- *Corresponding Author:
- Bhaskar Bagchi
Madhabnagar Badalmoni High School, Malda, West Bengal 732103,
India
E-mail: bhaskarbagchi@gmail.com
| Date of Received | 27 November 2021 |
| Date of Revision | 26 August 2022 |
| Date of Acceptance | 13 January 2023 |
| Indian J Pharm Sci 2023;85(1):44-52 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Severe acute respiratory syndrome coronavirus 2 poses a huge threat to humans. The β genus Severe acute respiratory syndrome coronavirus 2 has four structural proteins: Spike, envelope, membrane and nucleocapsid protein. Among them, spike plays an important role in the host cell attachment and fusion. The S1 subunit of the spike is responsible for bonding to the host and hence is the most crucial target for finding appropriate inhibitors. Molecular docking calculations at the receptor binding domain of S1 subunit have been carried out with twenty celastroid triterpenoids and found that they have very good binding energies. The interactions of these compounds with important amino acid residues were also thoroughly investigated. The docking poses were further validated by the farPPI web server.
Keywords
Severe acute respiratory syndrome coronavirus 2, celastroid triterpenoids, molecular docking, molecular mechanics with generalised born/Poisson-Boltzmann surface area
The recent pandemic of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) have caused human fatal pneumonia spread in a number of countries, resulting in more than 267 667 000 infections and ~5 291 200 deaths as of December 10, 2021. This disease spread rapidly from Wuhan, Hubei Province of China and World Health Organization (WHO) named the disease as Coronavirus Disease (COVID-19). Common symptoms of patients infected with coronavirus such as fever, cough, dyspnea and shortness of breath followed by severe acute respiratory syndrome, kidney failure and even death. Currently, there is no specific treatment or medicine for COVID-19 disease[1].
Coronaviruses are divided into four genera (α, β, γ and δ) and SARS-CoV-2 is a member of the β genus. Coronaviruses are usually constructed by four structural proteins, namely, Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) proteins[2]. During the virus infection, this S protein attaches to the surface of the host cell with the help of cellular receptor Angiotensin-Converting Enzyme 2 (ACE2) which is present in human cells[3-5]. S protein is cleaved into two functional subunits S1 and S2 where S1 is responsible for binding to the host cell and S2 contains the fusion machinery. The S1 subunits of different corona viruses use specific domains. The SARS related corona viruses use the SB domain by interacting ACE2 to enter the target cells[6]. Thus, any molecule that interacts with the spike protein at this time may inhibits viral infections[7]. Very recently, the Receptor Binding Domain (RBD) of SARS-CoV-2 have identified by Tai et al.[8]. They recognized residues 331 to 524 of the S glycoprotein bound strongly to the ACE2 receptors. Hence this RBD side is very important to explore potent and selective inhibitors.
Molecular docking is one of the powerful theoretical tools to identify potent inhibitors. Many theoretical and experimental studies suggested that when ligands are docked, the combination always diminishes the activity and never enhances[9-15]. In this study, we have performed docking with twenty celastroid triterpenoids[16]. These compounds are generally very toxic in nature. Celastroids are oxygenated and unsaturated 14-nor-D:A-friedo-oleanane triterpenoids (fig. 1), have shown antimicrobial and antineoplastic activities[16]. Finally, the farPPI web server (farppi) was used to validate the docking poses using the Molecular Mechanics energies combined with the Poisson Boltzmann or Generalized Born and Surface Area (MM/PB(GB)SA) methods.
