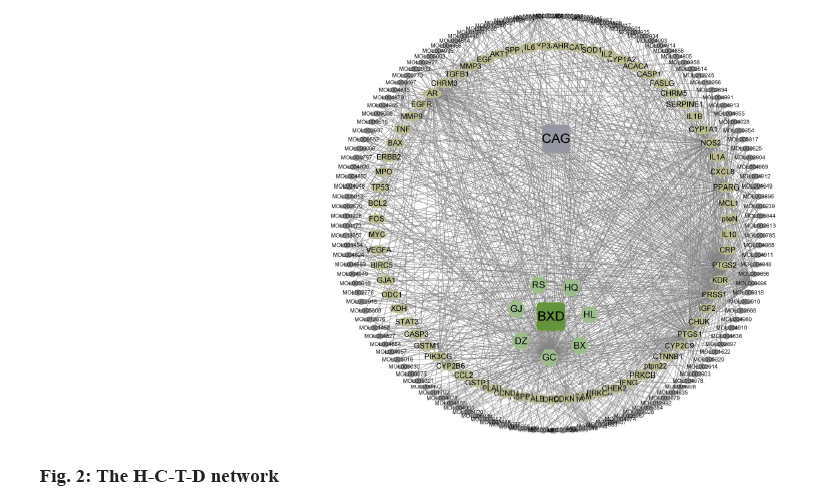
The targets overlapped between BXD and CAG clusters were considered as the therapeutic targets. Based on the data obtained above, we constructed the Herb-Compound-Targets-Disease (H-C-T-D) network, which consisted of 213 nodes (BXD, CAG, 7 herbals, 132 bioactive compounds and 71 targets) and 976 edges (fig. 2).
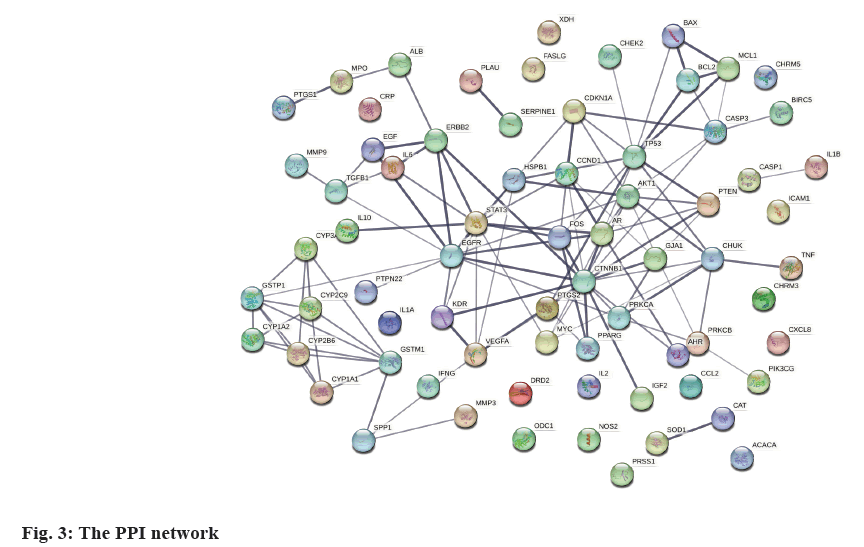
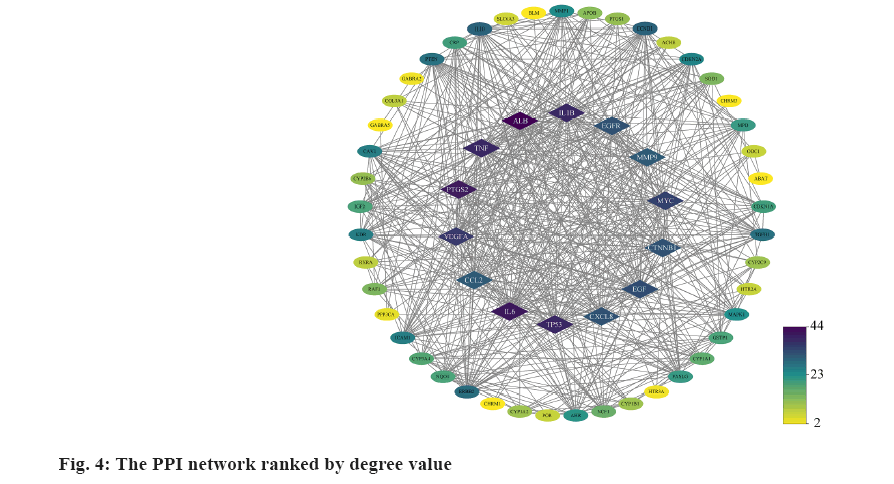
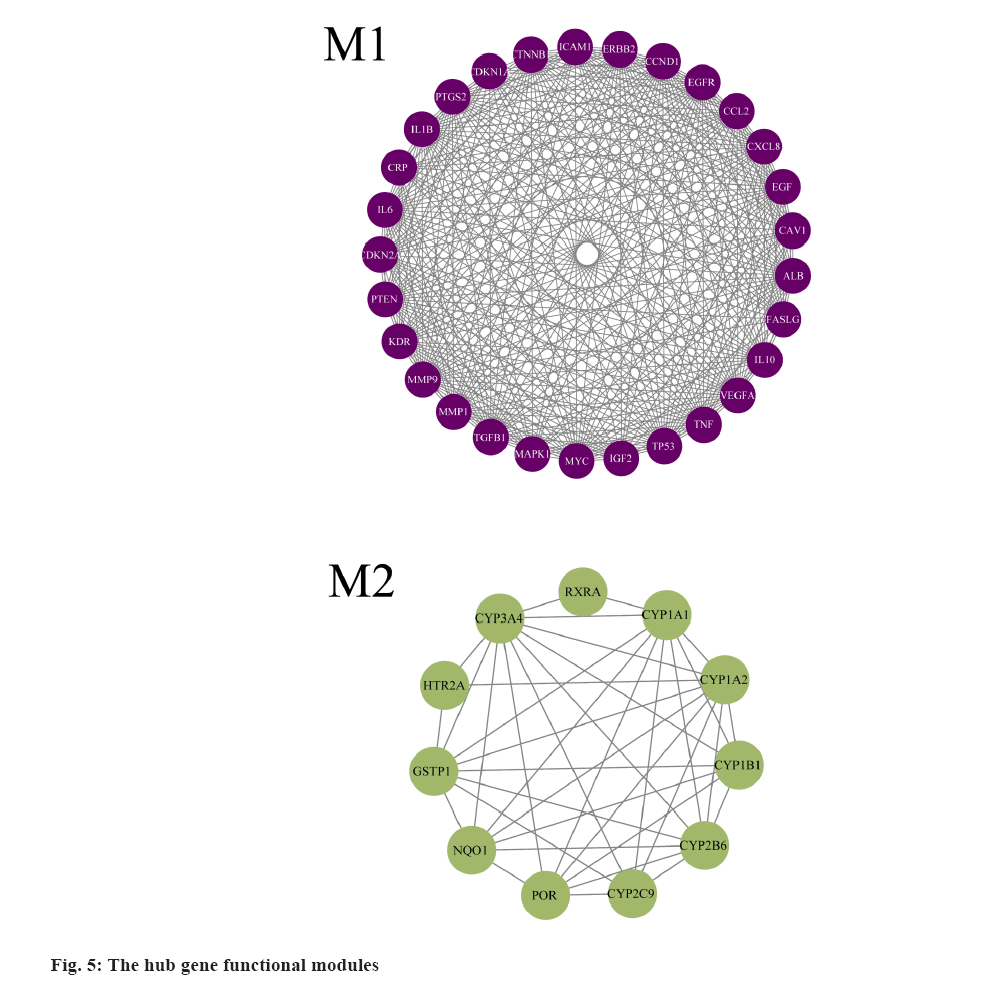
PPI network and hub-gene analysis was explained here. In further analysis, all 71 common targets were uploaded to STRING to construct the PPI network (fig. 3); the results were imported to Cytoscape to calculate the degree value of each gene using CytoNCA plugin and reconstruct the PPI network according to the degree value (Table 2), then, the PPI was reconstructed with the degree order (fig. 4). There were two possible biofunctional modules divided from the PPI network (fig. 5). After screening by CytoNCA, the top 10 targets were defined as hub targets.
| Genes | Degree | Betweenness | Closeness |
|---|---|---|---|
| ALB | 60 | 334.6169 | 0.864198 |
| TP53 | 59 | 180.34065 | 0.853659 |
| AKT1 | 58 | 208.1184 | 0.843374 |
| IL-6 | 55 | 370.48294 | 0.823529 |
| TNF | 54 | 116.79547 | 0.804598 |
| VEGFA | 54 | 90.39987 | 0.804598 |
| PTGS2 | 53 | 132.03102 | 0.795455 |
| EGFR | 52 | 118.89964 | 0.786517 |
