- *Corresponding Author:
- Min Zhang
Department of Endocrinology, Qingpu Branch of Zhongshan Hospital Affiliated to Fudan University, Qingpu District, Shanghai 200050, China
E-mail: mzhang202209@163.com
| Date of Received | 12 July 2023 |
| Date of Revision | 12 December 2023 |
| Date of Acceptance | 04 June 2024 |
| Indian J Pharm Sci 2024;86(3):940-949 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cnidium monnieri (L.) Cuss, belonging to the Umbelliferae family, is a widely used traditional herbal medicine in ameliorating nephritis. This study was to identify key targets and investigate the molecular mechanism of Cnidium, an extract of Cnidium monnieri that treats diabetic kidney disease, using network pharmacology and bioinformatics. Cnidium's active ingredients and target profiles were extracted from the traditional Chinese medicine systems pharmacology database and analysis platform database. The GEO2R online tool was utilized to mine the differentially expressed genes associated with diabetic kidney disease from the gene expression omnibus database. The diabetic kidney disease-related genes were also obtained from the GeneCards database and compared with the gene expression omnibus databases. Differentially expressed genes and drug targets were mapped to obtain common targets. The Cnidium-active ingredient-target-diabetic kidney disease network was constructed with the help of Cytoscape 3.7. As a result, we found that Cnidium has 19 compounds and 98 targets, among which beta-sitosterol and ostruthin played major roles in treating diabetic kidney disease. The key targets comprised prostaglandin-endoperoxide synthase 2, caspase-3, estrogen receptor 1, jun proto-oncogene, heat shock protein 90 alpha family class A member 1 (HSP90AA1), mammalian target of rapamycin, matrix metallopeptidase 9. The protein-protein interaction network of common targets was constructed using the search tool for the retrieval of interacting genes/proteins database. The David database was used for gene ontology and kyoto encyclopedia of genes and genomes pathway enrichment analysis for key targets. Gene ontology analysis revealed that these key targets regulated apoptosis and protein serine/threonine/tyrosine kinase activity. The kyoto encyclopedia of genes and genomes analysis indicated that the calcium signaling pathway, interleukin-17 signaling pathway, cyclic adenosine 3′,5′-monophosphate signaling pathway, and advanced glycation end products receptor for advanced glycation end products signaling pathway were involved in diabetic complications treatment. The key targets were verified by molecular docking with active ingredients. The results showed that beta-sitosterol and ostruthin bind to caspase-3 and estrogen receptor 1. From the perspective of network pharmacology, we found that the possible mechanism of Cnidium in treating diabetic kidney disease involves the interaction between beta-sitosterol and caspase-3, ostruthin and estrogen receptor 1.
Abstract
Cnidium monnieri (L.) Cuss, belonging to the Umbelliferae family, is a widely used traditional herbal medicine in ameliorating nephritis. This study was to identify key targets and investigate the molecular mechanism of Cnidium, an extract of Cnidium monnieri that treats diabetic kidney disease, using network pharmacology and bioinformatics. Cnidium's active ingredients and target profiles were extracted from the traditional Chinese medicine systems pharmacology database and analysis platform database. The GEO2R online tool was utilized to mine the differentially expressed genes associated with diabetic kidney disease from the gene expression omnibus database. The diabetic kidney disease-related genes were also obtained from the GeneCards database and compared with the gene expression omnibus databases. Differentially expressed genes and drug targets were mapped to obtain common targets. The Cnidium-active ingredient-target-diabetic kidney disease network was constructed with the help of Cytoscape 3.7. As a result, we found that Cnidium has 19 compounds and 98 targets, among which beta-sitosterol and ostruthin played major roles in treating diabetic kidney disease. The key targets comprised prostaglandin-endoperoxide synthase 2, caspase-3, estrogen receptor 1, jun proto-oncogene, heat shock protein 90 alpha family class A member 1, mammalian target of rapamycin, matrix metallopeptidase 9. The protein-protein interaction network of common targets was constructed using the search tool for the retrieval of interacting genes/proteins database. The David database was used for gene ontology and kyoto encyclopedia of genes and genomes pathway enrichment analysis for key targets. Gene ontology analysis revealed that these key targets regulated apoptosis and protein serine/threonine/tyrosine kinase activity. The kyoto encyclopedia of genes and genomes analysis indicated that the calcium signaling pathway, interleukin-17 signaling pathway, cyclic adenosine 3′,5′-monophosphate signaling pathway, and advanced glycation end products receptor for advanced glycation end products signaling pathway were involved in diabetic complications treatment. The key targets were verified by molecular docking with active ingredients. The results showed that beta-sitosterol and ostruthin bind to caspase-3 and estrogen receptor 1. From the perspective of network pharmacology, we found that the possible mechanism of Cnidium in treating diabetic kidney disease involves the interaction between beta-sitosterol and caspase-3, ostruthin and estrogen receptor 1.
Keywords
Cnidium, diabetic nephropathy, network pharmacology, bioinformatics, mechanism of action
With the rapid development of society and the economy, coupled with factors such as population aging and the emergence of various risk factors, the global number of individuals diagnosed with diabetes is on the rise. The mortality rate associated with diabetes is also increasing year by year. Diabetic Nephropathy (DN) is an inflammatory chronic End-Stage Renal Disease (ESRD) that occurs as a complication of diabetes[1,2]. The characteristic pathological features of DN include the proliferation of glomerular mesangial cells, the accumulation of extracellular matrix, the development of glomerular sclerosis and the occurrence of tubulointerstitial fibrosis[3,4]. It is currently believed that the occurrence of DN is related to various factors such as glucose and lipid metabolism disorders, oxidative stress and abnormal expression of cytokines[4] of which inflammatory response plays an essential role in accelerating the progression of DN. Pathological conditions such as hyperglycemia and hemodynamic disorders can activate multiple pro-inflammatory signaling pathways, leading to increased kidney inflammation. As a result, substantial macrophages infiltrated into the renal tissues. Tumor Necrosis Factor Alpha (TNF-α) is released in large quantities, which leads to glomerulosclerosis and tubulointerstitial fibrosis[5]. Studies have shown that Nuclear factor erythroid 2-related factor 2/Heme Oxygenase-1 (Nrf2/HO- 1) signaling is a potential target for reducing the damage to renal tubular epithelial cells induced by high glucose hypoxia and reoxygenation. Nrf2/ HO-1 is a crucial signal regulating oxidative stress, mitochondrial damage, inflammation and apoptosis. Upregulating the expression of Nrf2 and HO-1 can inhibit the peroxidation and inflammatory response induced by high glucose and oxygen-glucose deprivation/reoxygenation and reduce cell damage and apoptosis. Tubular epithelial cells Human Kidney-2 (HK-2) are protected from hypoxia/reoxygenation-induced cell damage by activating the Nrf2/HO-1 signaling pathway[6-8]. Currently, the treatment drugs and methods for DN are limited[9]. DN still has a high fatality rate and disability rate. Therefore, it is of great significance to find new drugs.