Materials and Methods
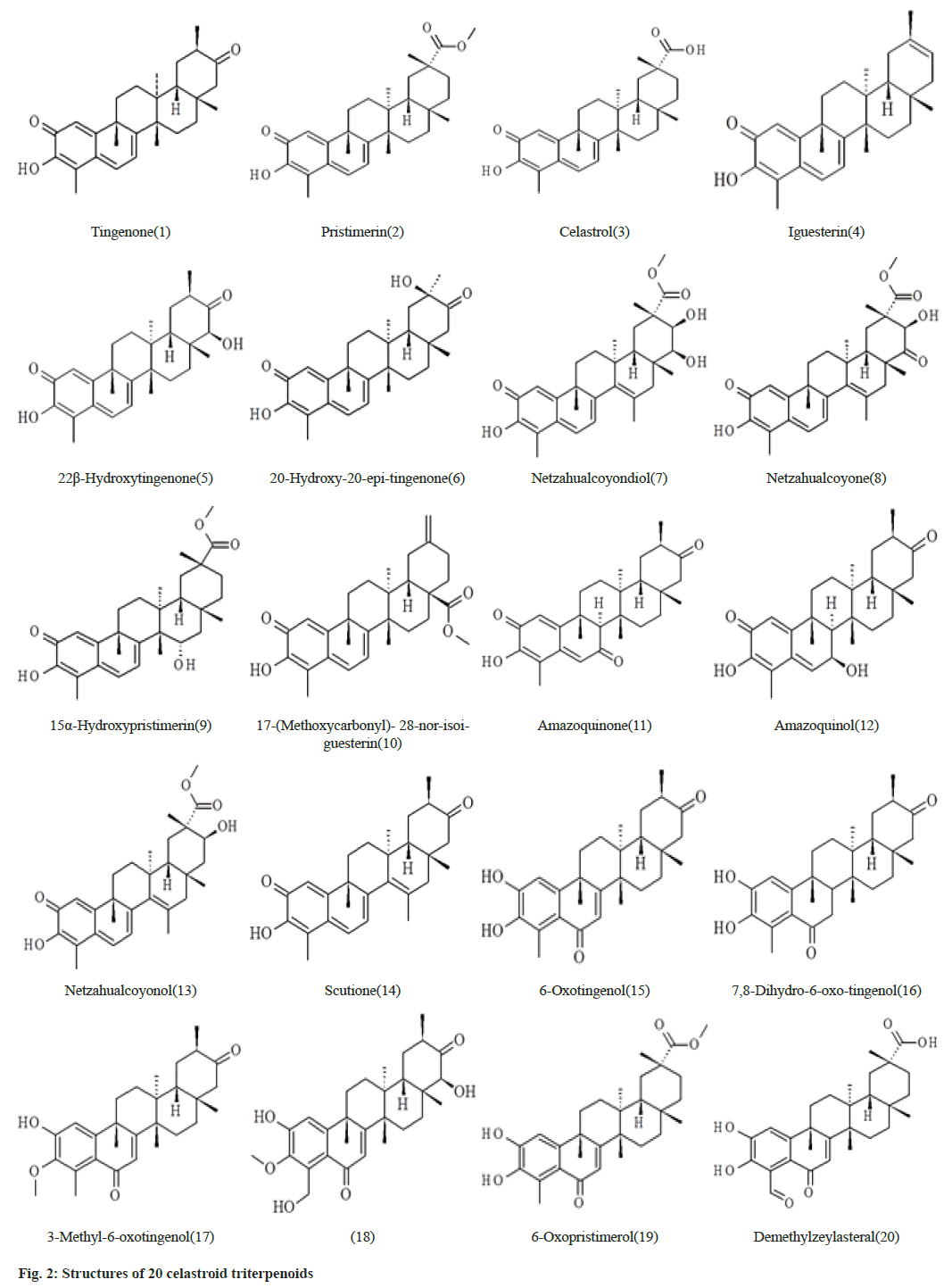
The data set which is selected from the literature contains 20 celastroid triterpenoids[16]. The initial structures of 20 compounds used in this study were constructed by ChemSketch (www.acdlabs.com) as shown in fig. 2. The coordinates of SARS-CoV-2 spike glycoprotein (6VXX.pdb)[6] were obtained from RCSB protein data bank (www.rcsb.org). The 20 celastroid triterpenoids were docked into the RBD site of the enzyme by using docking program Autodock 4.2[17- 19]. Initially the structures of the 20 compounds were optimized with the AM1 method and the hydrogen atoms were added to the enzyme. The Lamarckian Genetic Algorithm (LGA) was applied to search for the best conformers. A grid map with 120×120×120 points and 0.375 Å spacing was used in Autogrid program to evaluate the binding energies between the inhibitors and SARS-CoV-2 spike glycoprotein. For each compound, ten docking poses were saved and ranked by binding energy. The best docking poses of 20 celastroid triterpenoids were chosen for analyzing the type of interaction. The binding site was analyzed with Molegro molecular viewer software and Chimera[20,21]. Fast Amber Rescoring server provides seven MM/ PB(GB)SA methods and among them we have used PB3 methods with the combination of GAFF2 (for ligand) and ff14SB force field (for receptor) to calculate binding free energy of protein-ligand complex[22].
