- *Corresponding Author:
- Z. A. Khan
Faculty of Life Sciences, Baba Farid College, Bathinda, Punjab 151001, India
E-mail: khan.zaved@gmail.com
| Date of Received | 29 January 2022 |
| Date of Revision | 18 May 2023 |
| Date of Acceptance | 04 August 2023 |
| Indian J Pharm Sci 2023;85(4):1045-1067 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Phytoconstituents epigallocatechin gallate and withaferin A, found in Camellia sinensis (Kangra green tea) and Withania sominifera (Ashwagandha) respectively, were explored for their binding affinity towards various enzymes involved in the skin-aging process. Epigallocatechin gallate and withaferin A were analyzed for their physiochemical properties, drug-likeness and human intestinal absorptivity using Data Warrior, Molsoft and SwissADME (boiled egg model) respectively. Molecular docking analysis for different enzymes involved in aging (collagenase, elastase and hyaluronidase), antioxidant enzymes (superoxide dismutase, glutathione-s-transferase, glutathione peroxidase and catalase) and mitochondrial enzymes (nicotinamide adenine dinucleotide (NAD)+hydrogen (H) dehydrogenase, succinate dehydrogenase, cytochrome c oxidase and adenosine triphosphate synthase) was carried out for epigallocatechin gallate alone (1), withaferin A alone (2), epigallocatechin gallate and withaferin A in combination (3) and a reference molecule. Autodock Vina was employed to carry out individual molecular docking as well as multiple ligand simultaneous docking. The results were analyzed in terms of binding energy and different interacting residues. Interestingly, (3) displayed a higher binding affinity towards all the aging and antioxidant enzymes as compared to (1), (2) and the references. Moreover, the combination of the constituents exhibited better binding for most of the mitochondrial enzymes. Additionally, molecular dynamics simulations were performed to estimate stability and flexibility of best complexes, while collagenase activity colorimetric assay was carried out to study the effects of (1), (2) and (3) on collagenase. The in vitro analysis indicated a 1.5 times increase in collagenase inhibition upon using (3) as compared to ascorbic acid (standard). Overall, the results indicate that epigallocatechin gallate and withaferin A, in combination, may potentially inhibit skin-aging, while enhancing antioxidant effects of various enzymes, and warrant further experimental validation.
Keywords
Anti-aging, green tea, ashwgandha, withaferin A, epigallocatechin gallate, molecular docking
Skin aging is the result of complex intrinsic and extrinsic factors such as natural growth, exposure to environmental pollution, exposure to sun and or exposure to carcinogenic substances[1]. Ultraviolet radiation can cause change in physical texture and appearance of the skin via complex signaling cascades, which results in the production of free radicals. This, consequently, leads to oxidative stress, which further contributes to unhealthy skin, dryness, wrinkling and uneven pigmentation. Also, production of Matrix Metallo Proteinases (MMPs) of the skin is induced via activation of the Mitogen-activated Protein Kinases (MAPK) signaling cascade[2]. MMPs are a group of zinc-dependent extracellular proteinases which include collagenase and elastase, responsible for the degradation of collagen and elastin respectively[1]. In other words, overproduction of MMPs degrade collagen and elastin fibers required for skin elasticity, which might contribute to premature skin aging[1]. Production of several reactive oxygen species damages Deoxyribonucleic Acid (DNA), proteins and lipids[3].It may also cause premature aging, sunburn and or skin cancer[4].
Plants-based chemicals i.e. phytochemicals have become an efficient source to balance free radicals and consequently, to negate the age-related diseases caused by free radicals. Several fruits and vegetables contain antioxidants such as vitamin E, betacarotene, vitamin C, selenium, coenzyme Q10, and phytoconstituents such as flavonoids in green tea, onions, apples, etc. Antioxidants act via several chemical mechanisms like single electron transfer, hydrogen atom transfer and the ability to chelate transition metals[5,6].
Camellia sinensis (L.) O. Kuntze (Kangra green tea) is a local variety of Himachal Pradesh cultivated on the Western Himalayas at a sea level of 1290 m and 32°27’15.68” N latitude, 76°31’42.26” E longitude since the 1850s[7]. Epigallocatechin Gallate (EGCG) is the most active phytoconstituent of green tea, and is widely studied for its antiaging properties[8]. Green tea has been recognized for its anticancerous[ 9], anti-diabetic[10], anti-proliferative[11], anti-inflammatory[12], anti-atherosclerosis[13], neuroprotective[14], antioxidant[15], estrogenic[16] and anti-aging[17] properties. Similarly, Withania sominifera has also been reported for its medicinal, anti-aging, immunomodulatory, and rejuvenating properties in Ayurveda[18]. Off late, many in silico studies indicate at the potential of Withania sominifera in the fight against COVID-19, attributed to its antiviral and immunomodulatory activities[19-21].
Several recent studies indicate the beneficial synergistic effect of phytoconstituents. A combination of vitamin E with astaxanthin, a natural carotenoid that can scavenge free radicals, was reported to increase the effect of the latter[22]. Another study on ferulic acid in combination with resveratrol reported an increase in apoptotic cell death in cancer cell[23]. Moreover, tetrahydro curcumin in combination with Centella asiatica extract, helped to reduce the symptoms of aging[24]. Based on these synergistic reports, in silico analysis was conducted in the present study to estimate the effect of EGCG (Camellia sinensis) and withaferin A (Withania sominifera) on enzymes involved in skin-aging. Different aging, antioxidant and mitochondrial enzymes were targeted and the synergistic effect of both these components was observed. AutoDock tool was employed to conduct Multiple Ligand Simultaneous Docking (MLSD), to observe the combinatorial anti-aging potential of EGCG and withaferin A. Additionally, molecular dynamic simulations were performed to affirm the stability and flexibility of the complexes[25], as well as effect of the two compounds on collagenase inhibition was estimated under in vitro conditions,
Materials and Methods
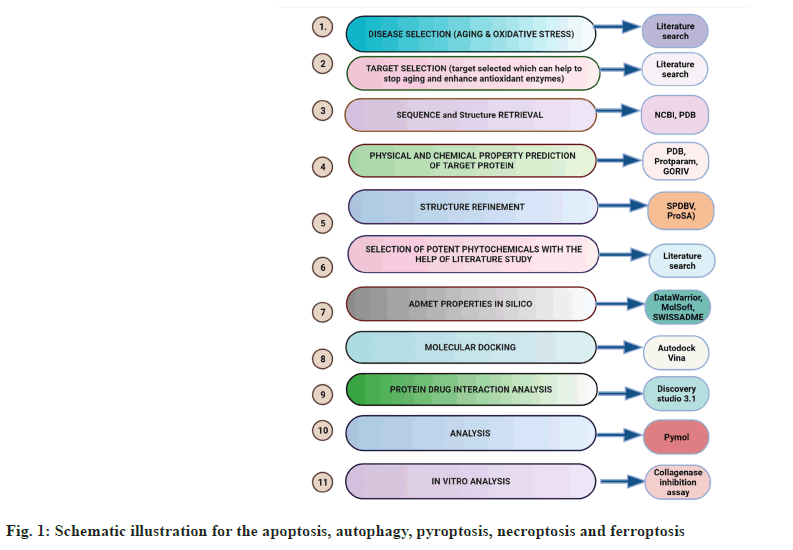
An overview of the methodology followed is presented in fig. 1.
Ligand preparation and Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) analysis:
Ligand structure: The Three-Dimensional (3D) structure of EGCG and withaferin A test ligands and reference molecules (ascorbic acid, ursolic acid, rosmarinic acid and resveratrol) were downloaded from PubChem in .sdf and .mol2 format and visualized in PyMOL[26]. The structures were then converted into .pdb format by using Open Babel GUI software[27]. These 3D structures were used during docking. A 2D view of these structures, obtained via ChemDraw, is presented in Table 1.
| Ligands | PubChem ID | IUPAC name | Structure |
|---|---|---|---|
| EGCG | 65064 | [(2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 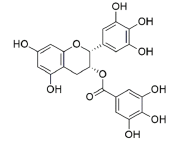 |
| Withaferin A | 265237 | ((1S,2R,6S,7R,9R,11S,12S,15R,16S)-6-hydroxy-15-[(1S)-1-[(2R)-5-(hydroxymethyl)-4-methyl-6-oxo-2,3-dihydropyran-2-yl]ethyl]-2,16-dimethyl-8-oxapentacyclo[9.7.0.02,7.07,9.012,16]octadec-4-en-3-one | 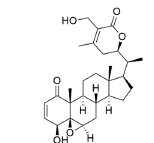 |
| Reference compounds | |||
| Ascorbic acid | 54670067 | (2R)-2-[(1S)-1,2-dihydroxyethyl]-3,4-dihydroxy-2H-furan-5-one | 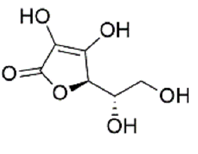 |
| Ursolic acid | 64945 | (1S,2R,4aS,6aR,6aS,6bR,8aR,10S,12aR,14bS)-10-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydro-1H-picene-4a-carboxylic acid | 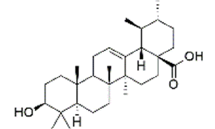 |
| Rosmarinic Acid | 5281792 | (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoic acid | 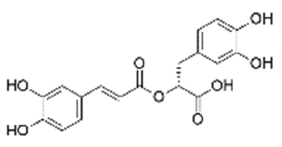 |
| Resveratrol | 445154 | 5-[(E)-2-(4-hydroxyphenyl)ethenyl]benzene-1,3-diol | 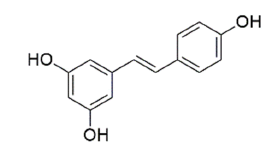 |
Table 1: The Structures of Various Ligands (Test and Reference)
Determination of ADMET properties: Ligands were analyzed for their toxicity, hydropathicity, mutagenicity, tumorigenicity, reproductive effect and irritant, clogP and isoelectric point using Data Warrior[28]. The drug-likeness score of an individual molecule was predicted using MolSoft (https://molsoft.com/mprop/). Molsoft analyses the ligands according to the Lipinski rule. Lipinski filters or Lipinski rule of five distinguishes between drug like and nondrug like molecules[29,30]. It is also known as Pfizer's rule of five. It is a thumb rule to predict the probability of a chemical to be orally active drug in humans. According to this rule molecular mass of molecule should be less than 500 Dalton, lipophilicity represented by LogP should be less than 5, Hydrogen Bond Donors (HBD) should be less than 5, Hydrogen Bond Acceptors (HBA) should be less than 10 and Molar Refractivity (MR) should be lies between 40-130. Molecule failing in more than 2 rules is considered as nondrug molecule. Further, boiled egg model was used to predict the human intestinal absorptivity of bio-actives using SwissADME[31].
Molecular docking: High-resolution crystal structures for aging (collagenase, elastase, and hyaluronidase), antioxidant (superoxide dismutase, catalase, glutathione peroxidase and glutathiones- transferase) and mitochondrial (Nicotinamide Adenine Dinucleotide (NAD)+Hydrogen (H) (NADH) dehydrogenase, succinate dehydrogenase, cytochrome c oxidase and Adenosine Triphosphate (ATP) synthase) enzymes were retrieved from Protein Data Bank (PDB)[32] (Table 2). Various enzymes were treated as receptors, while EGCG and withaferin A acted as ligands. Ascorbic acid was used as standard for all antioxidant enzymes[33], resveratrol was used as standard for all mitochondrial enzymes[34,35], while ascorbic acid, ursolic acid, and rosmarinic acid were used as standards for aging enzymes viz. collagenase, elastase and hyaluronidase respectively[36-38].
| Category | Enzyme | PDB ID | Resolution (Å) |
|---|---|---|---|
| Aging | Collagenase | 2CLT | 2.67 |
| Elastase | 7CBK | 2.7 | |
| Hyaluronidase | 2PE4 | 2 | |
| Antioxidant | Superoxide dismutase | 2JLP | 1.7 |
| Glutathione-s-transferase | 2VCT | 2.1 | |
| Glutathione peroxidase | 3KIJ | 1.8 | |
| Catalase | 1DGB | 2.2 | |
| Mitochondrial enzymes | NADH dehydrogenase | 5XTC | 3.7 |
| Succinate dehydrogenase | 1ZOY | 2.4 | |
| Cytochrome c oxidase | 5Z62 | 3.6 | |
| ATP synthase | 2XND | 3.5 |
Table 2: The Various Enzymes (Receptors) Selected For Molecular Docking Via Autodock Vina
Ligand molecules and receptor molecules were read separately using Autodock Tools (ADT). Water molecules and other atoms were deleted, and ADT measured the Gasteiger charges for protein atoms; Auto Grid was used with a grid box to create the grid map. The receptor and ligand files were saved with appropriate torsions and charges in Protein Data Bank, Partial Charge (Q), and Atom Type (T) (PDBQT) format. Docking was performed using Autodock Vina[39-41]. In the MLSD approach, we docked EGCG and withaferin A simultaneously with each enzyme.
Structures visualization was done by using Discovery Studio Visualizer and PyMOL. Analysis was done based on complex binding energy and interacting residues. The binding energies obtained on docking EGCG alone, withaferin A alone and a combination of these ligands were compared to the values obtained for standards.