| CASP3 | 51 | 74.19955 | 0.777778 |
| STAT3 | 51 | 66.56651 | 0.777778 |
| IL-1β | 51 | 63.150833 | 0.777778 |
| MYC | 50 | 91.731125 | 0.769231 |
| EGF | 48 | 50.615067 | 0.752688 |
| MMP9 | 47 | 58.15639 | 0.744681 |
| CTNNB1 | 46 | 62.132324 | 0.736842 |
| CXCL8 | 46 | 30.849913 | 0.736842 |
| CCND1 | 45 | 50.873108 | 0.729167 |
| CAT | 45 | 96.33492 | 0.729167 |
| PPARG | 45 | 69.95399 | 0.729167 |
| FOS | 44 | 59.804527 | 0.721649 |
| IL10 | 43 | 33.294266 | 0.714286 |
| CCL2 | 42 | 19.39998 | 0.707071 |
| PTEN | 42 | 59.528633 | 0.707071 |
| ERBB2 | 40 | 30.246576 | 0.693069 |
| ICAM1 | 39 | 9.656397 | 0.686275 |
| IL2 | 39 | 29.889345 | 0.686275 |
| TGFB1 | 37 | 6.63817 | 0.673077 |
| IFNG | 36 | 6.4944043 | 0.666667 |
| SERPINE1 | 35 | 24.655151 | 0.660377 |
| SPP1 | 34 | 13.671889 | 0.654206 |
| KDR | 34 | 24.73009 | 0.654206 |
| MMP3 | 33 | 22.96437 | 0.648148 |
| IL1A | 33 | 8.05034 | 0.648148 |
| MPO | 32 | 35.512 | 0.642202 |
| FASLG | 31 | 5.3879576 | 0.636364 |
| CASP1 | 30 | 8.180565 | 0.630631 |
| CDKN1A | 30 | 10.50631 | 0.630631 |
| MCL1 | 30 | 15.734691 | 0.630631 |
| CRP | 30 | 5.689564 | 0.630631 |
| NOS2 | 29 | 11.795232 | 0.625 |
| CHUK | 28 | 7.545771 | 0.619469 |
| AR | 28 | 24.579977 | 0.619469 |
| AHR | 27 | 40.32644 | 0.614035 |
| PLAU | 25 | 0.1359447 | 0.603448 |
| PRKCA | 24 | 5.8871307 | 0.598291 |
| HSPB1 | 24 | 3.353345 | 0.598291 |
| IGF2 | 23 | 0.6110213 | 0.59322 |