Cnidium is derived from the mature fruit of Cnidium monnieri (C. monnieri), a plant belonging to the Umbelliferae family. Its main structure comprises a benzene ring and a pyrone ring. Research has demonstrated that Cnidium exhibits a wide range of pharmacological and biological effects. These include anti-tumor, neuroprotective, anti-inflammatory, osteogenic and cardiovascular protective effects and antibacterial and antiparasitic activities[10-12]. In an in vitro study, Cnidium inhibited Lipopolysaccharide (LPS)- induced production of TNF-α, Nitric Oxide (NO), Prostaglandin E2 (PGE2) and Interleukin-6 (IL-6) in macrophages[13]. In the model of acute lung injury, Cnidium has exhibited anti-inflammatory activity by inhibiting various cell signaling pathways such as Janus kinase/Signal Transducers and Activators of Transcription (JAK-STAT), Mitogen-activated protein Kinase (MAPK), and Nuclear factor kappa B (NF-κB)[14,15]. In addition, in the focal segmental glomerulosclerosis model and the rat renal ischemia-reperfusion injury model, Cnidium was confirmed to have a protective effect on the kidneys[16]. However, Cnidium's role and related mechanisms in DN have not been reported in the literature.
This study used the Gene Expression Omnibus (GEO) database to mine the related chips of diabetes mellitus complicated with renal disease and the differentially expressed genes were analyzed. In Search Tool for the Retrieval of Interacting Genes/proteins (STRING) database, ingredients of Cnidium were mapped to relevant targets and a Protein-Protein Interaction (PPI) network was constructed to explore further Cnidium's biological functions and underlying mechanisms in treating diabetes complicated with renal complications. Our work will provide new research methods and therapeutic ideas for treating renal disease.
Materials and Methods
Databases and software:
The databases used in this study are Traditional Chinese Medicine Systems Pharmacology database and analysis platform (TCMSP) (https://tcmsp-e.com/tcmsp search.php), STRING (https:// cn.STRING-db.org/), DAVID (https://david.ncifcrf.gov/), UniProt (https://www.uniprot.org/), GeneCards (https://www.genecards.org/), GEO (https://www.ncbi.nlm.nih.gov/geo/), PubChem (https://pubchem.ncbi.nlm.nih.gov/) and Protein Data Bank (PDB) (https://www.rcsb.org/). The softwares used were RStudio, R3.5, Cytoscape3.7, AutoDock4.6, SangBox, PyMol and OpenBabel.
Active components and targets of Cnidium:
The chemical constituents of Cnidium were obtained by searching the TCMSP and the screening conditions were set as Oral Bioavailability (OB) ≥30 % and Drug-Like properties (DL) ≥0.18. The information regarding the targets of active ingredients was extracted and converted into corresponding targets within the UniProt database.
Screening of Diabetic Kidney Disease (DKD)-related targets and differential genes:
Search the GeneCards database for "DKD" to gather information on targets associated with DKD. Additionally, search for microarray chips in the GEO database related explicitly to DKD. Then, utilize the GEO2R online analysis tool to analyze the chip data and retrieve the dysregulated genes relevant to DKD. The screening condition was p<0.05 and log|FC|≥1.
Construction of Cnidium-DKD-active ingredient-target network:
The active ingredients obtained above and their corresponding targets were imported into Cytoscape 3.7 software to construct a network of "Cnidium-DKD-active ingredients-targets". The targets corresponding to the active ingredients were combined with those obtained from GeneCards and GEO databases. By finding the intersection of these targets, common targets were identified. Subsequently, a network comprising the common targets and their corresponding active ingredients was constructed.
Construction of common target PPI network and screening of key genes:
Upload the common targets to the STRING database, select the species as human and select the connection relationship with a combined score >0.4 to obtain the PPI network map of the common targets. The results obtained from the STRING platform were imported into Cytoscape3.7 software, and the "Cytohubba" and "MCODE" plug-ins were used to analyze the PPI network and screen critical targets.
GO analysis of key targets and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis:
Search the key targets in the DAVID database, set the species as human, and set the genotype as office gene symbol to obtain the GO and KEGG enrichment pathways of the key targets.
Molecular docking of key targets and active ingredients:
Use PyMol software to dehydrate molecules and ligands of key targets (proteins). Perform hydrogenation with AutoDock, calculate charges, set atomic properties and save them in pdbqt format. Obtain the structural formula of the active ingredient or the original ligand on the PubChem platform, save in the 2D SDF file and convert it to mol2 format with Open Babel. Use AutoDock to hydrogenate the active ingredient or original ligand, calculate the charge, set relevant parameters, and save it in pdbqt format. Set the grid size of Gridbox to include target and compound (original ligand), select Mark algorithm for operation mode and save as dpf format. Set the target as a rigid molecule and the compound (original ligand) as a docking molecule. By default, the docking method was saved in gpf format. Finally, check the dlg format file, open the target, perform energy rank and keep the molecular docking results in pdbqt format. Use Open Babel to convert it to pdb format and use PyMol to view the docking.