Molecular Dynamics (MD): MD simulation studies were carried out in order to determine the backbone configuration of target proteins bound to EGCG and withaferin A. MD simulation study was carried out in Desmond vs 2020-1[42]. To set up the initial parameters of an orthorhombic box of 10×10×10 Å, Desmond system builder was used. The target proteins and target-ligand complex were neutralized using NaCl (sodium chloride) by adding 0.15 M Na+ ions. The prepared systems were relaxed using the Desmond default protocol of relaxation[43]. An MDS run of 50 ns was set up at constant temperature and constant pressure (NPT) for the final production run. The NPT ensemble was set up using the Nosé-Hoover chain coupling scheme at a temperature of 300 K for final production and throughout the dynamics with relaxation time 1 ps. A Reversible Reference System Propagator Algorithms (RESPA) integrator was used to calculate the bonding interactions for a time step of 2 fs. All other parameters were associated in the settings followed as described by Shaw et al.[43]. After the final production run, the simulation trajectories of target proteins complexed with selected ligand molecules were analyzed for the final outcome of Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), and ligand RMSF derived from the simulation studies.
Sample collection: Leafy twigs of Camellia sinensis (Kangra green tea) and roots of Withania somnifera (Ashwagandha) were procured from Palampur tea gardens (Himachal Pradesh, India) (32°27’15.68” N latitude, 76°31’42.26” E) and Mohali (Punjab, India) (latitude 30.7046° N, longitude 76.7179° E) respectively. Plant samples were kept in a sterile bag and stored at 4° till further use.
Plant extract and stock solution preparation: Petroleum ether, methanol, ethanol and water (1:1) extracts of Camellia sinensis and Withania somnifera were prepared by the method described by Lunder et al.[15] with slight modifications. 20 g powdered roots of Withania somnifera and powdered leaves of Camellia sinensis were soaked in petroleum ether, methanol, ethanol and water (200 ml) each for one day at room temperature. The samples were vacuum filtrated by using Whatman filter paper and after filtration, the filtrate was subjected to evaporation with the help of a rotary evaporator. Dried crude samples were collected and stored at 4°.
Collagenase inhibitory assay: Inhibition of collagenase was estimated by using collagenase activity colorimetric assay kit (SIGMA). Collagenase provided in the kit was used, while a synthetic peptide 2-furanacryloyl-l-leucylglycyll- prolyl-l-alanine (FALGPA) mimics collagen, and acts as substrate. Crude extracts of Camellia sinensis (containing EGCG) and Withania somnifera (containing withaferin A) were dissolved in methanol at various concentrations (50, 100, 150, 200 μg/ml). A combination of the two extracts was prepared by dissolving equal concentrations of each extract (25+25, 50+50, 75+75 and 100+100 μg/ml). Ascorbic acid (a known inhibitor of collagenase) was used as a positive control.
The experiment was carried out in triplicates. 5 μl collagenase, 50 μl substrate and 5 μl diluted extract (Camellia sinensis, Withania somnifera and combined extracts, at varying concentrations) were added to their respective wells. For positive control, 5 μl ascorbic acid (50 μg/ml) was added. The volume was raised to 200 μl per well with the help of (Tris (hydroxymethyl) aminomethane (THAM) hydrochloride) Tris-HCL buffer. Absorbance was monitored at 340 nm after 30 mins of incubation.
The results were measured in terms of collagenase inhibition activity as (1-sample abs-blank abs/ positive control abs-negative control abs)×100
Results and Discussion
As per Lipinski rule of 5, EGCG has violated one rule of hydrogen bond donor and Ursolic acid violated two rules, i.e. molar refractrivity and Lipophilicity. Therefore, Ursolic acid fails in this category. According to bioavailablility radar of SwissAdme which consider six physicochemical properties; Lipophilicity, size, polarity, solubility, flexibility and saturation. Range of Lipophilicity: XLOGP3 between −0.7 and +5.0, size: Molecular weight between 150 and 500 g/mol, polarity: topological polar surface area between 20 and 130 Å2, solubility: log S not higher than 6, saturation: fraction of carbons in the sp3 hybridization not less than 0.25, and flexibility: Not more than 9 rotatable bonds[31]. Ursolic Acid violated lipophilicity, solubility and molar refractivity, EGCG violates saturation (i.e. sp3), similarly rosamarinic acid and reservatrol violates saturation rule.
SwissADME[31] considers various methods like Lipinski (Pfizer) filter, Ghose (Amgen), Veber (GSK), Egan (Pharmacia) and Muegge (Bayer) methods for calculation of drug likeness[30,44-47]. While among all of above rules Lipinski filter is pioneer rule and followed by all the scientists throughout world. As per Table 3 results, except Ursolic acid rest all molecules can be used as drug as they do not have any toxic effect and most of them are fit according to Lipinski rule.
| Properties | Withaferin A | EGCG | Ascorbic acid | Ursolic acid | Rosamarinic acid | Resveratrol | |
|---|---|---|---|---|---|---|---|
| Physicochemical properties | Formula | C28H38O6 | C22H18O11 | C6H8O6 | C30H48O3 | C18H16O8 | C14H12O3 |
| Molecular wt. | 470.60 g/mol | 458.37 g/mol | 176.12 | 456.70 g/mol | 360.31 g/mol | 228.24 g/mol | |
| Number of heavy atoms | 34 | 33 | 12 | 33 | 26 | 17 | |
| Number of aromatic heavy atoms | 0 | 18 | 0 | 0 | 12 | 12 | |
| Fraction csp3 | 0.79 | 0.14 | 0.5 | 0.9 | 0.11 | 0 | |
| Num. Of rotatable bonds | 3 | 4 | 2 | 1 | 7 | 2 | |
| No. Of H-bond acceptors | 6 | 10 | 6 | 3 | 8 | 3 | |
| No. Of H-bond donors | 2 | 8 | 4 | 2 | 5 | 3 | |
| Molar refractivity | 127.49 | 112.06 | 35.12 | 136.91 | 91.4 | 67.88 | |
| TPSA | 96.36 Ų | 197.37 Ų | 107.22 | 57.53 Ų | 144.52 Ų | 60.69 Ų | |
| Lipophilicity | Log Po/w (ilogp) | 3.39 | 1.83 | 0.39 | 4.01 | 1.17 | 1.71 |
| Log Po/w (XLOGP3) | 3.83 | 1.17 | -1.64 | 7.34 | 2.36 | 3.13 | |
| Log Po/w (WLOGP) | 3.35 | 1.91 | -1.41 | 7.09 | 1.65 | 2.76 | |
| Log Po/w (MLOGP) | 2.75 | -0.44 | -2.6 | 5.82 | 0.9 | 2.26 | |
| Log Po/w (SILICOS-IT) | 3.93 | 0.57 | -1.15 | 5.46 | 1.5 | 2.57 | |
| Consensus Log Po/w | 3.45 | 1.01 | -1.28 | 5.94 | 1.52 | 2.48 | |
| Solubility | Log S (ESOL) | -3.56 | 0.23 | -7.23 | -3.44 | -3.62 | |
| Solubility | 1.27e-01 mg/ml ; 2.76e-04 mol/l | 3.01e+02 mg/ml ; 1.71e+00 mol/l | 2.69e-05 mg/ml ; 5.89e-08 mol/l | 1.31e-01 mg/ml ; 3.63e-04 mol/l | 5.51e-02 mg/ml ; 2.41e-04 mol/l | ||
| Class | Soluble | Highly soluble | Soluble | Soluble | Soluble | ||
| Log S (Ali) | -4.91 | -0.1 | -8.38 | -5.04 | -4.07 | ||
| Solubility | 5.64e-03 mg/ml ; 1.23e-05 mol/l | 1.40e+02 mg/ml ; 7.93e-01 mol/l | 1.92e-06 mg/ml ; 4.21e-09 mol/l | 3.32e-03 mg/ml ; 9.22e-06 mol/l | 1.93e-02 mg/ml ; 8.44e-05 mol/l | ||
| Class | Moderately soluble | Very soluble | Soluble | Moderately soluble | Moderately soluble | ||
| Log S (SILICOS-IT) | -2.5 | 1.49 | -5.67 | -2.17 | -3.29 | ||
| Solubility | 1.46e+00 mg/ml ; 3.18e-03 mol/l | 5.46e+03 mg/ml ; 3.10e+01 mol/l | 9.72e-04 mg/ml ; 2.13e-06 mol/l | 2.41e+00 mg/ml ; 6.70e-03 mol/l | 1.18e-01 mg/ml ; 5.16e-04 mol/l | ||
| Class | Soluble | Soluble | Moderately soluble | Soluble | Soluble | ||
| Pharmakokinetic properties | GI absorption | High | Medium | High | Medium | Medium | High |
| BBB permeant | No | Yes | Yes | Medium | Medium | Medium | |
| Log Kp (skin permeation) | -6.45 cm/s | -8.54 | -3.87 cm/s | -6.82 cm/s | |||
| P-gp substrate | Yes | No | Yes | Yes | Yes | Yes | |
| Drug-likeness | Lipinski | Yes | Yes | Yes | Yes | Yes | Yes |
| Bioavailability score | 0.55 | 0.37 | 0.56 | 0.85 | 0.56 | ||
| Tumorgenicity | No | No | No | No | No | No | |
| Mutagenicity | No | No | No | No | No | No | |
| Reproductive Effect | No | No | No | No | No | No | |
| Irritant | No | No | No | No | No | No | |
| Mol log p | 3.26 | 1.44 | -1.59 | 5 | 1.54 | 2.88 | |
| Mol log PSA A2 | 75.66A2 | 158.72A2 | 85.73A2 | 44.14 A2 | 114.28 A2 | 52.82 A2 | |
| Mol Vol A3 | 564.08A3 | 404.49A3 | 161.80 A3 | 576.10 A3 | 339.44 A3 | 224.35 A3 |
Note: † Number of hydrogen bond donor, molpsa: Molecular Polar Surface Area (PSA), and Volume, BBB: Blood Brain Barrier, GI: gastrointestinal, P-gp: glycoprotein
Table 3: Admet Properties And Drug-Likeness Score Of EGCG, Withaferin A And Reference Compounds (Ascorbic Acid, Ursolic Acid, Rosmarinic Acid And Resveratrol)
EGCG (Pubchem CID-65064) and withaferin A (PubChem CID-265237) were analyzed for their physicochemical properties Table 3. Prediction of human intestinal absorptivity indicated that both, EGCG and withaferin A, are likely to be absorbed from the gastrointestinal tract. Further, drug-likeness prediction identified EGCG and withaferin A to possess the highest drug-likeness score, i.e. 0.55 and 0.37 respectively, suggesting their high oral bioavailability (Table 3). Both the phytochemicals were found to follow Lipinski’s rule with no tumorigenic, mutagenic and irritant effects (Table 3), which makes these compounds suitable to carry out the synergistic studies.
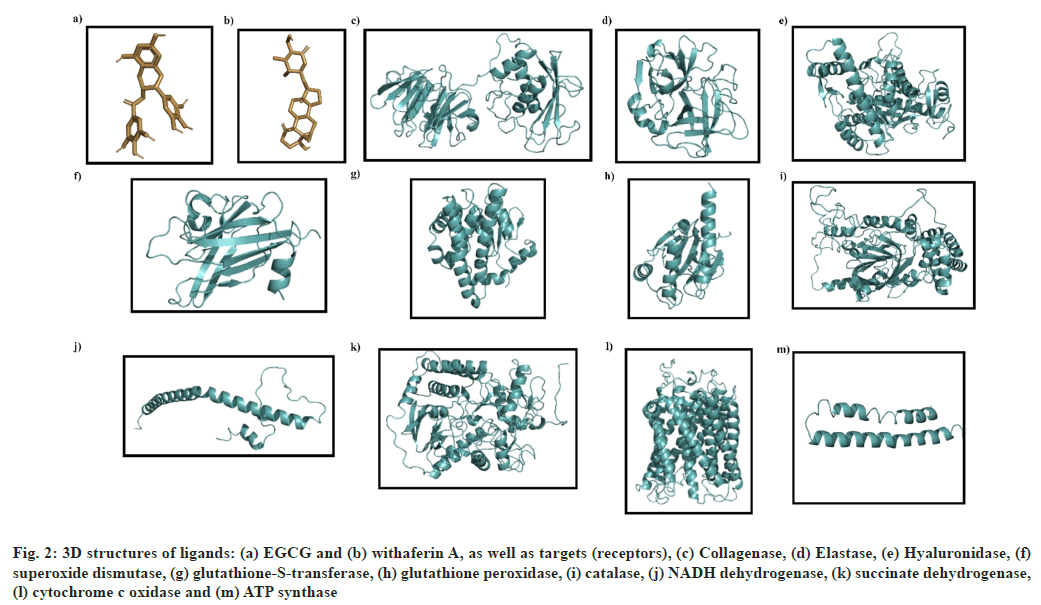
3D structures of various receptors, aging enzymes (collagenase, elastase and hyaluronidase), antioxidant enzymes (superoxide dismutase, catalase, glutathione peroxidase and glutathione-S-transferase) and mitochondrial enzymes (NADH dehydrogenase, succinate dehydrogenase, cytochrome c oxidase and ATP synthase) were downloaded from PDB. The 3D structures of both, the test ligands (EGCG and withaferin A) and standard molecules (ascorbic acid, ursolic acid, and rosmarinic acid) were obtained from PubChem. The various 3D structures have been presented in fig. 2.