| BAX | 20 | 4.339728 | 0.578512 |
| GJA1 | 20 | 3.928358 | 0.578512 |
| GSTP1 | 20 | 25.447138 | 0.569106 |
| SOD1 | 19 | 12.657464 | 0.57377 |
| CYP1A1 | 18 | 17.28925 | 0.564516 |
| PTGS1 | 17 | 13.424284 | 0.56 |
| CYP3A4 | 17 | 18.700836 | 0.564516 |
| BIRC5 | 14 | 1.4444444 | 0.522388 |
| GSTM1 | 14 | 9.805274 | 0.534351 |
| CHEK2 | 13 | 1.2958502 | 0.538462 |
| PIK3CG | 12 | 2.7351422 | 0.518519 |
| PRKCB | 12 | 2.64741 | 0.510949 |
| CYP2C9 | 11 | 3.9084525 | 0.507246 |
| BCL2 | 10 | 1.4067007 | 0.510949 |
| ODC1 | 10 | 0.46414414 | 0.522388 |
| CYP1A2 | 10 | 3.8603175 | 0.503597 |
| CYP2B6 | 10 | 2.8008583 | 0.503597 |
| XDH | 9 | 3.2610993 | 0.518519 |
| PTPN22 | 6 | 0 | 0.5 |
| DRD2 | 5 | 0.06451613 | 0.482759 |
| PRSS1 | 4 | 0 | 0.47619 |
| ACACA | 4 | 0 | 0.5 |
| CHRM3 | 2 | 138 | 0.460526 |
| CHRM5 | 1 | 0 | 0.316742 |
Table 2: The Related Information of the PPI Network
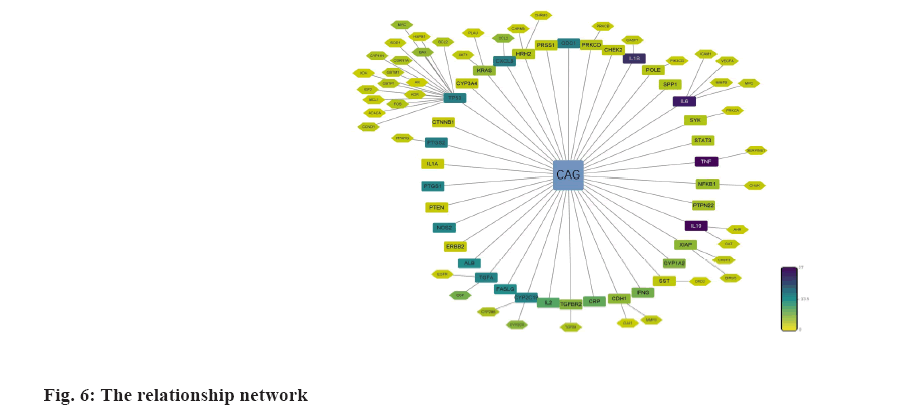
We analyzed the 71 common targets using VarElect to investigate the correlation between targets and CAG, and the results suggest that 26 targets were related to CAG directly, whereas 45 targets were related to CAG indirectly (fig. 6); among these targets, Tumor Protein P53 (TP53), AKT Serine/Threonine Kinase 1 (AKT1), Tumor Necrosis Factor (TNF), Epidermal Growth Factor Receptor (EGFR) and Interleukin-1 beta (IL-1β) have the highest score of correlation.