Results and Discussion
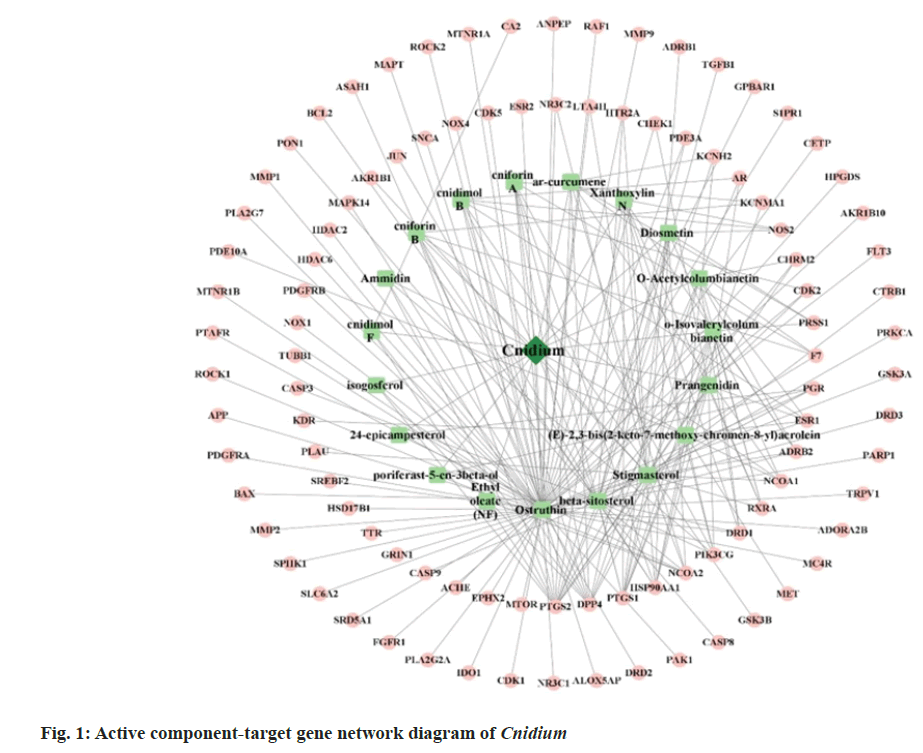
A search was conducted in the TCMSP database using the screening criteria of OB≥30 % and DL properties ≥0.18. This resulted in identifying 19 active ingredients from Cnidium, as listed in Table 1. Cytoscape 3.7 was used to construct the "active ingredient-target network" for Cnidium, as illustrated in fig. 1. A total of 98 targets were obtained. Then, the network was analyzed using the "Network Analyzer" plug-in to get the topological parameters of the network. Within the network, beta-sitosterol (β-sitosterol) and ostruthin ranked highest in degree, suggesting that these two components may be the primary active compounds in Cnidium.
| MOI ID | Name | OB (%) | DL (%) | Source |
|---|---|---|---|---|
| MOL003600 | Cnidimol B | 68.66 | 0.26 | Shechuangzi |
| MOL003624 | o-Isovalerylcolum bianetin | 64.03 | 0.36 | Shechuangzi |
| MOL003608 | O-Acetylcolumbianetin | 60.04 | 0.26 | Shechuangzi |
| MOL003605 | (E)-2,3-bis(2-keto-7-methoxy-chromen-8-yl) acrolein | 56.38 | 0.71 | Shechuangzi |
| MOL003606 | Cniforin A | 55.89 | 0.47 | Shechuangzi |
| MOL003604 | Cnidimol F | 54.43 | 0.28 | Shechuangzi |
| MOL003591 | ar-curcumene | 52.34 | 0.65 | Shechuangzi |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | Shechuangzi |
| MOL001510 | 24-epicampesterol | 37.58 | 0.71 | Shechuangzi |
| MOL001771 | Poriferast-5-en-3beta-ol | 36.91 | 0.75 | Shechuangzi |
| MOL000358 | β-sitosterol | 36.91 | 0.75 | Shechuangzi |
| MOL003607 | Cniforin B | 36.7 | 0.6 | Shechuangzi |