Fig. 2: 3D structures of ligands: (a) EGCG and (b) withaferin A, as well as targets (receptors), (c) Collagenase, (d) Elastase, (e) Hyaluronidase, (f) superoxide dismutase, (g) glutathione-S-transferase, (h) glutathione peroxidase, (i) catalase, (j) NADH dehydrogenase, (k) succinate dehydrogenase, (l) cytochrome c oxidase and (m) ATP synthase
The structure of collagenase (PDB ID: 2CLT), a human hydrolase, contains 367 residues with a resolution of 2.67Å (fig. 2c) while elastase structure (PDB ID: 7CBK) is composed of 267 residues with a resolution of 2.70 Å (fig. 2d). The hyaluronidase (a human hydrolase) structure (PDB ID: 2PE4) is composed of 424 aa with a resolution of 2.00 Å (fig. 2e).
The crystal structure of superoxide dismutase (2JLP), a human oxidoreductase, is composed of 222 aa with a resolution of 1.70Å (fig. 2f). Glutathione-s-transferase structure (2VCT) is 222 residues in length, and resolved at 2.10 Å (fig. 2g). Glutathione peroxidase structure (3KIJ) is composed of 180 aa with a resolution of 1.70Å (fig. 2h), while catalase structure (1DGB) is composed of 498 aa with a resolution of 2.20 Å (fig. 2i).
NADH dehydrogenase structure (5XTC) is composed of 320 aa with a resolution of 3.70 Å (fig. 2j). Succinate dehydrogenase complex II is also known as succinate ubiquinone oxidoreductase. Its structure (1ZOY) comprises of 622aa with a resolution of 2.40Å (fig. 2k). The structure of cytochrome c oxidase complex IV of mitochondrial electron transport chain (562) consists of 513 residues at a resolution of 3.70 Å (fig. 2l), while the ATP synthase structure (2XND) is composed of 492 aa, resolved at 3.70 Å (fig. 2m).
To improve our understanding of the protective role of EGCG and withaferin A in a synergistic mode, molecular interactions of EGCG and withaferin A with various enzymes were analyzed simultaneously via molecular docking and compared with reference (standard) compounds i.e. ascorbic acid, ursolic acid, and rosmarinic acid (Table 4). Comparison of molecular docking analysis for the EGCG alone (1), withaferin A alone (2), EGCG+withaferin A (3) and references indicate highest binding energy for (3) in majority of the cases (Table 4).
| All enzymes | Standard (kcal/mol) | Binding Site residues* | |
|---|---|---|---|
| Collagenase (2CLT) | Ascorbic acid | -4.3 | T264, F265, D266, A267, K279, L309, E310, A311, H357, I358, D359, K406, V407, D408, A409 |
| Elastase (7CBK) | Ursolic acid | -4.2 | L35, R36, F41, C42, H57, C58, N61, V62, G193, S195 |
| Hyaluronidase (2PE4) | Rosmarinic acid | -5.9 | N37, I73, Y75, V127, D129, E131, Y202, Y247, Y286, D292, W321 |
| Superoxide dismutase (2JLP) | Ascorbic acid | -4.5 | P122, Q123, H124, P125, G126, D127, F128, G129, R142, G144, L145, A146, A147 |
| Glutathione-s-transferase (2VCT) | -5.5 | R13, G14, R15, E17, I106, L107, L108, L109, P110, F111, Y166, E169, L170, P206, P207, M208 | |
| Glutathione peroxidase (3KIJ) | -5.4 | A76, S77, D78, C79, Q80, L81, T82, D83, F125 | |
| Catalase (1DGB) | -6.4 | R72, V73, V74, H75, R112, S114, G131, F132, A133, V146, G147, I332, A333, F334, Y358, T361, H362, R365 | |
| NADH dehydrogenase (5XTC) | Resveratrol | -4.9 | L84, L87, R88, L91, E108, S109, V110, F111, W116, V117, P118, P119, L120, E123 |
| Succinate dehydrogenase (1ZOY) | -5.4 | A24, Q25, D38, W40, M44, Y45, T47, V48, K49, D149, L153, T156, E161, R203, A204, G205, V206, F216, T219, G387, R388, A404, T408 | |
| Cytochrome c oxidase (5Z62) | -6 | I66, W126, W236, V243, Y244, L246, I247, H291, F348, H376, F377, V380, L381, G384, A385, R438, R439 | |
| ATP synthase (2XND) | -4.3 | V28, S31, L32, L46, F47, Y49, A50, G53, L56, S57 | |
| Enzymes (aging) | EGCG (kcal/mol) | Binding site residues* | |
| Collagenase | -6.6 | T270, I271, R272, G273, E274, E314, F315, A316, D317, S363, E364, E365, N366, T370, Y384, M412, K413, D414, G415 | |
| Elastase | -7.5 | L35, H40, F41, C42, H57, C58, V59, A60, N61, V62, N63, L143, I151, V190, C191, F192, G193, D194, S195, V216, C220 | |
| Hyaluronidase | -6.8 | R238, V279, Q280, I282, G283, V284, A287, L288, A290, A291, V326, A329, D348, P349, G350, Q351, M352, E353, L358, F370, S371 | |
| Withaferin A (kcal/mol) | |||
| Collagenase | -7.2 | S224, G225, D226, P303, Q304, L305, P306, N307, K325, G326, N327, K328, W330, P342 | |
| Elastase | -7.4 | A26, W27, P28, F29, Q119, V120, A121, Q122, Q135, C136, L137, V200, C201, N204, G205, L208, I 209 | |
| Hyaluronidase | -7.3 | G117, R118, R120, A121, Y122, A124, A125, A128, Q152, R155, R204, N205, Q207, L208, L211 | |
| EGCG-Withaferin A (kcal/mol) | |||
| Collagenase | -8.1 | F188, R189, E190, Y221, T222, F223, T270, I271, R272, G273, E274, S299, P303, Q304, E314, F315, A316, D317, N336, S363, E364, E365, N366, T370, Y384, M412, K413, D414, G415 | |
| Elastase | -7.8 | L35, G39, H40, F41, C42, H57, C58, V59, A60, N61, V62, N63, L73, W141, G142, L143, I151, A152, S153, V193, C194, F195, G196, D197, S198, V216, C220, | |
| Hyaluronidase | -7.9 | P1, Q2, L111, W112, G114, R118, R119, R120, A121, Y122, Q123, A124, W186, S188, N205, L208, H209, R246, L249, R253, V258, L259, A260, A291, G292, V293, R346, R347, D348, Y375, W376, G377, W378, T382 | |
| Enzymes (anti-oxidants) | EGCG (kcal/mol) | Binding site residues* | |
| Superoxide dismutase | -7.2 | Q68, P71, R72, A73, K74, Y114, P116, L117, V119, Q123, H124, P125, L145, A146, A147, S148, L149, A150, G151, H153 | |
| Glutathione-s-transferase | -6.1 | D85, K87, E88, A90, L91, M94, Y95, K138, V139, S142, H143, Q145, L148, V149, G150, N151 | |
| Glutathione peroxidase | -6.7 | S77, D78, C79, Q80, L81, T82, D83, R117, E121, F125, N129, Y130 | |
| Catalase | -7.3 | A117, G118, A123, T125, V126, R127, D128, Q168, K169, K177, F200, I205, V247, A250, A251, R252, S254, E461, N462, I463, A464, G465, H466, K468 | |
| Withaferin A (kcal/mol) | |||
| Superoxide dismutase | -6.9 | Q46, V47, Q48, P49, S50, A51, G101, L103, G106, G157, C189, C190, V191, V192, G193 | |
| Glutathione-s-transferase | -6.9 | K4, E29, F30, E31, E32, F34, K196, F197, Q199, P200, G201, S202, P203 | |
| Glutathione peroxidase | -8.3 | D78, C79, Q80, L81, G112, S114, R163, W164, W167, P183, E184 | |
| Catalase | -8 | A117, R127, D128, P129, Q168, K177, W186, L199, F200, V247, E461, N462, G465, H466, K468, Y500 | |
| EGCG-Withaferin A (kcal/mol) | |||
| Superoxide dismutase | -7.7 | R67, Q68, P71, R72, A73, K74, D76, Y114, P116, L117, V119, Q123, H124, P125, W139, R140, Y141, R142, A143, L145, A146, A147, S148, L149, A150, G151, H153, W200, E201, A204, R205 | |
| Glutathione-s-transferase | -9.2 | Y9, R15, R45, Q54, V55, P56, D85, K87, E88, A90, L91, M94, Y95, L108, F111, T112, Q113, E116, K138, V139, S142, H143, Q145, L148, V149, G150, N151, F220, R221, F222 | |
| Glutathione peroxidase | -8.8 | S77, D78, C79, Q80, L81, T82, D83, G112, E113, S114, R117, E121, F125, N129, Y130, R163, W164, W167, E184 | |
| Catalase | -8.3 | E67, R68, E71, R72, A76, K77, G78, R112, E119, S120, G121, S122, D157, P158, I159, D259, N324, Y325, F326, E330, P347, K349, Q352, S426, G427, E428, R430, R431, F432 | |
| Enzymes (mitochondrial | EGCG (kcal/mol) | Binding site residues* | |
| complex) | |||
| NADH dehydrogenase | -6.6 | L75, Q76, T79, D80, R82, T83, W116, P118, P119, L120, I121, L124, Y125, A136, S137 | |
| Succinate dehydrogenase | -7.9 | S51, M208, M211, E212, M213, W214, Q215, F216, H217, P218, R250, Y251, A252, P253, T337, Q338 | |
| Cytochrome c oxidase | -7.9 | Y260, Y261, S335, A336, A337, I394, H395, W396, P398, T404, L405, Q407, V482, L483, M484, V485, E486, W494 | |
| ATP synthase | -5.3 | A23, G24, I25, T27, V28, S31, L32, I33, G35, L46, F47, Y49, A50, I51, L52, G53, L56, S57, M60 | |
| Withaferin A (kcal/mol) | |||
| NADH dehydrogenase | -7 | E131, L134, H135, S137, H138, G139, W142, Y143, T144 | |
| Succinate dehydrogenase | -7.2 | Y45, G50, S51, Y53, D143, K146, W214, Q215, F216, H217, A252, P253, N254, Y330, G334, Q338 | |
| Cytochrome c oxidase | -6.9 | F268, G269, G272, M273, W275, A276, S279, L283, I311, I314, P315, G317, V318, K319, F321, S322 | |
| ATP synthase | -6.3 | G20, S21, A23, G24, I25, T27, V28, S31, L32, G35, Q45, L46, Y49, A50, L52, G53, L56, S57 | |
| EGCG-Withaferin A (kcal/mol) | |||
| NADH dehydrogenase | -9.1 | L75, Q76, T79, D80, R82, T83, L87, N90, L91, E94, G107, E108, S109, V110, F111, R115, W116, V117, P118, P119, L120, I121, G122, E123, L124, Y125, G126, L127, A136, S137 | |
| Succinate dehydrogenase | -9.3 | S51, M208, M211, E212, M213, W214, Q215, F216, H217, P218, D243, R250, Y251, A252, P253, H292, D296, V297, E299, S300, T337, Q338, K351, D352, R468, A472, H473, L474, D476 | |
| Cytochrome c oxidase | -7.5 | L18, L21, F22, W25, F102, W103, P106, L109, L110, L113, Y260, Y261, S335, A336, A337, I394, H395, W396, P398, T404, L405, Q407, V482, L483, M484, V485, E486, W494 | |
| ATP synthase | -6.8 | A23, G24, I25, T27, V28, S31, L32, I33, G35, R38, N39, P40, S41, L42, Q45, L46, F47, Y49, A50, I51, L52, G53, L56, S57, M60 | |
Note: *All residues within 5Å of the ligand have been enlisted. The hydrogen bond forming residues are presented in bold
Table 4: Binding Energies and Interacting Residues Observed Upon Molecular Docking Studies
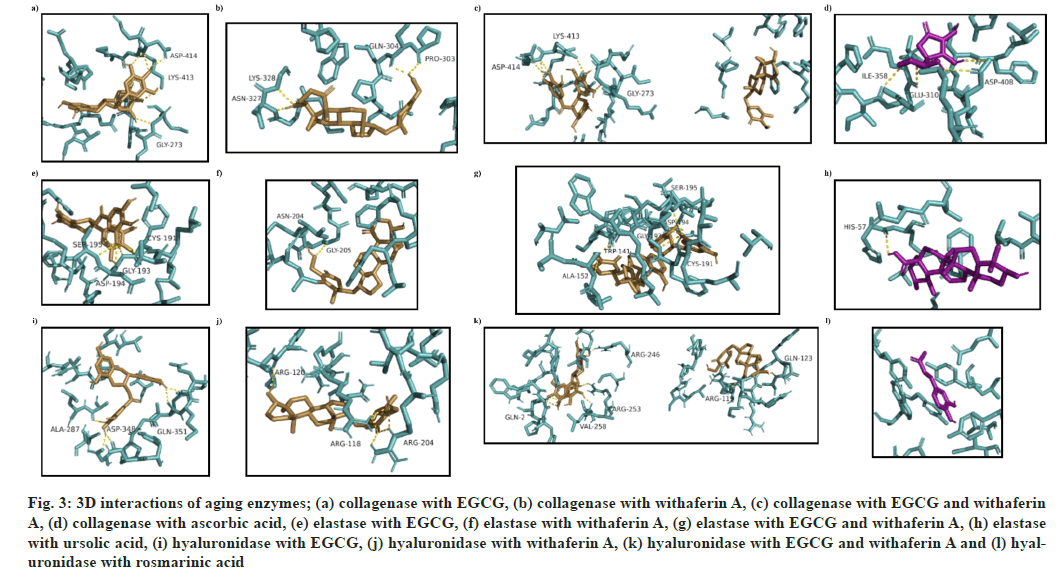
In case of collagenase, (1) was found to form a hydrogen bond (h-bond) with G273, K413 and D414 residues (fig. 3a), while (2) was found to form h-bonds with residues P303, Q304, N327 and K328 (fig. 3b). (3) was found to interact with receptor binding site via h-bonds at G273, K413 and D414 residues (fig. 3c). Ascorbic acid was marked with three strong h-bonds at E310, I358 and D408 residues, as shown in (fig. 3d).