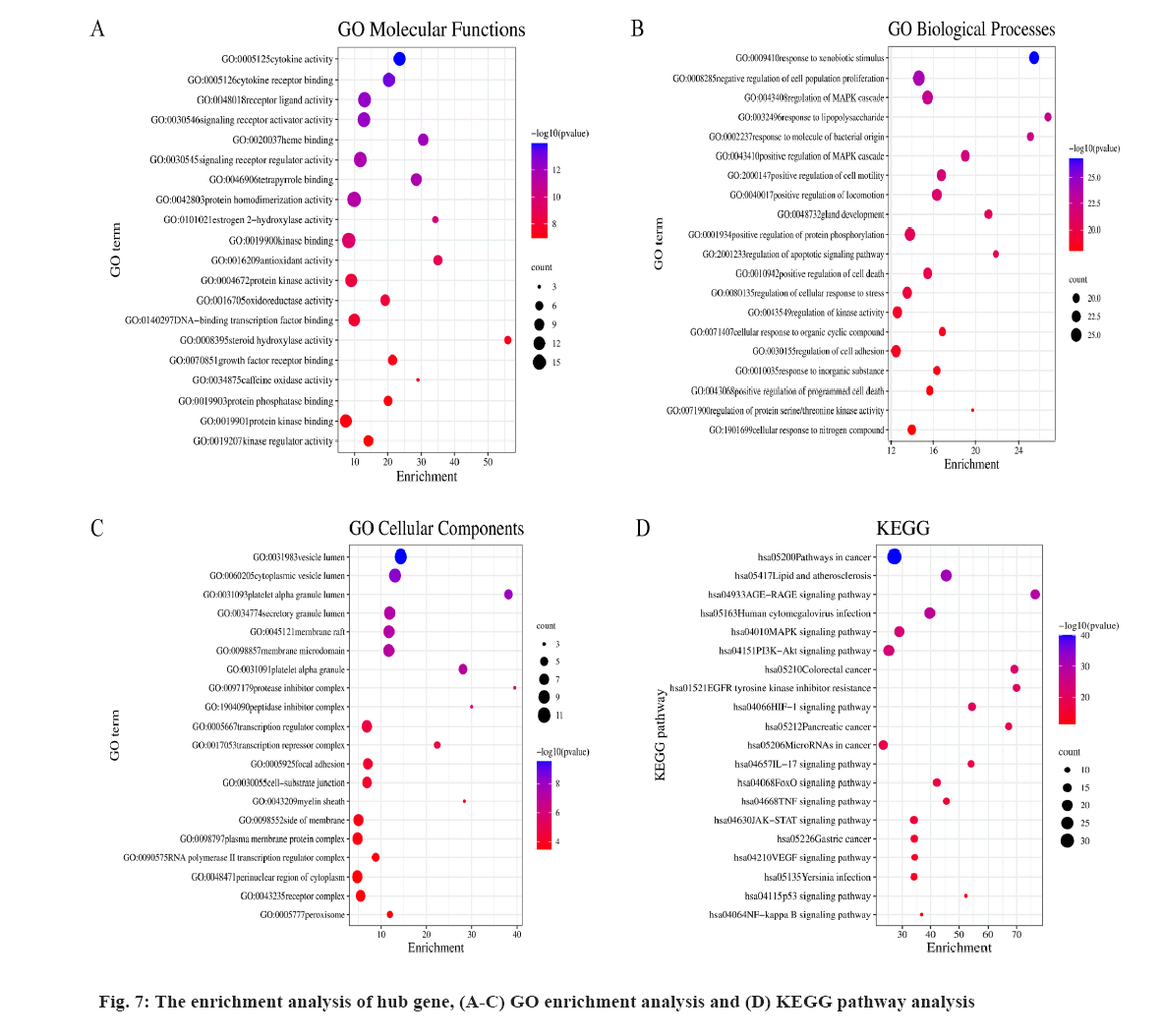
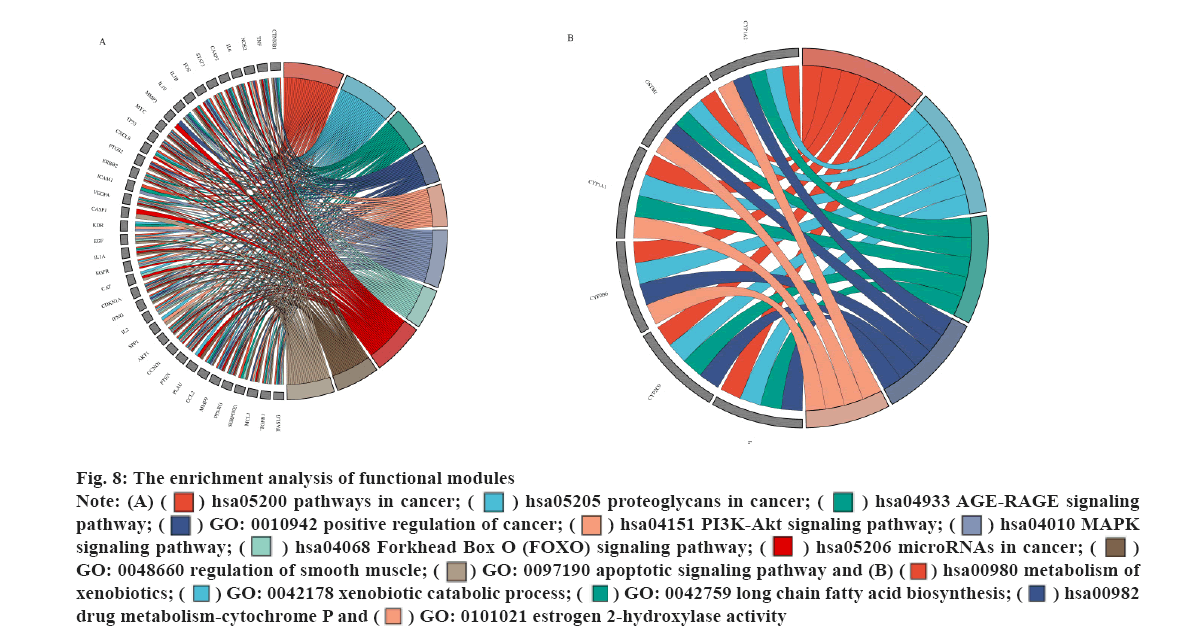
The enrichment analysis of GO and KEGG of the 21 common targets was showcased in fig. 7A-fig. 7D, the results were ranked by −log 10 (p value). Besides, the functional analysis of the two potential biofunctional modules divided from the PPI network is shown in fig. 8. In GO and KEGG enrichment analysis, terms with high enrichment scores suggest that Mitogen-Activated Protein Kinase (MAPK) signaling pathway, TNF signaling pathway and Phosphatidylinositol 3-Kinase (PI3K)/Protein Kinase B (Akt) signaling pathway could be the most possible mechanisms of BXD in treating CAG. For the 2 protein modules, module 1 can regulate the MAPK signaling pathway, PI3K-Akt signaling pathway, Advanced Glycation Endproducts-Receptor for Advanced Glycation Endproducts (AGE-RAGE) signaling pathway and apoptotic signaling pathway. Module 2 can regulate the metabolism of xenobiotics by xenobiotic catabolic process, long-chain fatty acid biosynthesis, drug metabolism-cytochrome P and estrogen 2-hydroxylase activity, which suggested BXD can act on CAG through multiple pathways.