| MOL003588 | Prangenidin | 36.31 | 0.22 | Shechuangzi |
| MOL003584 | Xanthoxylin N | 35.51 | 0.21 | Shechuangzi |
| MOL001941 | Ammidin | 34.55 | 0.22 | Shechuangzi |
| MOL002883 | Ethyl oleate (NF) | 32.4 | 0.19 | Shechuangzi |
| MOL002881 | Diosmetin | 31.14 | 0.27 | Shechuangzi |
| MOL003626 | Ostruthin | 30.65 | 0.23 | Shechuangzi |
| MOL003617 | Isogosferol | 30.07 | 0.25 | Shechuangzi |
Note: Acquisition of DKD-associated Differentially Expressed Genes (DEGs) with volcano plots
Table 1: Active Ingredients of Cnidium
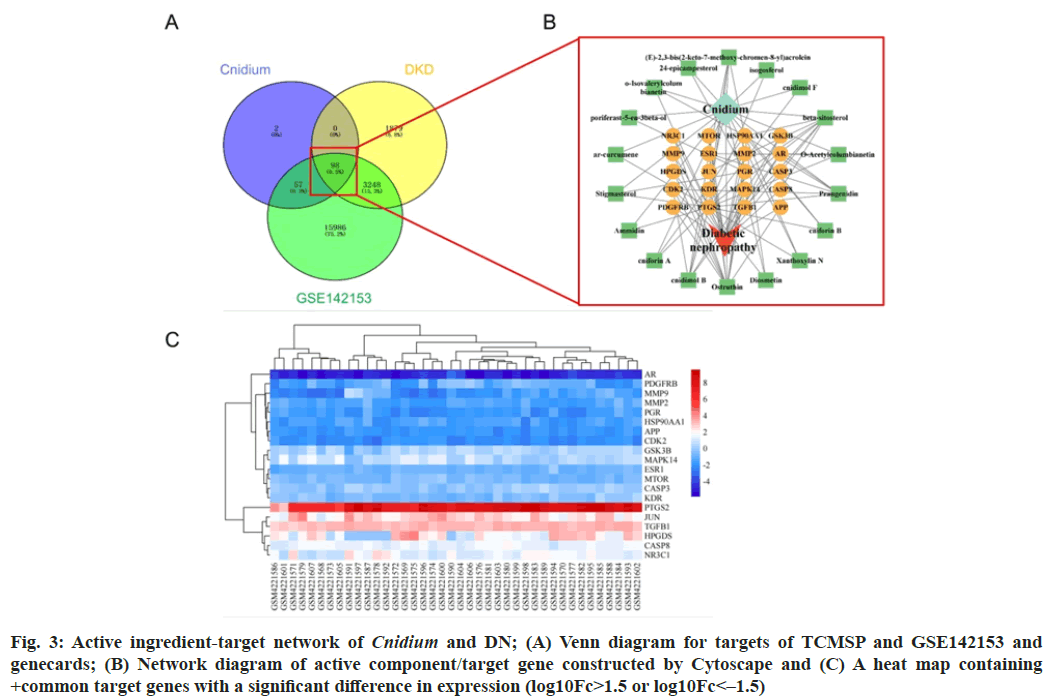
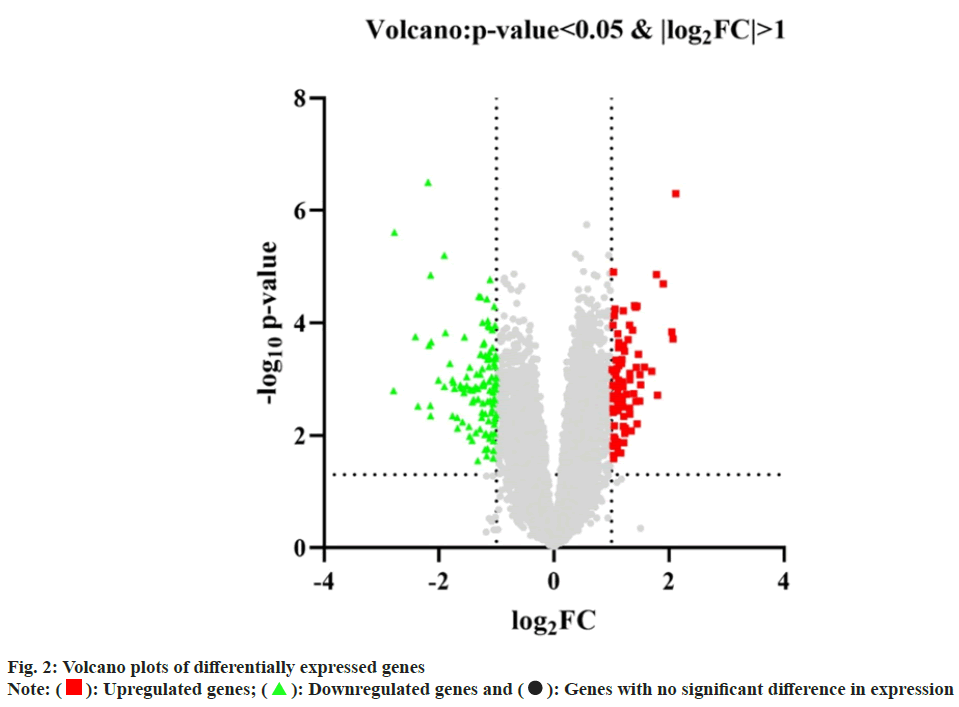
The GEO2R online analysis platform was utilized to analyze the two sets of data (DKD group and control group) from the GSE142153 chip. The "volcano" function in the R language was employed to generate volcano plots depicting the results, as shown in fig. 2. A total of 3346 genes were found to be associated with DKD in the GSE142153 chip. Common targets were obtained by taking the intersection of targets acquired from the TCMSP, GeneCards and GEO databases, as illustrated in fig. 3A. The shared targets corresponding to the compounds were selected, and Cytoscape 3.7 was utilized to construct the "active ingredient-DKD- common target" network (fig. 3B). The graph contained 98 nodes and 578 edges. The common targets were subjected to cluster analysis using the "heatmap" function in the R language. The resulting heat map of the common targets was shown in fig. 3C. The heat map showed the expression of common target genes in each sample.