Encouragingly, the docked complex of elastase and (3) showed the highest docking energy (-7.8 kcal/mol) as compared to a complex with (1), (2) or the standard (ursolic acid) (Table 4). Four h-bond interactions were found in EGCG-elastase docked complex i.e. C191, G193, D194 and S195 (fig. 3e), while two were found in withaferin A-elastase complex i.e. N204 and G205 (fig. 3f). The number of residues involved increased in EGCG-withaferin A-elastase complex i.e. W141, A152, C194, G196, D197 and S198 (fig. 3g), while H57 was the only receptor residue involved in the ursolic acid-elastase docking complex (fig. 3h).
The binding energies obtained after docking of hyaluronidase with (1), (2), (3) and rosmarinic acid (standard) were -6.8, -7.3, -7.9 and -5.9 kcal/ mol respectively. Once again, the two ligands proved to be more effective in combination. (1) exhibited binding interactions with A287, D348 and Q351 (fig. 3i), while a docked complex of hyaluronidase and (2) showed binding interactions with arginine residues at locations R118, R120 and R204 (fig. 3j). A complex of (3)-hyaluronidase showed the highest docking energy of -7.9 kcal/mol, with more h-bonds (Q2, R119, Q123, R246, R253 and V258) than other complexes (fig. 3k). Rosmarinic acidhyaluronidase docking complex showed a binding energy of -5.9 kcal/mol, with receptor binding site residues consisting of N37, I73, Y75, V127, D129, E131, Y202, Y247, Y286, D292 and W321 (fig. 3l).
Fig. 3: 3D interactions of aging enzymes; (a) collagenase with EGCG, (b) collagenase with withaferin A, (c) collagenase with EGCG and withaferin A, (d) collagenase with ascorbic acid, (e) elastase with EGCG, (f) elastase with withaferin A, (g) elastase with EGCG and withaferin A, (h) elastase with ursolic acid, (i) hyaluronidase with EGCG, (j) hyaluronidase with withaferin A, (k) hyaluronidase with EGCG and withaferin A and (l) hyaluronidase with rosmarinic acid
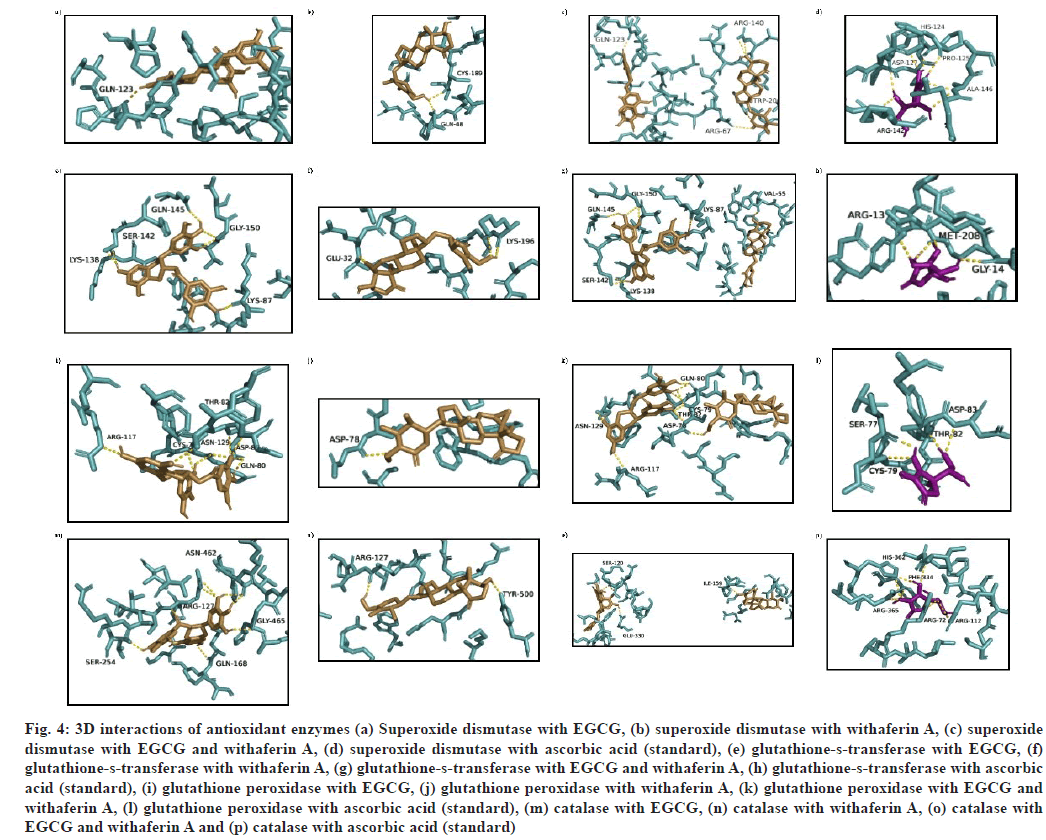
(1) when docked with superoxide dismutase (SOD) (PDB ID: 2JLP) showed that three residue stretches, viz. 68-74 (Q68, P71, R72, A73, K74), 114-125 (Y114, P116, L117, V119, Q123, H124, P125) and 145-153 (L145, A146, A147, S148, L149, A150, G151 and H153) were involved in receptor binding site (fig. 4a). The binding energy shown was -7.2 kcal/mol. Docking of (2) with SOD showed h-bonds at Q48 and C189 (fig. 4b), with binding energy -6.9 kcal/mol. When SOD was docked with multiple ligands i.e. (3), an increase in binding energy was found (-7.7 kcal/ mol) with R67, Q123, R140 and W200 as h-bond interacting residues (fig. 4c). Binding energy with ascorbic acid (reference molecule) was found to be -4.5 kcal/mol, making the binding energy order for SOD as (3)>(1)>(2)>ascorbic acid (fig. 4d).
Interaction of (1) with glutathione-s-transferase (PDB ID: 2VCT) showed binding energy as -6.1 kcal/mol, with K87, K138, S142, Q145 and G150 residues involved in h-bond formation, H143 causing a basic environment while E88 causing an acidic environment (fig. 4e). When (2)-glutathione-s-transferase docking complex was analyzed, two h-bond forming residues (E32 and K196) were found, while the acidic environment was provided by E29, E31, E32 residues, and the basic environment was provided by K4, K196 and G201 residues (fig. 4f). During MLSD analysis with glutathione-s-transferase, interactions were found at V55, K87, K138, S142, Q145 and G150 residues, with a binding energy of -9.2 kcal/mol (fig. 4g). On the other hand, glutathione-s-transferase interaction with ascorbic acid generates -5.5 kcal/mol binding energy with R13, G14 and M208 as interacting residues (fig. 4h). Therefore, glutathiones- transferase activity has been found to be enhanced when interacting with multiple ligands i.e. EGCG and withaferin A in combination, because binding energy was significantly high as compare to single ligand docking and reference molecule.
Fig. 4: 3D interactions of antioxidant enzymes (a) Superoxide dismutase with EGCG, (b) superoxide dismutase with withaferin A, (c) superoxide dismutase with EGCG and withaferin A, (d) superoxide dismutase with ascorbic acid (standard), (e) glutathione-s-transferase with EGCG, (f) glutathione-s-transferase with withaferin A, (g) glutathione-s-transferase with EGCG and withaferin A, (h) glutathione-s-transferase with ascorbic acid (standard), (i) glutathione peroxidase with EGCG, (j) glutathione peroxidase with withaferin A, (k) glutathione peroxidase with EGCG and withaferin A, (l) glutathione peroxidase with ascorbic acid (standard), (m) catalase with EGCG, (n) catalase with withaferin A, (o) catalase with EGCG and withaferin A and (p) catalase with ascorbic acid (standard)
Interaction of (1) with glutathione peroxidase (PDB ID: 3KIJ) was found to show binding energy of -6.7 kcal/mol with binding site residues as S77, D78, C79, Q80, L81, T82, D83, R117, E121, F125, N129 and Y130 (fig. 4i). (2)-glutathione peroxidase docking complex showed h-bond at a lone residue, D78, with binding energy as -8.3 kcal/mol (fig. 4j). During docking with (3), a slight increase in binding energy (-8.8 kcal/mol) was found, with a high number of interacting residues at S77, D78, C79, Q80, L81, T82, D83, S114, R117, E121, F125, N129, Y130, R163, W164 and W167 (fig. 4k). On the other hand, ascorbic acid was found to show interactions at C79, T82 and D83 residues with -5.4 kcal/mol binding energy (fig. 4l).
While interacting with catalase (CAT) (PDBID: 1DGB), (1) presented R127, Q168, S254, N462 and G465 as h-bond residues. H466 provided with a basic environment, while A117, A123, A250, A251 and A464 residues (fig. 4m) contributed to acidic environment. The binding energy of interaction was -7.3 kcal/mol (Table 4). CAT interactions with (2) displayed two h-bonds (R127, Y500 residues) amongst 16 binding site residues. The basic environment was provided by Q168, H466, and N462 residues, while an acidic environment was provided by D128 (fig. 4n). The binding energy of interaction was -8.0 kcal/mol. On the other hand, the interaction of (3) with CAT showed the interaction at residues S120, I159 and E330 (fig. 4o). The energy of reference molecule ascorbic acid with catalase was -6.4 kcal/mol with h-bond interactions at R72, R112, F334, H362 and R365 residues (fig. 4p).
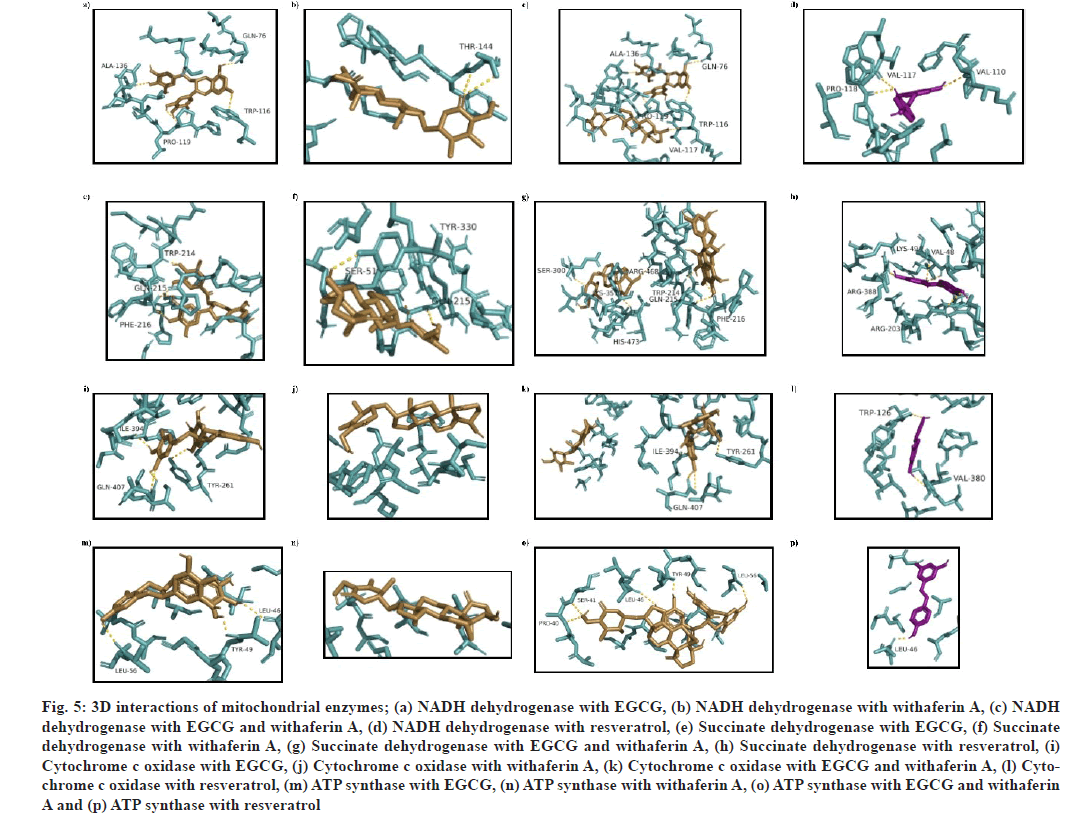
Mitochondrial complexes were docked to analyze the interaction of EGCG and withaferin A with them, and perform MLSD with these two ligands. The docked complex of NADH dehydrogenase (PDB ID: 5XTC) with (1) was found to exhibit affinity at binding site consisting of L75, Q76, T79, D80, R82, T83, W116, P118, P119, L120, I121, L124, Y125, A136 and S137 residues. The binding energy was found to be -6.6 kcal/mol (fig. 5a). (2)-NADH dehydrogenase docking complex showed a binding energy of -7 kcal/mol, with a single h-bond at T144 (fig. 5b). During MLSD analysis, binding energy was found to be -9.1 kcal/mol, with interacting residues at Q76, W116, V117, P119, and L136 (fig. 5c).