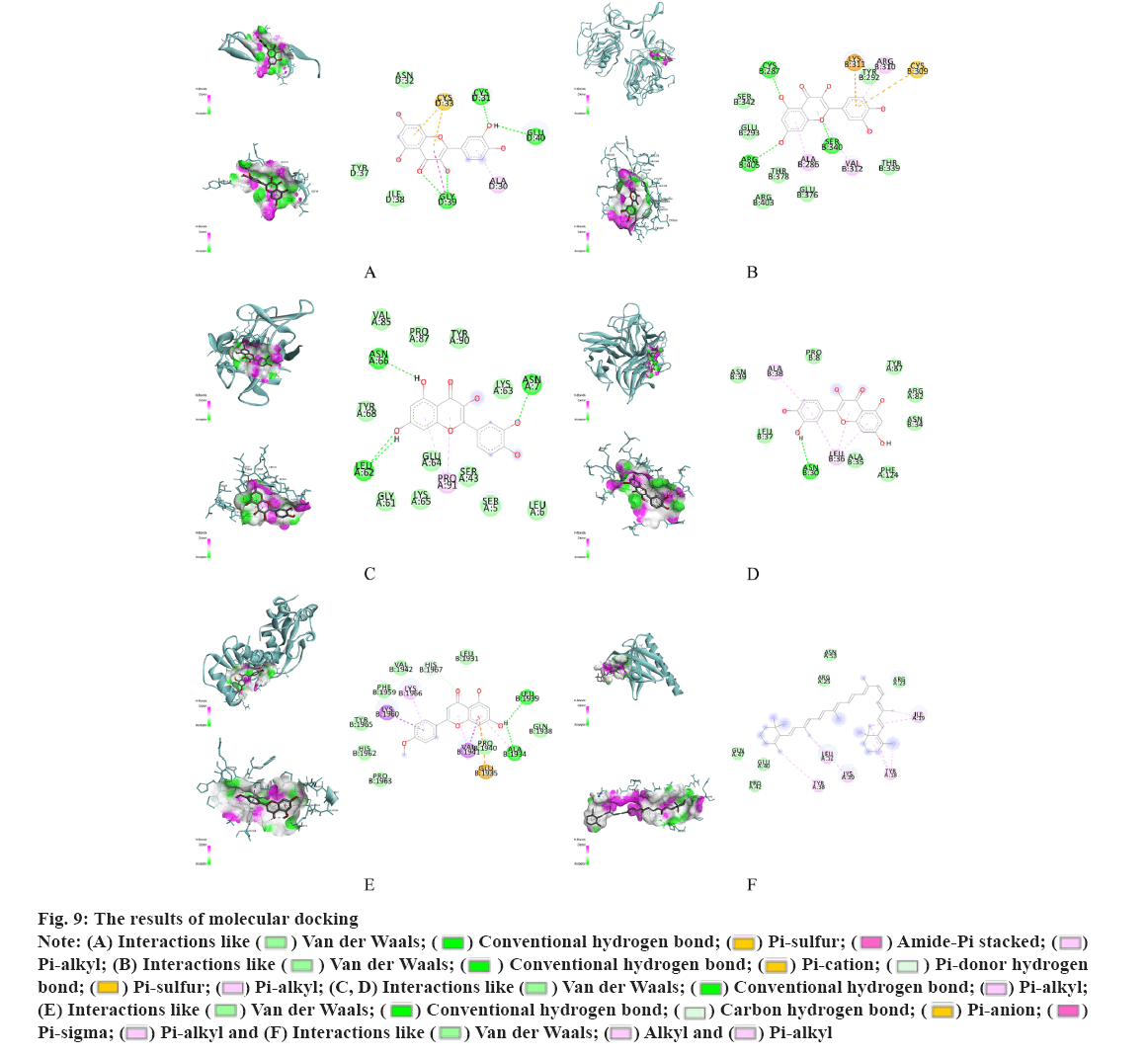
The molecular structure were downloaded from Research Collaboratory for Structural Bioinformatics (RCSB) database, TNF (PDB ID: 1A8M), TP53 (PDB ID: 1GZH), AKT1 (PDB ID: 1h10), IL-1β (PDB ID: 1hib), EGF (PDB ID: 1IVO) and EGFR (PDB ID: 1IVO). As shown in fig. 9, the molecular docking results demonstrated the binding of hub targets and corresponding components. The docking information of components and hub targets was listed in Table 3 and the results suggested that components in Data Flow Diagram (DFD) could interact with the hub targets against Ventricular Arrhythmia (VA), following the principle that the lower the binding energy, the more stable the docking modules. The docking results mainly show quercetin has the highest binding energy connected with TNF (fig. 9A), quercetin connected with TP53 (fig. 9B), beta- carotene connected with AKT1 (fig. 9C), quercetin connected with IL-1β (fig. 9D), quercetin connected with EGF (fig. 9E) and quercetin connected with EGFR (fig. 9F).