Fig. 3: Active ingredient-target network of Cnidium and DN; (A) Venn diagram for targets of TCMSP and GSE142153 and genecards; (B) Network diagram of active component/target gene constructed by Cytoscape and (C) A heat map containing +common target genes with a significant difference in expression (log10Fc>1.5 or log10Fc<–1.5)
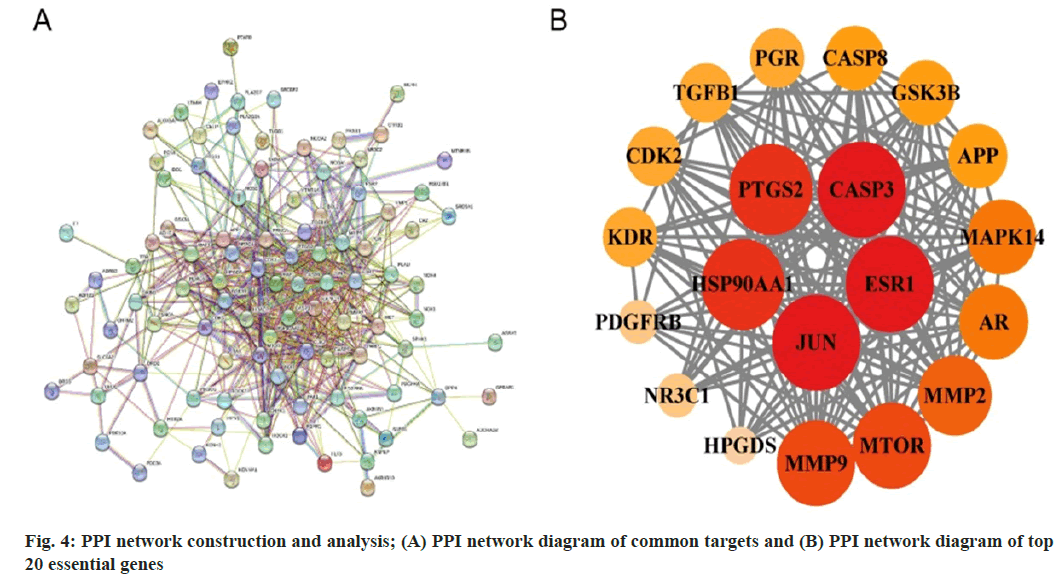
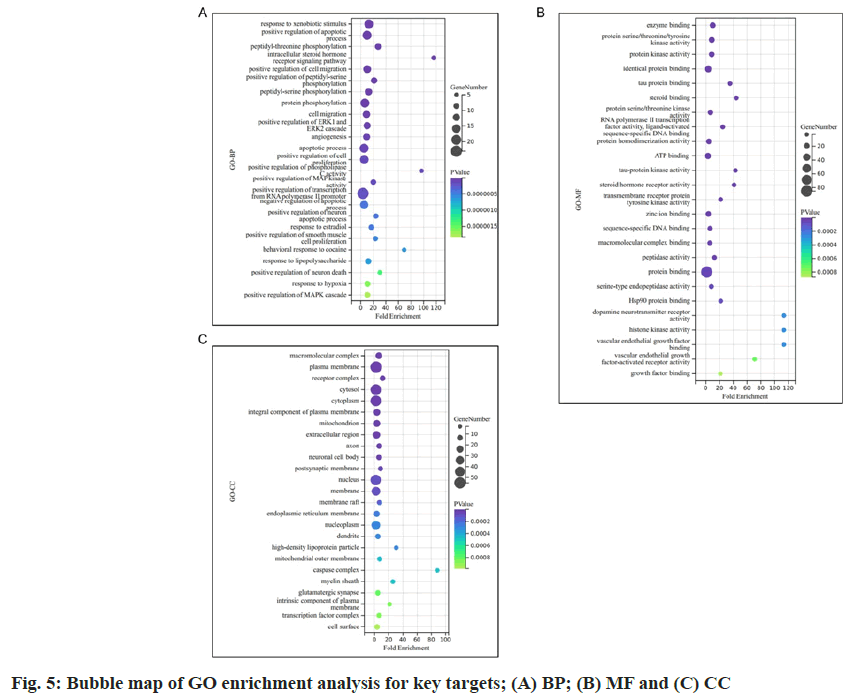
The common targets obtained previously were utilized to query the STRING database, generating a PPI network for the common targets (fig. 4A). The network density was calculated to be 0.629, with an average node degree of 11.7, indicating a strong correlation in the network, as it surpassed the threshold values of 0.5 for network density and 3 for average node degree, indicative of a well- connected network. Fig. 4B shows the PPI network of the top 20 target genes with high degrees of freedom. The target exhibits a darker color and a larger circle size, corresponding to a higher degree value. Finally, the data demonstrated that the key targets of Cnidium in treating DKD comprised HSP9001A, prostaglandin-endoperoxide synthase 2 (PTGS 2), Cysteine Aspartate Specific Protease 3 (CASP 3), Estrogen Receptor 1 (ESR1) and Jun proto-oncogene (JUN). We utilized the DAVID database to perform GO and KEGG pathway enrichment analysis on the genes corresponding to the 20 target proteins in the PPI network. GO pathway enrichment encompasses three components, viz., biological process, cellular component and molecular function. The biological processes contained cellular responses to xenobiotic stimulus, positive regulation of the apoptotic process, intracellular steroid hormone- receptor signaling pathway, etc., (fig. 5A) cellular components included the macromolecular receptor, plasma membrane, receptor complex, etc., (fig. 5B) and molecular functions involved enzyme binding, protein serine/threonine/tyrosine kinase activity, etc. (fig. 5C).
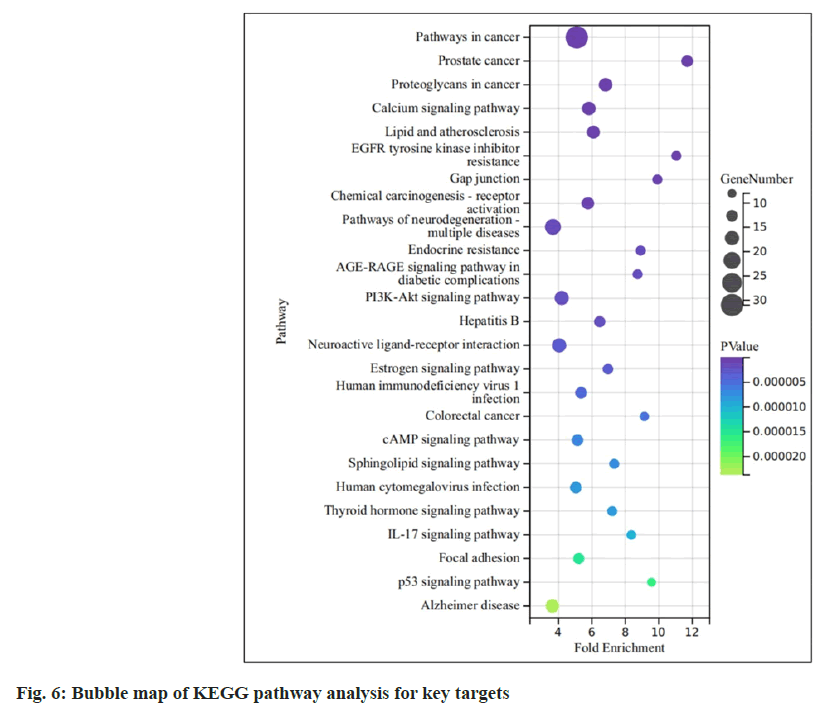
The KEGG analysis revealed the enrichment of 25 signaling pathways (p<0.05). After excluding cancer-related pathways, the prominently enriched pathways included the calcium signaling pathway, Interleukin-17 (IL-17) signaling pathway, cyclic Adenosine 3′,5′-Monophosphate (cAMP) signaling pathway, Advanced Glycation Endproduct-Receptor for Advanced Glycation Endproducts (AGE-RAGE) signaling pathway in diabetic complications pathway and several others (fig. 6).