Succinate dehydrogenase docking with (1) was found to show h-bonds at W214, Q215 and F216, with binding energy as -7.9 kcal/mol (fig. 5e). (2) succinate dehydrogenase docking complex was shown to form h-bonds at S51, Q215 and Y330, with binding energy of -7.2 kcal/mol (fig. 5f). While (3) succinate dehydrogenase complex was found to show interact at a binding site consisting of S51, M208, M211, E212, M213, W214, Q215, F216, H217, P218, D243, R250, Y251, A252, P253, H292, D296, V297, E299, S300, T337, Q338, K351, D352, R468, A472, H473, L474 and D476 residues, with highest binding energy at -9.3 kcal/mol (fig. 5g)..
Fig. 5: 3D interactions of mitochondrial enzymes; (a) NADH dehydrogenase with EGCG, (b) NADH dehydrogenase with withaferin A, (c) NADH dehydrogenase with EGCG and withaferin A, (d) NADH dehydrogenase with resveratrol, (e) Succinate dehydrogenase with EGCG, (f) Succinate dehydrogenase with withaferin A, (g) Succinate dehydrogenase with EGCG and withaferin A, (h) Succinate dehydrogenase with resveratrol, (i) Cytochrome c oxidase with EGCG, (j) Cytochrome c oxidase with withaferin A, (k) Cytochrome c oxidase with EGCG and withaferin A, (l) Cytochrome c oxidase with resveratrol, (m) ATP synthase with EGCG, (n) ATP synthase with withaferin A, (o) ATP synthase with EGCG and withaferin A and (p) ATP synthase with resveratrol
Cytochrome c oxidase docking with (1) was found to show interact with Y261, I394 and Q407 residues. The binding energy was found as -7.9 kcal/mol (fig. 5i). (2) was found to show a binding energy of -6.9 kcal/mol (fig. 5j). Cytochrome c oxidase interaction with (3) was found to show interaction at Y261, I394 and Q407 residues, with binding energy of -7.5 kcal/ mol, which is less than single ligand interaction with (1), therefore, no activity enhancement is seen in case of cytochrome c oxidase (fig. 5k).
ATP synthase docking with (1) was found to show h-bond interactions at L46, Y49 and L56 residues, with a binding energy of -5.3 kcal/mol (fig. 5m). On the other side, (2)-ATP synthase docked complex was found to interact at binding site consisting of G20, S21, A23, G24, I25, T27, V28, S31, L32, G35, Q45, L46, Y49, A50, L52, G53, L56, S57 residues, with a binding energy of -6.3 kcal/mol (fig. 5n). While in multiple ligand simultaneous docking, (3) showed interactions at P40, S41, L46, Y49 and L56, with a binding energy of -6.8 kcal/mol (fig. 5o), which is slightly high as compared to single ligand docking.
Overall, the binding energy for all mitochondrial enzymes was found to be better for the (1), (2) and (3) as compared to resveratrol (standard). Interestingly, the binding energy for (3) was better than all cases for all mitochondrial enzymes (except cytochrome c oxidase).
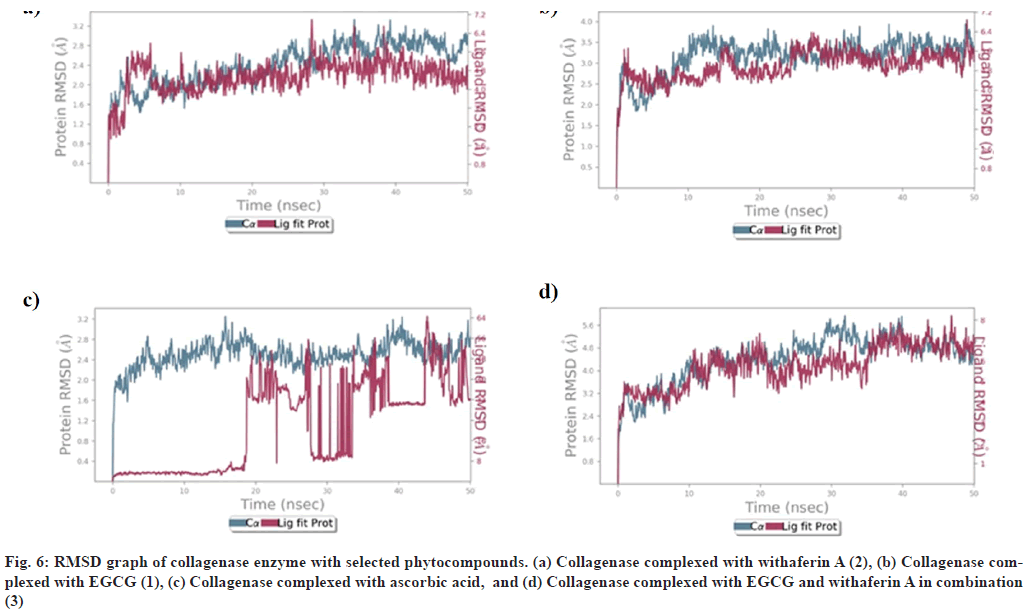
On the basis of molecular docking results, collagenase and glutathione-s-transferase complexes, showing high binding energy, were selected for MD simulation studies for a time period of 50 ns.
RMSD of collagenase, in complex with (1), (2), (3), and ascorbic acid, is depicted in (fig. 6). It was observed that collagenase complex with (1), (2), and (3) was found to be stabilized at 10 ns, between 1.8-2.8 Å, 2.5-3.5 Å and 3.2- 4.8 Å respectively, with little fluctuations in acceptable range. (fig. 6). However, RMSD of ascorbic acid-collagenase complex was found to be stabilized at 20-22 ns, between 1.6-2.4 Å, at 44-46 ns between 2.0-2.4 Å, which may be due to the instability of complex (fig. 6).
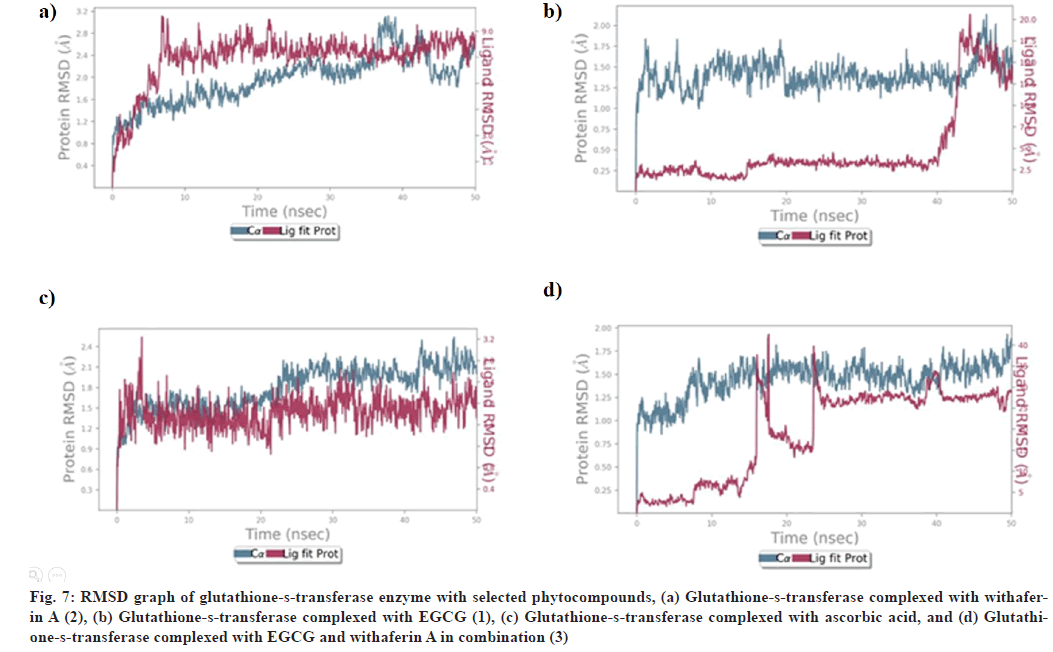
RMSD of glutathione-s-transferase complex with (1) showed stabilization after 40 ns, with slight fluctuation within 1.6-2.8 Å and remains stable upto 50 ns (fig. 7). Glutathione-s-transferase-(2) also revealed the stability of protein-ligand complex after 45 ns between 1.0-1.75 Å and remains stable upto 50 ns (fig. 7). RMSD analysis of glutathione-s-transferase complexed with (3) showed stability after 25 ns within acceptable range of 1.0-1.75 Å and remains stable upto 50 ns (fig. 7). However, RMSD of glutathione-stransferase- ascorbic acid complex was found to be stabilized from 0-50 ns within acceptable range of 1.2-2.1 Å (fig. 7).
Fig. 7: RMSD graph of glutathione-s-transferase enzyme with selected phytocompounds, (a) Glutathione-s-transferase complexed with withaferin A (2), (b) Glutathione-s-transferase complexed with EGCG (1), (c) Glutathione-s-transferase complexed with ascorbic acid, and (d) Glutathione- s-transferase complexed with EGCG and withaferin A in combination (3)
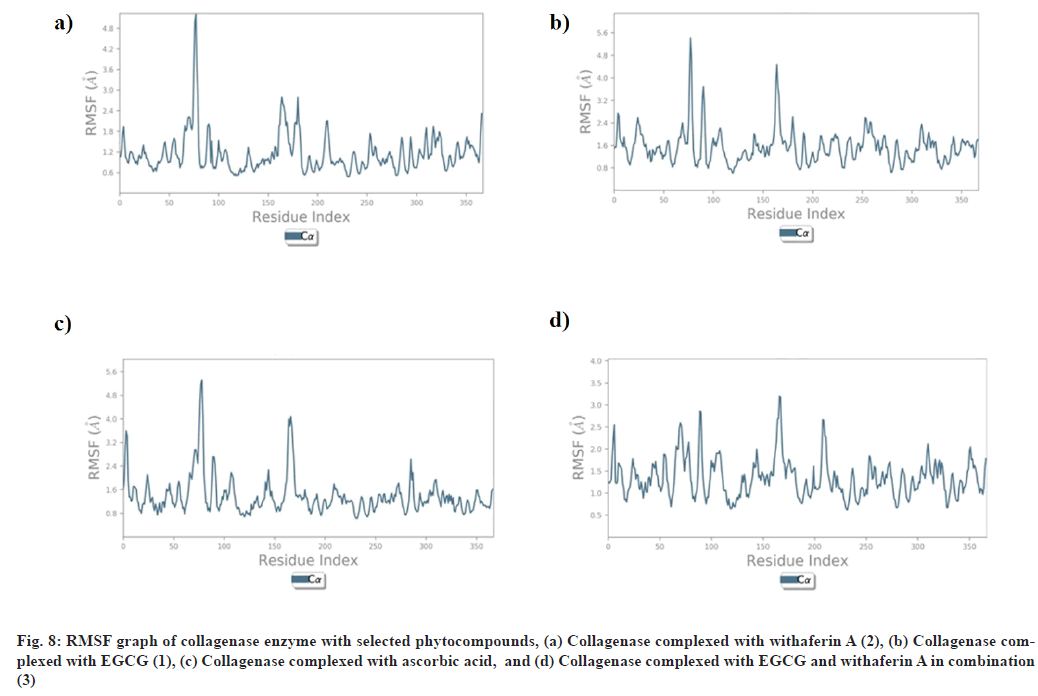
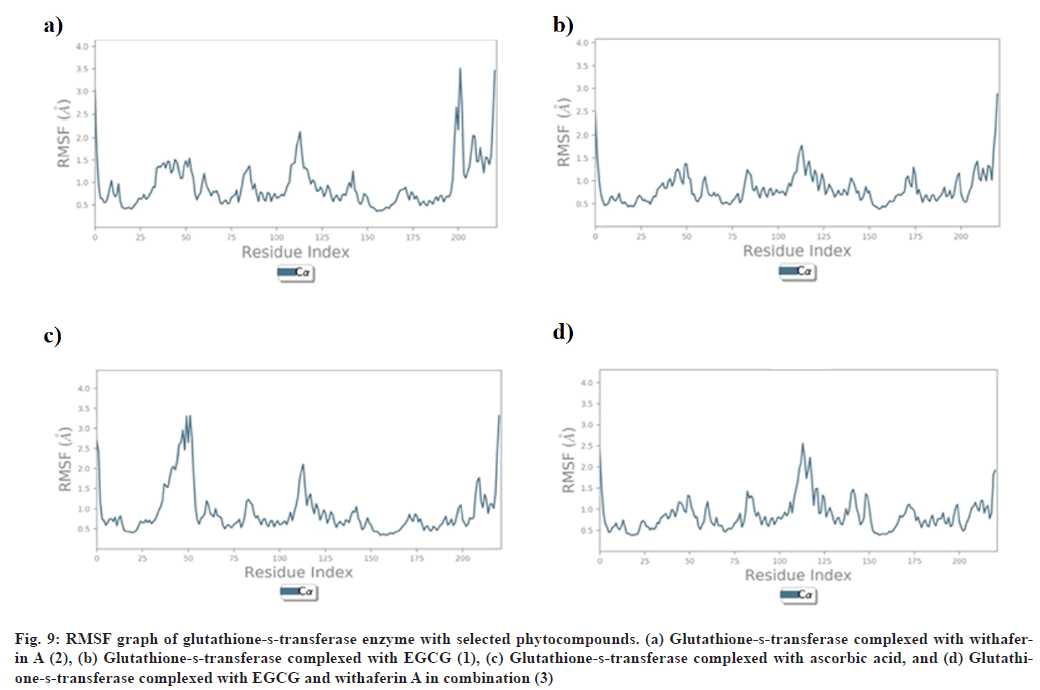
RMSF explains the flexibility of protein structures and fluctuations of interactive residues in secondary structure elements of target proteins. RMSF plot of collagenase with (1), (2), (3), and ascorbic acid was found to show less fluctuation during stimulation in α-helical and β-strands within acceptable range of 0.6-2.4 Å, 0.8-4.8 Å, 0.8-3.0 Å, and 0.8-4.8 Å respectively (fig. 8). RMSF plot of glutathione-s-transferase with (1), (2), (3), and ascorbic acid was found to show less fluctuation during stimulation in α-helical and β-strands within acceptable range of 0.5-2.0 Å (fig. 9).