| Ligand | Mol ID | PubChem CID | Molecular targets | Binding energy |
|---|---|---|---|---|
| Quercetin | MOL000098 | 5280343 | TNF | -7.1 |
| Wogonin | MOL000173 | 5281703 | TNF | -6.3 |
| Kaempferol | MOL000422 | 5280863 | TNF | -6.8 |
| Ginsenoside Rh2 | MOL005344 | 119307 | TNF | -7.1 |
| Quercetin | MOL000098 | 5280343 | TP53 | -7.9 |
| Wogonin | MOL000173 | 5281703 | TP53 | -7.8 |
| Acacetin | MOL001689 | 5280442 | TP53 | -7.7 |
| Baicalein | MOL002714 | 5281605 | TP53 | -7.8 |
| Quercetin | MOL000098 | 5280343 | AKT1 | -6.1 |
| Wogonin | MOL000173 | 5281703 | AKT1 | -6 |
| Kaempferol | MOL000422 | 5280863 | AKT1 | -6.1 |
| Baicalein | MOL002714 | 5281605 | AKT1 | -6.7 |
| Beta-carotene | MOL002773 | 5280489 | AKT1 | -6.9 |
| Naringenin | MOL004328 | 439246 | AKT1 | -6.2 |
| Quercetin | MOL000098 | 5280343 | IL-1β | -7.3 |
| Ginsenoside Rh2 | MOL005344 | 119307 | IL-1β | -7.2 |
| Quercetin | MOL000098 | 5280343 | EGF | -8.6 |
| Quercetin | MOL000098 | 5280343 | EGFR | -6.4 |
Table 3: The Details of Molecular Docking
BXD is a classical formula of traditional Chinese medicine, which has been widely used to treat CAG in China and achieved satisfactory effects. Nevertheless, the identification of active ingredients and pharmacological molecular mechanism of Metachromatic Leukodystrophy (MLD) is unclear and needs to be solved urgently. In this study, the bioactive components and underlying molecular mechanisms of BXD in the treatment of CAG were analyzed systematically.
Through related information, collection and primary screening, we identified 21 potential targets of BXD in the treatment of CAG. A PPI network was constructed with STRING and Cytoscape 3.8.0, the top 10° value genes were selected as hub genes and 2 functional modules were divided based on their interactions. All potential genes were analyzed using VarElect, all 10 hub genes are suggested directly related to the treatment of CAG and among these genes, TP53, AKT1, TNF, EGFR as well as IL-1β, have the highest scores of correlations; in other words, these genes are the most promising targets for BXD against CAG. Previous results demonstrate that TP53 expression is significantly correlated as independent prognostic factors with tumor cell proliferation and might be associated with relevant events involved in gastrointestinal tumor biology[21]. One research suggested that TP53 plays an important role in the development of CAG, intestinal metaplasia, dysplasia and the intestinal type of gastric cancer. Accumulation of molecular abnormalities may increase the susceptibility to the inflammatory response of the gastric mucosa and the severity of gastric mucosal damage correlates with the presence of mutations in the gastric mucosa and the age of patients[22]. Meanwhile, another research observed that expression of TP53 continually increased along the normal epithelium-atrophic gastritis-dysplasia- carcinoma sequence[23]. A study conducted using an ethanolic extract of Panax ginseng Berry Calyx (Pg- C-EE) suggested that the Pg-C-EE may have Nuclear- Factor-kappa B (NF-κB)-targeted anti-inflammatory properties through suppression of AKT1 and play an anti-inflammatory role in a variety of inflammatory diseases such as gastritis[24]. And a study reported that Akt rs1130233 polymorphisms were associated with increased CAG risk in Helicobacter pylori (H. pylori)-negative individuals. Akt rs1130233 genotype has a significant interaction between the H. pylori infection in the progression from healthy controls to CAG, even gastric cancer influenced phosphorylated Akt (p-Akt) protein expression in H. pylori-infected individuals[25].