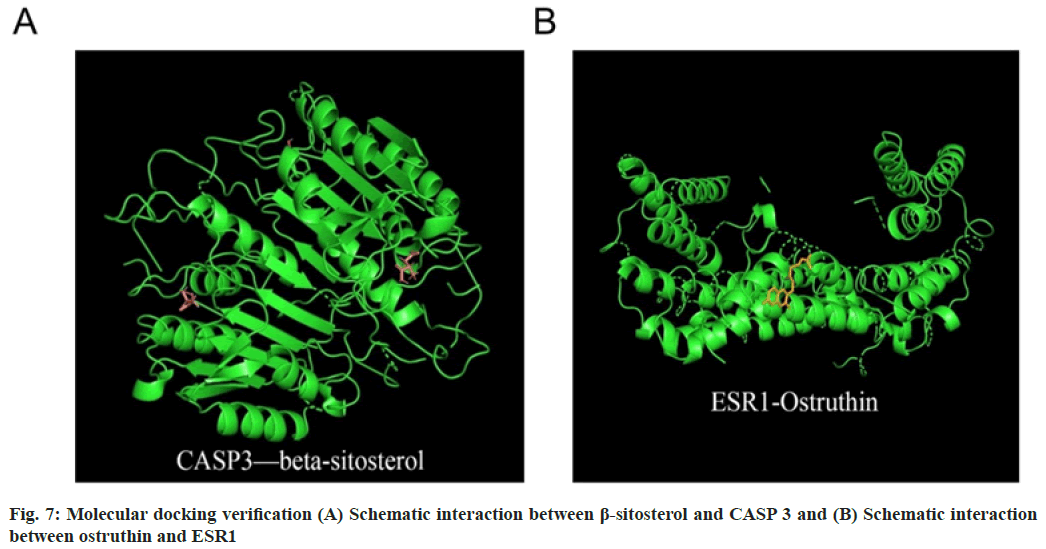
Based on the analysis mentioned above, it was evident that β-sitosterol and ostruthin played significant roles in the functional activity of Cnidium. The two components, β-sitosterol and ostruthin were queried in the PubChem database. Molecular docking was performed with AutoDock Vina software and visually displayed. We found that β-sitosterol and ostruthin exhibit the strongest binding affinity to the proteins associated with the CASP 3 and ESR1 genes, respectively, compared to the original ligand. The docking binding energy of CASP 3 protein and β-sitosterol was -6.45 kcal·mol-1. ESR1 protein and ostruthin's docking binding energy was -4.72 kcal·mol-1 (fig. 7A and fig. 7B). These results suggested that Cnidium mainly regulates these two targets in DKD treatment.
Traditional Chinese medicine has been reported to have the potential not only to reduce urine protein levels in patients with DKD but also reduce blood sugar levels and serum creatinine levels and improve lipid metabolism in patients with DKD. Moreover, it has shown potential in delaying the occurrence and progression of DKD. Due to these beneficial effects, traditional Chinese medicine is widely utilized to prevent and treat chronic kidney disease[17-19]. Cnidium is a coumarin compound extracted from the fruit of the Umbelliferae Cnidium plant. It is recognized for containing a high concentration of hydrocarbon coumarin chemicals. Modern pharmacological studies have yielded results indicating that Cnidium possesses a range of beneficial effects. These include anti- arrhythmic, anti-inflammatory, anti-tumor, anti- hypertensive and anti-allergic properties[20]. These findings highlight the potential of Cnidium as a valuable therapeutic agent in various medical applications.
Recent research findings have demonstrated that Cnidium is effective in preventing ischemia- reperfusion injury. Additionally, it has been shown to effectively inhibit the levels of pro- inflammatory cytokines in the body and reduce the volume of cerebral infarction lesions[21]. These results highlight the potential neuroprotective properties of Cnidium and its potential application in treating and managing conditions related to ischemic events in the brain. Hyperglycemia in patients can lead to an elevation of abnormal active oxidative groups in vascular cells, contributing to various complications. In this regard, Cnidium demonstrates its beneficial effects by scavenging free radicals, exerting antioxidant properties, and controlling blood lipid metabolism. These actions of Cnidium can effectively combat the various diseases induced by diabetes and its associated complications. This study combined network pharmacology and bioinformatics to analyze and predict the potential pharmacological mechanism of Cnidium in treating DKD. The study discovered that Cnidium contained 19 active components regulating the disease. Among these, the main active components associated with DKD were β-sitosterol and ostruthin. Furthermore, the analysis identified 20 critical targets involved in the regulation of DKD. The target genes identified in the study were primarily concentrated in pathways such as calcium signaling pathway, IL-17 signaling pathway, cAMP signaling pathway and AGE-RAGE signaling pathway in diabetic complications pathway. These pathways are relevant to the growth and apoptosis of glomerular epithelial cells. Besides, these pathways are closely associated with the onset and progression of inflammation, indicating that Cnidium may intervene in the development of DKD through a multi-component, multi-target and multi-pathway approach. These findings provided valuable insights into the potential therapeutic effects of Cnidium in treating DKD.
β-sitosterol is considered a typical representative of phytosterols. β-sitosterol exits in abundance in the deodorized distillate of various plants and during the refining process of vegetable oils. It has multiple biological activities such as antioxidant, anti-tumor and anti-inflammatory[22-24]. Furthermore, studies have revealed that β-sitosterol could help to rectify insulin secretion defects, increase total insulin levels and promote the reorganization of agonist proteins[25]. In in vitro experiments, treatment of L6 cells with different concentrations of β-sitosterol significantly promoted glucose uptake[26]. In the in vivo test, after oral administration of β-sitosterol, the fasting blood glucose and blood lipid indexes (such as triacylglycerol and cholesterol) of KK-Ay mice were significantly reduced, and the insulin resistance and oral glucose tolerance of KK-Ay mice were alleviated considerably. The mechanism may be related to the increased translocation and expression of Glucose Transporter 4 (GLUT4), suggesting the potential benefit of β-sitosterol for treating type 2 diabetes[27].