Fig. 9: RMSF graph of glutathione-s-transferase enzyme with selected phytocompounds. (a) Glutathione-s-transferase complexed with withaferin A (2), (b) Glutathione-s-transferase complexed with EGCG (1), (c) Glutathione-s-transferase complexed with ascorbic acid, and (d) Glutathione- s-transferase complexed with EGCG and withaferin A in combination (3)
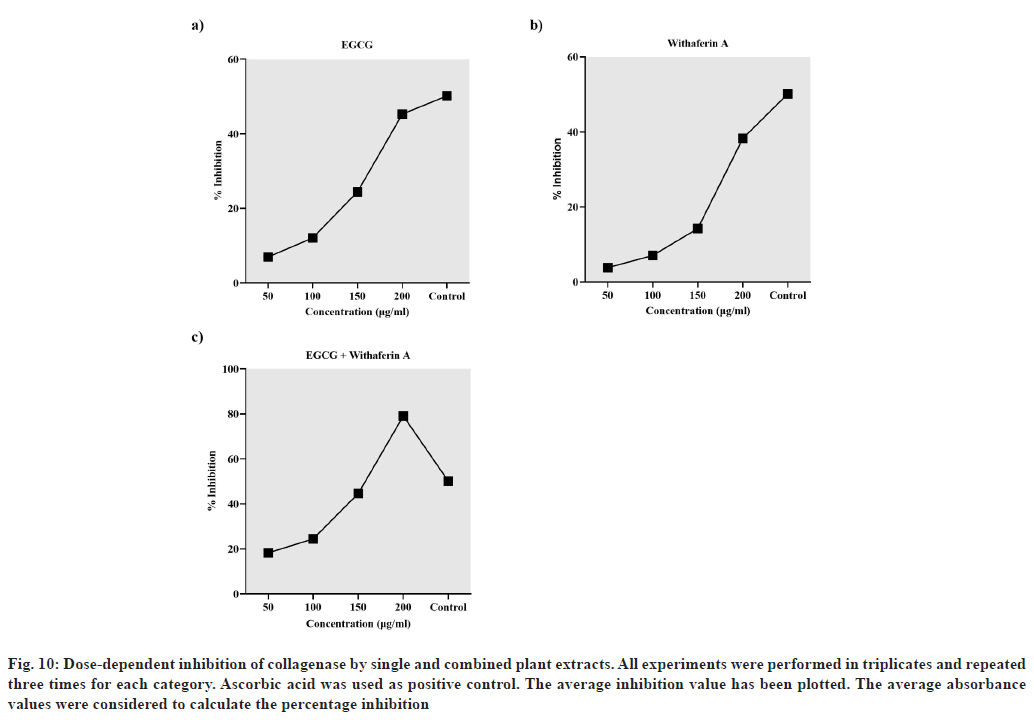
The dose-dependent inhibition of collagenase was estimated via collagenase activity colorimetric assay. The results are presented in terms of % inhibition of collagenase activity by Camelia sinensis extract (containing EGCG), Withania somnifera extract (containing withaferin A), combined plant extract (containing EGCG and withaferin A) and ascorbic acid (control) (fig. 10). The results indicate that a combination of EGCG and withaferin A inhibited collagenase 1.5 times more (79.08 % inhibition) than the positive control (50.15 % inhibition). Also, the inhibition by the combination was greater than that displayed by EGCG (45.27 %) and withaferin A (38.31 %) alone, even at the highest concentration (200 μg/ml) (fig. 10). Overall, the in vitro results support the molecular docking analysis.
Fig. 10: Dose-dependent inhibition of collagenase by single and combined plant extracts. All experiments were performed in triplicates and repeated three times for each category. Ascorbic acid was used as positive control. The average inhibition value has been plotted. The average absorbance values were considered to calculate the percentage inhibition
Proteome and its analysis is a hotspot of research for therapeutic and drug development studies. Drug discovery is very expensive and time-consuming, thus, computational support is required to efficiently, hasten the process. Molecular docking studies enable an acceptable prediction of binding sites[48]. MLSD technique is employed to compare the interaction of single and multiple ligands with a particular receptor molecule[49]. MLSD predicts the synergistic effect of two or more compounds simultaneously docked onto a receptor molecule[50].
Molecular docking has been reported as an effective method to predict the regulatory impact of biomolecules on skin photo-aging[51,52]. Also, an additive effect of phytochemicals has been observed in previous in vitro studies[53-55] . The present study compared the binding energy of EGCG alone (1), withaferin A alone (2) and multiple ligand simultaneous docking of EGCG+withaferin A (3) with aging-related metalloproteinase (collagenase, elastase and hyaluronidase), antioxidant enzymes (superoxide dismutase, glutathione-s-transferase, glutathione peroxidase and catalase) and mitochondrial enzymes (NADH dehydrogenase, succinate dehydrogenase, cytochrome c oxidase and ATP synthase). The purpose of the study was to estimate the combined effect of both, EGCG and withaferin A, against various enzymes involved in the aging process.
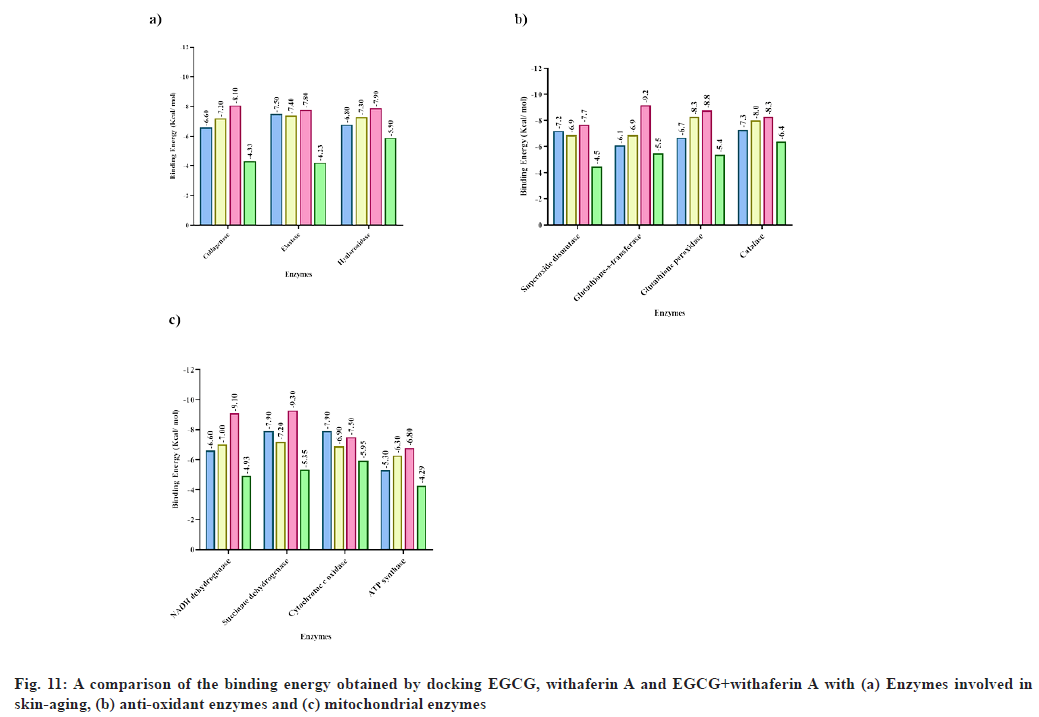
Docking of collagenase, an enzyme responsible for aging, with (3) was found to show binding energy -8.1 kcal/mol, which is higher compared to the binding energy exhibited by caffeine (-6.16 kcal/mol)[51], kaempferol-3-O-glucoside (-7.97 kcal/mol), gallic acid methyl ester (-7.15 kcal/mol), quercetin-3-O-glucoside (-7.37 kcal/ mol) and quercetin-3-O-galactoside (-7.84 kcal/ mol)[56] . Also, this result was better than the binding energy obtained upon docking (1) (-6.6 kcal/mol) and (2) (-7.2 kcal/mol) individually with collagenase (fig. 11). Moreover, the phytoconstitutents displayed a higher binding energy than the reference molecule (ascorbic acid; -4.33 kcal/mol) in both, individual and multiple ligand docking (fig. 11). Based on the molecular docking results, we expected a higher experimental inhibition of collagenase by (1), (2) and (3) versus the standard. The in vitro analysis suggested that though, the maximum collagenase inhibition for (1) (45.27 %) and (2) (38.31 %) was less than ascorbic acid (50.15 %), a greater inhibition of collagenase activity was achieved upon addition of (3) (79.08 %) at a concentration of 200 μg/ml. Also, molecular dynamic simulations further affirmed the stability and flexibility of collagenase-(3) complex.
Docking of elastase with (3) also exhibited a slightly higher binding energy (-7.8 kcal/mol) than (1) (-7.5 kcal/mol) and (2) alone (-7.4 kcal/mol). Moreover, the significance of these phytoconstituents is highlighted by the fact that caffeine has been reported to show only -3.36 kcal/mol binding energy with elastase[51] , while the reference molecule used in this study, ursolic acid, also exhibited a low binding energy (-4.23 kcal/mol). Similarly, interaction of (1), (2) and (3) with hyaluronidase was found to show -6.8, -7.3, -7.9 kcal/mol binding energy respectively, while reference molecule (rosmarinic acid) exhibited a binding energy of -5.9 kcal/mol (fig. 10). Also, hyaluronidase interaction with apigenin has been reported to show a binding energy of -7.2 kcal/mol[57]. Therefore, once again, a combination of the two ligands may be thought to bind with more affinity to the enzyme.
In case of the antioxidant enzymes, ascorbic acid was used a reference for all cases[33]. The binding energy of superoxide dismutase with (1), (2) and (3) was -7.2, -6.9 and -7.7 kcal/mol respectively. The reference exhibited a binding energy of -4.5 kcal/mol (fig. 10). Also, piperine interaction with superoxide dismutase has been reported with -4.2 kcal/mol binding energy[58]. The binding energy of glutathione-s-transferase with EGCG, withaferin A, EGCG+withaferin A and reference was -6.1, -6.9, -9.2 and -5.5 kcal/mol respectively (fig. 10). Capsicin interaction with glutathione-s-transferase has been previously reported to exhibit a -7.9 kcal/mol binding energy[58], which is greater than the energy obtained for individual ligands, but less than the energy obtained upon MLSD analysis in the current study. The binding energy of glutathione peroxidase with (1), (2), (3) and reference was -6.7, -8.3, -8.8 and -5.4 kcal/mol respectively. The binding energy of glutathione peroxidase with piperine has been reported as -6.2 kcal/mol[58]. Further, the binding energy of catalase with (1), (2), (3) and reference was -7.3, -8.0, -8.3 and -6.4 kcal/mol respectively. In a recent research, binding energy of catalase with capsaicin was found to be -7.3 kcal/mol[58] . Overall, it may be concluded that for all antioxidant enzymes, docking EGCG+withaferin A simultaneously resulted in a better docking complex, with higher binding affinity, as compared to previous reports and standard molecules.
In case of mitochondria, complex I (CI) plays a crucial role in maintaining mitochondrial homeostasis, not only through its role in energy metabolism and the Reactive Oxygen Species (ROS) production, but also by regulating the NAD+/NADH ratio[59]. The simultaneous effects of phytochemicals on NADH ubiquinone oxidoreductase and F0F1-ATPase/ATP synthase could significantly affect mitochondrial function and alter ATP levels, mitochondrial transmembrane potential and generation of reactive oxygen species, which are implicated in many cellular processes such as cellular protection, apoptosis, O2 sensing and aging[60,61].
Therefore, (1), (2) and (3) were analyzed for binding affinity towards various mitochondrial enzymes (NADH dehydrogenase, succinate dehydrogenase, cytochrome c oxidase and ATP synthase). Resveratrol has been studied in vitro with complex I and reportedly increases its activity, leading to an increase in NAD+ levels. Ultimately, SIRT1 activity is increased, which further delays the onset of aging[34]. Therefore, resveratrol was used as a standard for all mitochondrial enzymes.