Further, the results of GO and KEGG analysis elucidated the MAPK signaling pathway, TNF signaling pathway and PI3K-Akt signaling pathway, which revealed the potential mechanisms of BXD in treating CAG. According to the study of Jianmei Zhang, Hymenocallis littoralis (Jacq.) Salisb, also known as Pancratium littorale Jacq, exerts eminently anti-inflammatory activities in Lipopolysaccharide (LPS)-stimulated RAW264.7 cells in vitro and in Hydrochloric acid (HCl)/Ethanol (EtOH)-induced gastritis mice models in vivo and these activities could be attributed to its modulatory effects on the MAPK signaling pathway[26]. One study showed that Berberine (BBR) can improve CAG by reducing the expression of inflammatory factors TNF-α and IL- 1β, down-regulating the expression of Transforming Growth Factor-beta (TGF-β) axis-related signals such as TGF-β1, PI3K, p-Akt/Akt and improving the gastric tissue injury of CAG rats[27]. Similarly, there is a study which reported that Zuojin pill has a good therapeutic effect on CAG induced by N-methyl-N'-Nitro-N-Nitrosoguanidine (MNNG), which could significantly reduce the content of G-17 and inflammatory factors IL-8, TNF-α, IL-6 and IL-1β, inhibit the expression of TGF-β1, PI3K and their downstream signals p-Akt, phosphorylated mammalian Target of Rapamycin (p-mTOR), P70S6K[28]. Reports also suggested that inhibition of heparanase may reduce pro-inflammatory and pro-tumorigenic cytokines (i.e., IL-1, IL-6, IL-1β, TNF-α, Macrophage Inflammatory Protein-2 (MIP- 2), inducible Nitric Oxide Synthase (iNOS)), block a vicious circle driven by enhanced NF-κB and p38-MAPK signaling and thereby reduce the risk of gastritis and gastric cancer[29].
Besides, the potential targets were divided into 2 function modules, as shown in fig. 5. The enrichment analysis results indicate module 1 can regulate the MAPK signaling pathway, PI3K-Akt signaling pathway, AGE-RAGE signaling pathway and apoptotic signaling pathway, which suggests that module 1 has great potential in the anti-inflammatory response. Similarly, the results of enrichment analysis indicate that module 2 was involved in the regulation of the metabolism of xenobiotics by xenobiotic catabolic process, long-chain fatty acid biosynthesis, drug metabolism-cytochrome P and estrogen 2-hydroxylase activity, which has illustrated the anti- inflammatory potential of module 2. Furthermore, as shown in fig. 5, the multiple regulations in different aspects may benefit patients suffering from related diseases such as gastric ulcers, cancer and other diseases.
Nevertheless, the present study has some limitations. First, as a traditional Chinese decoction, BXD treats CAG through multicomponent and multitarget, indicating that the underlying mechanisms are complex. Although we have made some identification on the mechanisms, there is still a lot to explore. Here, this study is designed to understand the mechanism profoundly through network pharmacology as well as molecular docking technology. Like other network pharmacology analyses, taking the intersection of targets about BXD and CAG in this study not only follows the operating processes of network pharmacology but also makes the result of virtual screening more reliable[30]. Undoubtedly, targets that involve either BXD or CAG might be ignored and missed, a common and inevitable issue in network pharmacology. And for this reason, in the collection of components and targets, despite BXD and CAG, we tried our best to reduce bias by searching as much more databases as we can. Finally, so many components are boiled together and more research is needed to detect whether there are some newly formed compounds.
As we mentioned above, BXD could be employed for CAG through mechanisms, including complex interactions between related components and targets, as predicted by network pharmacology and molecular docking. This work confirmed that BXD could apply for the treatment of VA and promote the explanation of BXD for CAG in the molecular mechanisms. The systematic analysis in this work can provide a comprehensive consideration for further studies.
Author’s contributions:
Xin Rui Wu and Long Zhao conceived, designed and planned the study. Xin Rui Wu and Long Zhao acquired and analyzed the data. Long Zhao and Guo Hui Xiao interpreted the results. Xin Rui Wu drafted the manuscript and Long Zhao and Guo Hui Xiao contributed to the critical revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was funded by the General Project of Sichuan Provincial Administration of Traditional Chinese Medicine. This work was funded by the General Project of Sichuan Provincial Administration of Traditional Chinese Medicine [2021MS085].
Conflict of Interests
The authors declare that they have no conflicts of interest.
References
- Shah SC, Piazuelo MB, Kuipers EJ, Li D. AGA clinical practice update on the diagnosis and management of atrophic gastritis: Expert review. Gastroenterology 2021;161(4):1325-32.
[Crossref] [Google scholar] [PubMed]
- Annibale B, Esposito G, Lahner E. A current clinical overview of atrophic gastritis. Expert Rev Gastroenterol Hepatol 2020;14(2):93-102.
[Crossref] [Google scholar] [PubMed]
- Du Y, Bai Y, Xie P, Fang J, Wang X, Hou X, et al. Chronic gastritis in China: A national multi-center survey. BMC Gastroenterol 2014;14(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 2019;47(D1):D976-82.
[Crossref] [Google scholar] [PubMed]
- Chen ZZ, Liu ZH. Research progress of traditional Chinese medicine in the treatment of chronic atrophic gastritis. J Chin Folk Ther 2022;30(15):117-9.
- Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J. Network pharmacology in research of Chinese medicine formula: Methodology, application and prospective. Chin J Integr Med 2020;26(1):72-80.