According to the analysis conducted, it has been identified that targets such as PTGS 2, CASP 3, ESR1, JUN and Heat Shock Protein 90 Alpha family class A member 1 (HSP90AA1) may serve as key targets for Cnidium in the treatment of DN. These targets were associated with the potential therapeutic effects of Cnidium. Furthermore, Cnidium contained several active ingredients that interact with these targets, suggesting their importance in the pharmacological actions of Cnidium for treating DN. Indeed, studies have indicated that PTGS 2 and Cyclooxygenase-2 (COX-2) was upregulated in diabetes-related kidney diseases, including DN[28]. These results suggested that targeting PTGS 2 could hold therapeutic potential for treating DN. By modulating PTGS 2 activity, it may be possible to mitigate the inflammatory response and associated pathological processes in the kidneys. Therefore, PTGS 2 could be a promising target for interventions to manage or treat DN[29,30]. It appears that CASP 3 may be associated with the pathogenesis of renal disease. Research suggested that CASP 3-KO mice developed immune abnormality-associated renal phenotypes[31]. Another critical target found in our study, ESR1 was shown to be a potential candidate underlying type 2 diabetes[32]. Furthermore, in silico analysis demonstrated that ESR1 was a hub gene regulating mesangial cell proliferation in Immunoglobulin A (IgA) nephropathy and serves as a potential target for IgA nephropathy treatment[33].
In the conducted study, a combination of network pharmacology and bioinformatics analysis was employed to analyze the mechanism of action of Cnidium in treating DKD. The aim was to explore potential active components and targets involved in the therapeutic effects of Cnidium. Additionally, molecular docking was utilized to verify the reliability of the data. Notably, the data obtained from this analysis were consistent with relevant literature reports, thus reinforcing the credibility and validity of this study's findings. Currently, limited clinical research is available on β-sitosterol, and further investigation is required to determine its potential clinical applications. The screening of network pharmacology is based on oral availability and drug-like properties, which may ignore other active ingredients. With the development of computer technology and bioinformatics technology, potential molecular mechanisms and targets will be gradually discovered.
Conflict of interests:
The authors declared that there is no conflict of interests.
References
- Alicic RZ, Rooney MT, Tuttle KR. Diabetic kidney disease: Challenges, progress, and possibilities. Clin J Am Soc Nephrol 2017;12(12):2032-45.
[Crossref] [Google Scholar] [PubMed]
- Khan NU, Lin J, Liu X, Li H, Lu W, Zhong Z, et al. Insights into predicting diabetic nephropathy using urinary biomarkers. Biochim Biophys Acta Proteins Proteom 2020;1868(10):140475.
[Crossref] [Google Scholar] [PubMed]
- Li X, Lu L, Hou W, Huang T, Chen X, Qi J, et al. Epigenetics in the pathogenesis of diabetic nephropathy: Epigenetics in the pathogenesis of diabetic nephropathy. Acta Biochim Biophys Sin (Shanghai) 2022;54(2):163.
[Crossref] [Google Scholar] [PubMed]
- Li KX, Ji MJ, Sun HJ. An updated pharmacological insight of resveratrol in the treatment of diabetic nephropathy. Gene 2021;780:145532.
[Crossref] [Google Scholar] [PubMed]
- Kushwaha K, Sharma S, Gupta J. Metabolic memory and diabetic nephropathy: Beneficial effects of natural epigenetic modifiers. Biochimie 2020;170:140-51.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Duan JZ, He Q, Wang CQ. miR-155 modulates high glucose‑induced cardiac fibrosis via the Nrf2/HO-1 signaling pathway. Mol Med Rep 2020;22(5):4003-16.
[Crossref] [Google Scholar] [PubMed]
- Zhi SM, Fang GX, Xie XM, Liu LH, Yan J, Liu DB, et al. Melatonin reduces OGD/R-induced neuron injury by regulating redox/inflammation/apoptosis signaling. Eur Rev Med Pharmacol Sci 2020;24(3):1524-36.
[Crossref] [Google Scholar] [PubMed]
- Feng J, Kong R, Xie L, Lu W, Zhang Y, Dong H, et al. Clemaichinenoside protects renal tubular epithelial cells from hypoxia/reoxygenation injury in vitro through activating the Nrf2/HO‐1 signalling pathway. Clin Exp Pharmacol Physiol 2020;47(3):495-502.
[Crossref] [Google Scholar] [PubMed]
- Nogueira A, Pires MJ, Oliveira PA. Pathophysiological mechanisms of renal fibrosis: A review of animal models and therapeutic strategies. In vivo 2017;31(1):1-22.
[Crossref] [Google Scholar] [PubMed]
- Wang R, Kong J, Wang D, Lien LL, Lien EJ. A survey of Chinese herbal ingredients with liver protection activities. Chin Med 2007;2:1-8.
[Crossref] [Google Scholar] [PubMed]
- Wang XL, Shang X, Cui Y, Zhao X, Zhang Y, Xie ML. Osthole inhibits inflammatory cytokine release through PPAR α/γ-mediated mechanisms in LPS-stimulated 3T3-L1 adipocytes. Immunopharmacol Immunotoxicol 2015;37(2):185-92.
[Crossref] [Google Scholar] [PubMed]
- Li K, Ding D, Zhang M. Neuroprotection of osthole against cerebral ischemia/reperfusion injury through an anti-apoptotic pathway in rats. Biol Pharm Bull 2016;39(3):336-42.
[Crossref] [Google Scholar] [PubMed]
- He YQ, Zhou CC, Yu LY, Wang L, Deng JL, Tao YL, et al. Natural product derived phytochemicals in managing acute lung injury by multiple mechanisms. Pharmacol Res 2021;163:105224.