Deficiency of NADH, ubiquinone oxidoreductase or NADH dehydrogenase has been associated with mitochondrial diseases in populations belonging to different age groups[62]. It has been reported that upregulation of NADH dehydrogenase increases longevity in round worms[63]. The binding energy of NADH dehydrogenase with (1), (2) and (3) was -6.6, -7.0, -9.1 kcal/mol respectively. A significant increase in binding energy confirms the additive effect of these phytochemicals on the activity of NADH dehydrogenase.
Succinate dehydrogenase is situated in the Inner Mitochondrial Membrane (IMM) and is the only enzyme sharing in both the tricarboxylic acid cycle and the Electron Transport Chain (ETC)[64]. Succinate dehydrogenase facilitates oxidation of succinate in the Tricarboxylic Acid (TCA) cycle, which is linked with ubiquinone reduction to ubiquinol in the ETC[64]. It is reported that with aging, the level of succinate decreases, which can be restored by phytochemical supplementation[65,66]. In this study, the binding energy of succinate dehydrogenase with (1), (2) and (3) was found to be -7.9, -7.2, -9.3 kcal/ mol respectively, which shows the energy of combination is quite high as compared to a single ligand.
Cytochrome C Oxidase (COX) is the highly regulated enzyme complex IV, and a key player in aerobic mitochondrial energy metabolism[67]. It helps in controlling age-related neurodegeneration and aids in neuroprotection[68]. COX plays a central role in energy metabolism and is reported to protect the aging nerve cells from damage, but with aging, its low expression becomes the reason for neurodegeneration[68]. Age-related deterioration in the COX function was linked with corresponding variations in mitochondrial membrane potential, respiration and an upsurge in hydrogen peroxide production[69]. The agerelated deterioration of COX function could be re-established, in vivo, by exogenously added cardiolipin. Therefore, COX may be considered a potential target in drug discovery to delay aging[70]. Phytochemicals including curcumin and EGCG has been reported to modulate mitochondrial complexes[71]. In our study, the binding energy of COX with (1), (2) and (3) was -7.9, -6.9 and -7.5 kcal/mol respectively, which shows that combined treatment of phytochemicals may be efficient, though further validation is required in this case.
ATPase/ATP synthase (F-type ATPase, complex V) is present in the inner membrane of eukaryotic mitochondria and acts as the powerhouse of the cell by synthesizing ATP[72]. Mitochondrial proton ATPase/ATP synthase synthesizes ATP during oxidative phosphorylation. Several groups of polyphenolic phytochemicals, including stilbenes, isoflavones, flavones, catechins and chalcones, have been reported to be important in human diets, for their effect on the activity of mitochondrial ATPase/ATP synthase[73]. A study demonstrated that ATPase/ATP synthase is a common target site for resveratrol from red wine, aglycone isoflavones (genistein, biochanin A and daidzein) from soybean, gallate esters of catechins from many sources, and several other polyphenolic compounds[60]. Therefore, ATP synthase can be a potential target in age-related pathology. In this study, we have reported the binding energy of ATP synthase with (1), (2) and (3) as -5.3, -6.3, -6.8 kcal/mol respectively.
Overall, (1), (2) and (3) exhibited a better binding affinity towards mitochondrial enzymes as compared to standard (resveratrol). Also, a significant increase in binding energy has been observed in combination docking of EGCG with withaferin A with mitochondrial enzymes, which shows a significant additive effect of EGCG and withaferin A.
Additionally, collagenase activity colorimetric assay was performed to estimate the collagenase inhibitory potential of crude plant extracts. Collagenase is a Matrix MetalloProteinase-1 (MMP-1) which is responsible for cleaving amino acid linkage present in collagen, ultimately leading to wrinkle formation and skin damage. Therefore, collagenase can be targeted to delay the onset of aging skin. Previously, multiple studies have reported an inhibitory effect of plant phenolics on collagenase. In 2011, Ganesan et al. contemplated a role of Withania somnifera in rheumatoid arthritis and osteoarthritis after observing a dose-dependent inhibition of Clostridium histolyticum collagenase by the plant extract[74]. Another study, based on mouse model, reported the potential of withaferin A in treatment of cartilage degenerative diseases[75]. Recently, Won et al. observed inhibitory effect of EGCG on MMP-1 gene expression and secretion in Hs68 cells (human skin fibroblasts) and contemplated it as a solution to skinaging symptoms[76]. Moreover, a recent review brilliantly summarized the role of natural compounds, including polyphenols from green tea, in inhibition of skin aging[77]. In the current study, the results indicate a greater inhibition of collagenase activity upon interaction with (3), as compared to (1), (2) and standard (ascorbic acid).
Many previous works support the findings of our study. CD (Circular dichroism) spectral studies showed significant changes in the secondary structure of collagenase on treatment with higher concentration of catechin and EGCG. Higher inhibition of EGCG compared to catechin has been attributed to the ability of EGCG to exhibit better hydrogen bonding and hydrophobic interaction with collagenase[78]. EGCG has been found to inhibit elastase and hyaluronidase via non-covalent binding[51]. EGCG has been reported to enhance the activity of antioxidant enzymes by stimulating ( protein kinase B ) PKB or Akt pathway, blocking Nf-κB (Nuclear Factor Kappa B ) and by inducing (nuclear factor erythroid 2–related factor 2 ) Nrf-2-mediated gene expression of SOD, CAT, glutathione peroxidase, glutathione-S-transferase[79]. Phytochemicals like EGCG, withaferin A, resveratrol, curcumin etc. has been found to enhance antioxidant enzymes through the Keap1–Nrf2 pathway[80,81]. Mitochondria targeted phytochemicals like EGCG, oleuropein, curcumin, withaferin A have been found to increase mtDNA expression, PGC- 1α (peroxisome proliferation-activated receptor γ coactivator 1α), complex I, II, III and IV, regulated mitochondrial function, mitogenesis, and dynamics through mitofusion 1 (Mfn1) and dynamin related protein 1 (Drp1) and attenuated oxidative stress in the hypothalamic paraventricular nucleus of spontaneously hypertension rats[82,83].
Overall, it can be said that enzymes which are upregulated during aging are responsible for the degradation of the dermal matrix, which makes skin sensitive to free radicals. Therefore, inhibition of metalloproteinases can be useful to delay the effects of aging. Antioxidant enzymes help to scavenge free radicals, therefore, enhancing the function of antioxidant enzymes (superoxide dismutase, glutathione- S-transferase, glutathione peroxidase and catalase) can delay aging by neutralizing the effect of toxic free radicals. Mitochondrial complexes are involved in energy generation in oxidative phosphorylation and their modulation with phytochemicals can be a potential way in fighting aging problems. Therefore, in this study, we analyzed the binding affinity of EGCG and withaferin A towards various aging related enzymes. Results showed that our combinations (EGCG+withaferin A) were more potent when they were docked as multiple ligands since they had the highest binding affinity value as compared to binding with a single ligand (EGCG or withaferin A). Moreover, ADMET data also supports the potent application of withaferin A and EGCG as antiaging agent as they follow Lipinski rule of 5, and are non-mutagenic, non-irritant, and non- tumorgenic in nature. They also showed high bioavailability and did not show any effect on reproductive system. In other words, natural phenolics, when used in combination, were found to significantly inhibit aging enzymes, enhance the function of antioxidant enzymes and modulate the function of mitochondrial complexes as compared to their individual interaction with different enzymes such as collagenase, hyaluronidase, elastase etc. Furthermore, a combination of plant extracts resulted in a higher inhibition of collagenase activity under in vitro conditions. On the other hand, our study is limited in terms of computational and in vitro validation of other enzymes such as elastase and hyaluronidase. Further studies and clinical trials of combination of these compounds will provide clues about their possible pharmaceutical exploration in the field of medicine.
Acknowledgements:
The authors thank Chandigarh University for providing the infrastructure to carry out the research work. We also thank ADI BIOSOLUTIONS, who assisted us is performing molecular docking analysis.
Conflict of interest:
The authors have no competing interests to declare.
References
- Pientaweeratch S, Panapisal V, Tansirikongkol A. Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and silymarin: An in vitro comparative study for anti-aging applications. Pharm Biol 2016;54(9):1865-72.
- Jiratchayamaethasakul C, Ding Y, Hwang O, Im ST, Jang Y, Myung SW,et al. In vitro screening of elastase, collagenase, hyaluronidase, and tyrosinase inhibitory and antioxidant activities of 22 halophyte plant extracts for novel cosmeceuticals. Fish Aquat Sci 2020;23:1-9.
- D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV radiation and the skin. Int J Mol Sci 2013;14(6):12222-48.
[Crossref] [Google Scholar] [PubMed]
- Gonzaga ER. Role of UV light in photodamage, skin aging, and skin cancer: Importance of photoprotection. Am J Clin Dermatol 2009;10:19-24.
[Crossref] [Google Scholar] [PubMed]
- Kandaswami C, Middleton Jr E. Free radical scavenging and antioxidant activity of plant flavonoids. Free radicals in diagnostic medicine: A systems approach to laboratory technology. Clin Correlat Antioxidant Ther 1994:351-76.
[Crossref] [Google Scholar] [PubMed]
- Santos-Sánchez NF, Salas-Coronado R, Villanueva-Cañongo C, Hernández-Carlos B. Antioxidant compounds and their antioxidant mechanism. Antioxidants 2019;10:1-29.
- Sourabh A, Kanwar SS, Sud RG, Ghabru A, Sharma OP. Influence of phenolic compounds of Kangra tea [Camellia sinensis (L) O Kuntze] on bacterial pathogens and indigenous bacterial probiotics of Western Himalayas. Braz J Microbiol 2013;44:709-15. .
[Crossref] [Google Scholar] [PubMed]
- Singh BN, Shankar S, Srivastava RK. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem Pharmacol 2011;82(12):1807-21.
[Crossref] [Google Scholar] [PubMed]
- Cooper R, Morré DJ, Morré DM. Medicinal benefits of green tea: Part II review of anticancer properties. J Altern Complement Med 2005;11(4):639-52. .
[Crossref] [Google Scholar] [PubMed]
- Sabu MC, Smitha K, Kuttan R. Anti-diabetic activity of green tea polyphenols and their role in reducing oxidative stress in experimental diabetes. J Ethnopharmacol 2002;83(1-2):109-16.
[Crossref] [Google Scholar] [PubMed]
- Wang P, Heber D, Henning SM. Quercetin increased the antiproliferative activity of green tea polyphenol (-)-epigallocatechin gallate in prostate cancer cells. Br J Nutr 2012;64(4):580-7.
[Crossref] [Google Scholar] [PubMed]
- Ohishi T, Goto S, Monira P, Isemura M, Nakamura Y. Anti-inflammatory action of green tea. Antiinflamm Antiallergy Agents Med Chem 2016;15(2):74-90.
[Crossref] [Google Scholar] [PubMed]
- Lee W, Min WK, Chun S, Lee YW, Park H, Lee DH, et al. Long-term effects of green tea ingestion on atherosclerotic biological markers in smokers. Ann Clin Biochem 2005;38(1):84-7.
[Crossref] [Google Scholar] [PubMed]
- Mandel SA, Avramovich-Tirosh Y, Reznichenko L, Zheng H, Weinreb O, et al,. Multifunctional activities of green tea catechins in neuroprotection. Neurosignals 2005;14(1-2):46-60.
[Crossref] [Google Scholar] [PubMed]
- Lunder TL. Catechins of green tea: Antioxidant activity. Am Chem Soc 1992;9:114-120.
[Crossref] [Google Scholar] [PubMed]
- Fuhrman BJ, Pfeiffer RM, Wu AH, Xu X, Keefer LK, Veenstra TD, et al. Green tea intake is associated with urinary estrogen profiles in Japanese-American women. Nutr J 2013;12(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Hsu S. Green tea and the skin. J Am Acad Dermatol 2005;52(6):1049-59.
[Crossref] [Google Scholar] [PubMed]
- Singh N, Bhalla M, de Jager P, Gilca M. An overview on ashwagandha: A Rasayana (rejuvenator) of Ayurveda. Afr J Tradit Complement Altern Med 2011;8(5S):208-13.
[Crossref] [Google Scholar] [PubMed]
- Shree P, Mishra P, Selvaraj C, Singh SK, Chaube R, Garg N, et al.Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants–Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi): A molecular docking study. J Biomol Struct Dyn 2022;40(1):190-203.
[Crossref] [Google Scholar] [PubMed]
- Dhawan M, Parmar M, Sharun K, Tiwari R, Bilal M, Dhama K. Medicinal and therapeutic potential of withanolides from Withania somnifera against COVID-19. J Appl Pharm Sci 2021;11(4):6-13.
- Sudeep HV, Gouthamchandra K, Shyamprasad K. Molecular docking analysis of Withaferin A from Withania somnifera with the Glucose regulated protein 78 (GRP78) receptor and the SARS-CoV-2 main protease. Bioinformation 2020;16(5):411.
[Crossref] [Google Scholar] [PubMed]
- Sorrenti V, Davinelli S, Scapagnini G, Willcox BJ, Allsopp RC, Willcox DC. Astaxanthin as a putative geroprotector: Molecular basis and focus on brain aging. Mar Drugs 2020;18(7):351.