[Crossref] [Google scholar] [PubMed]
- Zhang GB, Li QY, Chen QL, Su SB. Network pharmacology: A new approach for Chinese herbal medicine research. Evid Based Complement Alternat Med 2013;2013:1-9.
[Crossref] [Google scholar] [PubMed]
- Liang Y, Liang B, Chen W, Wu XR, Liu-Huo WS, Zhao LZ. Potential mechanism of Dingji Fumai decoction against atrial fibrillation based on network pharmacology, molecular docking and experimental verification integration strategy. Front Cardiovasc Med 2021;8:712398.
[Crossref] [Google scholar] [PubMed]
- Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13(6):6964-82.
[Crossref] [Google scholar] [PubMed]
- Tian S, Wang J, Li Y, Li D, Xu L, Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev 2015;86:2-10.
[Crossref] [Google scholar] [PubMed]
- Che FF, Liang Y, Li H, Chen W, Li XL, Zhao LZ. Potential mechanism of maslinic acid against hypolipidemia based on network pharmacology and molecular docking integration strategy. Indian J Pharm Sci 2022:162-71.
- Liang Y, Liang B, Wu XR, Chen W, Zhao LZ. Network pharmacology-based systematic analysis of molecular mechanisms of Dingji Fumai decoction for ventricular arrhythmia. Evid Based Complement Alternat Med 2021;2021:1-12.
[Crossref] [Google scholar] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43(D1):D447-52.
[Crossref] [Google scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
[Crossref] [Google scholar] [PubMed]
- Stelzer G, Plaschkes I, Oz-Levi D, Alkelai A, Olender T, Zimmerman S, et al. VarElect: The phenotype-based variation prioritizer of the GeneCards suite. BMC Genomics 2016;17(2):195-206.
[Crossref] [Google scholar] [PubMed]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Goodsell DS, Zardecki C, di Costanzo L, Duarte JM, Hudson BP, Persikova I, et al. RCSB protein data bank: Enabling biomedical research and drug discovery. Protein Sci 2020;29(1):52-65.
[Crossref] [Google scholar] [PubMed]
- Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ 2016;44(5):433-7.
[Crossref] [Google scholar] [PubMed]
- Goodsell DS, Sanner MF, Olson AJ, Forli S. The AutoDock suite at 30. Protein Sci 2021;30(1):31-43.
[Crossref] [Google scholar] [PubMed]
- Montero E, Abreu C, Tonino P. Relationship between VEGF and p53 expression and tumor cell proliferation in human gastrointestinal carcinomas. J Cancer Res Clin Oncol 2008;134(2):193-201.
[Crossref] [Google scholar] [PubMed]
- Hnatyszyn A, Szalata M, Skrzypczak-Zielinska M, Wielgus K, Stanczyk J, Dziuba I, et al. DNA variants in Helicobacter pylori infected patients with chronic gastritis, dysplasia and gastric cancer. Adv Med Sci 2019;64(1):79-84.
[Crossref] [Google scholar] [PubMed]
- Kim SH, Lee SH, Choi YL, Wang LH, Park CK, Shin YK. Extensive alteration in the expression profiles of TGFB pathway signaling components and TP53 is observed. Histol Histopathol 2008;23(12):1439-52.
[Crossref] [Google scholar] [PubMed]
- Han SY, Kim J, Kim E, Kim SH, Seo DB, Kim JH, et al. AKT-targeted anti-inflammatory activity of Panax ginseng calyx ethanolic extract. J Ginseng Res 2018;42(4):496-503.
[Crossref] [Google scholar] [PubMed]
- Piao Y, Li Y, Xu Q, Liu JW, Xing CZ, Xie XD, et al. Association of MTOR and AKT gene polymorphisms with susceptibility and survival of gastric cancer. PLoS One 2015;10(8):e0136447.
[Crossref] [Google scholar] [PubMed]
- Zhang J, Sayakoummane S, Kim SA, Lee JS, Choung ES, Kim ES, et al. Hymenocallis littoralis ameliorates inflammatory responses in LPS-stimulated RAW264. 7 cells and HCl/EtOH-induced gastric mucosal injury via targeting the MAPK pathway. J Ethnopharmacol 2022;295:115400.
[Crossref] [Google scholar] [PubMed]
- Tong Y, Liu L, Wang R, Yang T, Wen J, Wei S, et al. Berberine attenuates chronic atrophic gastritis induced by MNNG and its potential mechanism. Front Pharmacol 2021 5;12:644638.
[Crossref] [Google scholar] [PubMed]
- Tong Y, Wang R, Liu X, Tian M, Wang Y, Cui Y, et al. Zuojin Pill ameliorates chronic atrophic gastritis induced by MNNG through TGF-β1/PI3K/Akt axis. J Ethnopharmacol 2021;271:113893.
[Crossref] [Google scholar] [PubMed]
- Tang L, Tang B, Lei Y, Yang M, Wang S, Hu S, et al. Helicobacter pylori-induced heparanase promotes H. pylori colonization and gastritis. Front Immunol 2021;12:675747.
[Crossref] [Google scholar] [PubMed]
- Shao LI, Zhang B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin J Nat Med 2013;11(2):110-20.
[Crossref] [Google scholar] [PubMed]