[Crossref] [Google Scholar] [PubMed]
- Luo LN, Xie DQ, Zhang XG, Jiang R. Osthole decreases renal ischemia-reperfusion injury by suppressing JAK2/STAT3 signaling activation. Exp Ther Med 2016;12(4):2009-14.
[Crossref] [Google Scholar] [PubMed]
- Fan H, Gao Z, Ji K, Li X, Wu J, Liu Y, et al. The in vitro and in vivo anti-inflammatory effect of osthole, the major natural coumarin from Cnidium monnieri (L.) Cuss, via the blocking of the activation of the NF-κB and MAPK/p38 pathways. Phytomedicine 2019;58:152864.
[Crossref] [Google Scholar] [PubMed]
- Yang SM, Chan YL, Hua KF, Chang JM, Chen HL, Tsai YJ, et al. Osthole improves an accelerated focal segmental glomerulosclerosis model in the early stage by activating the Nrf2 antioxidant pathway and subsequently inhibiting NF-κB-mediated COX-2 expression and apoptosis. Free Radic Biol Med 2014;73:260-9.
[Crossref] [Google Scholar] [PubMed]
- Xue H, Li P, Luo Y, Wu C, Liu Y, Qin X, et al. Salidroside stimulates the Sirt1/PGC-1α axis and ameliorates diabetic nephropathy in mice. Phytomedicine 2019;54:240-7.
[Crossref] [Google Scholar] [PubMed]
- Hu Q, Qu C, Xiao X, Zhang W, Jiang Y, Wu Z, et al. Flavonoids on diabetic nephropathy: advances and therapeutic opportunities. Chin Med 20217;16(1):74.
[Crossref] [Google Scholar] [PubMed]
- Yu Z, Zhang W, Li B, Bao P, Wang F, Sun J, et al. Efficacy and safety of acupuncture combined with Chinese herbal medicine for diabetic nephropathy: A protocol for systematic review and meta-analysis. Medicine 2021;100(35):e27087.
[Crossref] [Google Scholar] [PubMed]
- Tziastoudi M, Tsezou A, Stefanidis I. Cadherin and Wnt signaling pathways as key regulators in diabetic nephropathy. PloS One 2021;16(8):e0255728.
[Crossref] [Google Scholar] [PubMed]
- Typiak M, Piwkowska A. Antiinflammatory actions of klotho: Implications for therapy of diabetic nephropathy. Int J Mol Sci 2021;22(2):956.
[Crossref] [Google Scholar] [PubMed]
- Zhang P, Liu N, Xue M, Zhang M, Liu W, Xu C, et al. Anti-inflammatory and antioxidant properties of β-sitosterol in copper sulfate-induced inflammation in Zebrafish (Danio rerio). Antioxidants 2023;12(2):391.
[Crossref] [Google Scholar] [PubMed]
- Abeesh P, Guruvayoorappan C. Preparation and characterization of beta sitosterol encapsulated nanoliposomal formulation for improved delivery to cancer cells and evaluation of its anti-tumor activities against daltons lymphoma ascites tumor models. Int J Drug Deliv Technol 2022;70:102832.
- Hooriyah h, Priya VV, Selvaraj J, Gayathri R, Kavitha S. Comparative evaluation of the antiinflammatory potential of three different anti diabetic natural compounds (piperine, lupeol, beta sitosterol). J Res Med Dent Sci 2021;9(11):230-34.
- Gupta R, Sharma AK, Dobhal MP, Sharma MC, Gupta RS. Antidiabetic and antioxidant potential of β‐sitosterol in streptozotocin‐induced experimental hyperglycemia. J Diabetes 2011;3(1):29-37.
[Crossref] [Google Scholar] [PubMed]
- Hwang SL, Kim HN, Jung HH, Kim JE, Choi DK, Hur JM, et al. Beneficial effects of β-sitosterol on glucose and lipid metabolism in L6 myotube cells are mediated by AMP-activated protein kinase. Biochem Biophys Res Commun 2008;377(4):1253-8.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Huang M, Yang J, Ma X, Zheng S, Deng S, et al. Anti-diabetic activity of stigmasterol from soybean oil by targeting the GLUT4 glucose transporter. Food Nutr Res 2017;61(1):1364117.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Hu J, Zhu J, Li Q, Fang L. Machine learning-based metabolism-related genes signature and immune infiltration landscape in diabetic nephropathy. Front Endocrinol 2022;13:1026938.
[Crossref] [Google Scholar] [PubMed]
- Das RR, Rahman MA, Al-Araby SQ, Islam MS, Rashid MM, Babteen NA, et al. The antioxidative role of natural compounds from a green coconut mesocarp undeniably contributes to control diabetic complications as evidenced by the associated genes and biochemical indexes. Oxid Med Cell Longev 2021;2021:1-22.
[Crossref] [Google Scholar] [PubMed]
- Ming J, Sana SR, Deng X. Identification of copper-related biomarkers and potential molecule mechanism in diabetic nephropathy. Front Endocrinol 2022;13:978601.
[Crossref] [Google Scholar] [PubMed]
- Suzuki T, Ichii O, Nakamura T, Horino T, E lewa YH, Kon Y. Immune-associated renal disease found in caspase 3-deficient mice. Cell Tissue Res 2020;379:323-35.
[Crossref] [Google Scholar] [PubMed]
- Gallagher CJ, Keene KL, Mychaleckyj JC, Langefeld CD, Hirschhorn JN, Henderson BE, et al. Investigation of the estrogen receptor-α gene with type 2 diabetes and/or nephropathy in African-American and European-American populations. Diabetes 2007;56(3):675-84.
[Crossref] [Google Scholar] [PubMed]
- Mirfazeli ES, Marashi SA, Kalantari S. In silico prediction of specific pathways that regulate mesangial cell proliferation in IgA nephropathy. Med Hypotheses 2016;97:38-45.
[Crossref] [Google Scholar] [PubMed]
 Genes with no significant difference in expression
Genes with no significant difference in expression