[Crossref] [Google Scholar] [PubMed]
- Kumar CS, Thangam R, Mary SA, Kannan PR, Arun G, Madhan B. Targeted delivery and apoptosis induction of trans-resveratrol-ferulic acid loaded chitosan coated folic acid conjugate solid lipid nanoparticles in colon cancer cells. Chem Rev 2020 Mar 1;231:115682.
[Crossref] [Google Scholar] [PubMed]
- Astuti IY, Yupitawati A, Nurulita NA. Anti-aging activity of tetrahydrocurcumin, Centella asiatica extract, and its mixture. Adv Tradit Med 2021;21:57-63.
- Kaushal N, Jain S, Baranwal M. Computational design of immunogenic peptide constructs comprising multiple HLA restricted Dengue virus envelope epitopes. J Mol Recognit 2022:e2961.
[Crossref] [Google Scholar] [PubMed]
- DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40(1):82-92.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: An open chemical toolbox. J Cheminform 2011 ;3(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 2015;55(2):460-73.
[Crossref] [Google Scholar] [PubMed]
- Singhal D, Saxena S. Screening and toxicity analysis of catechin isomers against femA protein. Indian J Pharm Sci 2015;77(6):758.
[Crossref] [Google Scholar] [PubMed]
- Lipinski C. A. Lead-and drug-like compounds: The rule-of-five revolution. Curr Top Med Chem 2004;1(4):337-41.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Ulster Med J 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res 2000;28(1):235-42.
[Crossref] [Google Scholar] [PubMed]
- Nandi A, Chatterjee IB. Scavenging of superoxide radical by ascorbic acid. Int J Biol Sci 1987;11:435-41.
- Desquiret-Dumas V, Gueguen N, Leman G, Baron S, Nivet-Antoine V, Chupin S, et al., Resveratrol induces a mitochondrial complex I-dependent increase in NADH oxidation responsible for sirtuin activation in liver cells. J Biol Chem 2013;288(51):36662-75.
[Crossref] [Google Scholar] [PubMed]
- Huang TC, Chang HY, Hsu CH, Kuo WH, Chang KJ, Juan HF. Targeting therapy for breast carcinoma by ATP synthase inhibitor aurovertin B. J Proteome Res 2008;7(4):1433-44.
[Crossref] [Google Scholar] [PubMed]
- Bodiga VL, Bodiga S. Ascorbic acid is a potential inhibitor of collagenases-In silico and in vitro biological studies. In Silico Drug Design 2019; pp. 649-77. Academic Press.
- Donarska B, Z Łączkowski K. Recent advances in the development of elastase inhibitors. Future Med Chem 2020;12(20):1809-13.
[Crossref] [Google Scholar] [PubMed]
- Li HZ, Ren Z, Reddy NV, Hou T, Zhang ZJ. In silico evaluation of antimicrobial, antihyaluronidase and bioavailability parameters of rosmarinic acid in Perilla frutescens leaf extracts. SN Appl Sci 2020;2:1-4.
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Dhiman G, Lohia N, Jain S, Baranwal M. Metadherin peptides containing CD4+ and CD8+ T cell epitopes as a therapeutic vaccine candidate against cancer. Med Microbiol Immunol 2016;60(9):646-52.
[Crossref] [Google Scholar] [PubMed]
- Jain S, Baranwal M. Conserved peptide vaccine candidates containing multiple Ebola nucleoprotein epitopes display interactions with diverse HLA molecules. Med Microbiol Immunol 2019;208:227-38.
[Crossref] [Google Scholar] [PubMed]
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing 2006: pp. 84-es.
- Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM et al,. Millisecond-scale molecular dynamics simulations on Anton. In: Proceedings of the conference on high performance computing networking, storage and analysis 2009; (pp. 1-11).
- Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem 1999;1(1):55-68.
[Crossref] [Google Scholar] [PubMed]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45(12):2615-23.
[Crossref] [Google Scholar] [PubMed]
- Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem 2000;43(21):3867-77.
[Crossref] [Google Scholar] [PubMed]
- Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem 2001;44(12):1841-6.
[Crossref] [Google Scholar] [PubMed]
- Jain S, Baranwal M. Computational analysis in designing T cell epitopes enriched peptides of Ebola glycoprotein exhibiting strong binding interaction with HLA molecules. J Theor Biol. 2019;465:34-44.
[Crossref] [Google Scholar] [PubMed]
- Li H, Li C. Multiple ligand simultaneous docking: orchestrated dancing of ligands in binding sites of protein. J Comput Chem 2010;31(10):2014-22.
- Li H, Xiao H, Lin L, Jou D, Kumari V, Lin J, Li C. Drug design targeting protein–protein interactions (PPIs) using multiple ligand simultaneous docking (MLSD) and drug repositioning: Discovery of raloxifene and bazedoxifene as novel inhibitors of IL-6/GP130 interface. J Med Chem 2014;57(3):632-41.
[Crossref] [Google Scholar] [PubMed]
- Eun Lee K, Bharadwaj S, Yadava U, Gu Kang S. Evaluation of caffeine as inhibitor against collagenase, elastase and tyrosinase using in silico and in vitro approach. J Enzyme Inhib Med Chem 2019;34(1):927-36.
[Crossref] [Google Scholar] [PubMed]
- Widowati W, Ginting CN, Lister IN, Girsang E, Amalia A, Wibowo SH et al. Anti-aging effects of mangosteen peel extract and its phytochemical compounds: Antioxidant activity, enzyme inhibition and molecular docking simulation. Trop Life Sci Res 2020;31(3):127.
[Crossref] [Google Scholar] [PubMed]
- Liu RH. Health benefits of fruit and vegetables are from additive and synergistic combinations of phytochemicals. Am J Clin Nutr 2003;78(3):517S-20S.
[Crossref] [Google Scholar] [PubMed]
- Jayaraman P, Sakharkar MK, Lim CS, Tang TH, Sakharkar KR. Activity and interactions of antibiotic and phytochemical combinations against Pseudomonas aeruginosa in vitro. Int J Biol Sci 2010;6(6):556.
[Crossref] [Google Scholar] [PubMed]
- Mikstacka R, Rimando AM, Ignatowicz E. Antioxidant effect of trans-resveratrol, pterostilbene, quercetin and their combinations in human erythrocytes in vitro. Proc Nutr Soc 2010;65:57-63.
[Crossref] [Google Scholar] [PubMed]
- Deniz FS, Salmas RE, Emerce E, Cankaya II, Yusufoglu HS, Orhan IE. Evaluation of collagenase, elastase and tyrosinase inhibitory activities of Cotinus coggygria Scop. through in vitro and in silico approaches. South Afr J Botany 2020;132:277-88.
- Tan H, Sonam T, Shimizu K. The potential of triterpenoids from loquat leaves (Eriobotrya japonica) for prevention and treatment of skin disorder. Int J Mol Sci 2017;18(5):1030.
[Crossref] [Google Scholar] [PubMed]
- Hejazi II, Khanam R, Mehdi SH, Bhat AR, Rizvi MM, Thakur SC, et al. Antioxidative and anti-proliferative potential of Curculigo orchioides Gaertn in oxidative stress induced cytotoxicity: In vitro, ex vivo and in silico studies. Food Chem Toxicol 2018;115:244-59.
- Sherratt HS. Mitochondria: structure and function. Rev Neurol 1991;147(6-7):417-30.
[Google Scholar] [PubMed]
- Zheng J, Ramirez VD. Inhibition of mitochondrial proton F0F1‐ATPase/ATP synthase by polyphenolic phytochemicals. Br J Pharmacol 2000;130(5):1115-23.
[Crossref] [Google Scholar] [PubMed]
- Patro S, Ratna S, Yamamoto HA, Ebenezer AT, Ferguson DS, Kaur A, et al. ATP synthase and mitochondrial bioenergetics dysfunction in Alzheimer’s disease. Int J Mol Sci 2021;22(20):11185.
[Crossref] [Google Scholar] [PubMed]
- Weiss H, Friedrich T, Hofhaus G, Preis D. The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur J Biochem 1991. 1992:55-68.
[Crossref] [Google Scholar] [PubMed]
- Pujol C, Bratic-Hench I, Sumakovic M, Hench J, Mourier A, Baumann L, et al. Succinate dehydrogenase upregulation destabilize complex I and limits the lifespan of gas-1 mutant. PloS one. 2013;8(3):e59493.
[Crossref] [Google Scholar] [PubMed]
- Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase–assembly, regulation and role in human disease. Mitochondrion 2010;10(4):393-401.
[Crossref] [Google Scholar] [PubMed]
- Beal MF. Mitochondria take center stage in aging and neurodegeneration. Annals of Neurology. Ann Neurol 2005;58(4):495-505.
[Crossref] [Google Scholar] [PubMed]
- Fogarty MJ, Marin Mathieu N, Mantilla CB, Sieck GC. Aging reduces succinate dehydrogenase activity in rat type IIx/IIb diaphragm muscle fibers. Appl Physiol 2020;128(1):70-7.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Park JS, Deng JH, Bai Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr 2006;38:283-91.
[Crossref] [Google Scholar] [PubMed]
- Arnold S. Cytochrome c oxidase and its role in neurodegeneration and neuroprotection. Mitochondrial oxidative phosphorylation. Adv Exp Med Biol 2012:305-39.
[Crossref] [Google Scholar] [PubMed]
- Grimm A, Eckert A. Brain aging and neurodegeneration: from a mitochondrial point of view. J Neurochem 2017;143(4):418-31.
[Crossref] [Google Scholar] [PubMed]
- Nicoletti VG, Tendi EA, Lalicata C, Reale S, Costa A, Villa RF, et al. Changes of mitochondrial cytochrome c oxidase and FoF1 ATP synthase subunits in rat cerebral cortex during aging. J Neurochem 1995;20:1465-70.
[Crossref] [Google Scholar] [PubMed]
- Calzia D, Oneto M, Caicci F, Bianchini P, Ravera S, Bartolucci M, Diaspro A, Degan P, Manni L, Traverso CE, Panfoli I. Effect of polyphenolic phytochemicals on ectopic oxidative phosphorylation in rod outer segments of bovine retina. Br J Pharmacol 2015;172(15):3890-903.
[Crossref] [Google Scholar] [PubMed]
- Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis 2012;35:211-25.
[Crossref] [Google Scholar] [PubMed]
- Ahmad Z, Hassan SS, Azim S. A therapeutic connection between dietary phytochemicals and ATP synthase. Curr Med Chem 2017;24(35):3894-906.
[Crossref] [Google Scholar] [PubMed]
- Ganesan K, Sehgal PK, Mandal AB, Sayeed S. Protective effect of Withania somnifera and Cardiospermum halicacabum extracts against collagenolytic degradation of collagen. Biotechnol Appl Biochem 2011;165:1075-91.
[Crossref] [Google Scholar] [PubMed]
- Choudhary D, Adhikary S, Ahmad N, Kothari P, Verma A, Trivedi PK, et al. Prevention of articular cartilage degeneration in a rat model of monosodium iodoacetate induced osteoarthritis by oral treatment with Withaferin A. Biomed Pharmacother 2018;99:151-61.
[Crossref] [Google Scholar] [PubMed]
- Won HR, Lee P, Oh SR, Kim YM. Epigallocatechin-3-gallate suppresses the expression of TNF-α-induced MMP-1 via MAPK/ERK signaling pathways in human dermal fibroblasts. Biol Pharm Bull 2021;44(1):18-24.
[Crossref] [Google Scholar] [PubMed]
- Michalak M. Plant-derived antioxidants: Significance in skin health and the ageing process. Int J Mol Sci 2022;23(2):585.
[Crossref] [Google Scholar] [PubMed]
- Madhan B, Krishnamoorthy G, Rao JR, Nair BU. Role of green tea polyphenols in the inhibition of collagenolytic activity by collagenase. Int J Biol Macromol 2007;41(1):16-22.
[Crossref] [Google Scholar] [PubMed]
- Mihailović M, Dinić S, Arambašić Jovanović J, Uskoković A, Grdović N, Vidaković M. The influence of plant extracts and phytoconstituents on antioxidant enzymes activity and gene expression in the prevention and treatment of impaired glucose homeostasis and diabetes complications. Antioxidants 2021;10(3):480.
[Crossref] [Google Scholar] [PubMed]
- Lü JM, Lin PH, Yao Q, Chen C. Chemical and molecular mechanisms of antioxidants: experimental approaches and model systems. J Cell Mol Med 2010;14(4):840-60.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Tsao R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr Opin Food Sci 2016;8:33-42.
- Naoi M, Wu Y, Shamoto-Nagai M, Maruyama W. Mitochondria in neuroprotection by phytochemicals: Bioactive polyphenols modulate mitochondrial apoptosis system, function and structure. Int J Mol Sci 2019;20(10):2451.
[Crossref] [Google Scholar] [PubMed]
- Surapaneni DK, Adapa SR, Preeti K, Teja GR, Veeraragavan M, Krishnamurthy S. Shilajit attenuates behavioral symptoms of chronic fatigue syndrome by modulating the hypothalamic–pituitary–adrenal axis and mitochondrial bioenergetics in rats. J Ethnopharmacol 2012 Aug 30;143(1):91-9.
[Crossref] [Google Scholar] [PubMed]