Results and Discussion
The binding energies of 20 celastroid triterpenoids range from -10.01 to -6.69 kcal/mol and their MM/ PB(GB)SA score ranges from -16.31 to -6.01 kcal/mol. The SA and SB domain of S1 subunit are represented in fig. 3. Interaction of the 20 compounds with different amino acid residues of S glycoprotein are presented in Table 1.
| Compound number | Interaction with different amino acid residues of S glycoprotein |
|---|---|
| 1 | Trp353, Asp420, Tyr421, Asn422, Lys424, Lys462, Glu465, Asp467, Pro491 |
| 2 | Glu340, Ala344, Arg346, Phe347, Ala348, Ser349, Asn354, Ser399, Asn450 |
| 3 | Leu335, Cys336, Phe338, Gly339, Phe342, Asn343, Val362, Asp364, Val367, Leu368, Ser371, Ser373, Phe374 |
| 4 | Phe338, Gly339, Phe342, Asn343, Val367,Leu368, Ser371, Ser373, Trp436 |
| 5 | Tyr369, Ser371, Ala372, Phe374, Phe377, Lys378, Cys379, Pro384 |
| 6 | Arg355, Tyr396, Asp428, Phe464, Ser414, Thr430, Phe515, Glu516, Leu517 |
| 7 | Glu340, Ala344, Arg346, Phe347, Ala348, Ser349, Asn354, Ser399, Asn450 |
| 8 | Phe329, Pro330, Asn331, Ile332, Leu335, Val362, Asp364, Gly524, Lys528, Pro527, Lys529, Ser530, Gln580 |
| 9 | Arg355, Tyr396, Asp428, Phe429, Thr430, Phe464, Ser514, Phe515, Glu516 |
| 10 | Phe338, Gly339, Phe342, Asn343, Asp364, Val367, Leu368, Ser371, Ser373 |
| 11 | Trp353, Lys362, Glu365, Asp420, Tyr421, Asn422, Lys424,Asp467, Pro491 |
| 12 | Arg355, Asp428, Thr430, Phe464, Ser414, Phe515, Leu517 |
| 13 | Leu335, Cys336, Gly339, Asn343, Val362, Asp364, Val367, Ser371, Ser373, Phe374 |
| 14 | Asn334, Leu335, Pro337, Arg357, Ile358, Ser359, Asn360, Cys361 |
| 15 | Trp353, Asp420, Tyr421, Asn422, Lys424, Lys462, Glu465, Asp467, Pro491 |
| 16 | Trp353, Asp420, Tyr421, Asn422, Lys424, Lys462, Glu465, Arg466, Asp467, Pro491 |
| 17 | Arg355, Asp428, Thr430, Phe464, Ser514, Phe515, Leu517 |
| 18 | Cys336, Gly339, Phe338, Phe342, Asn343, Val367, Leu368, Ser371, Ser373, Phe374, Trp436 |
| 19 | Cys336, Phe338, Gly339, Glu340, Asn343, Asp364, Val362, Val367 |
| 20 | Tyr369, Ser371, Phe374, Thr376, Phe377, Lys378, Cys379, Ser383, Pro384 |
Table 1: Interaction of the 20 Different Celastroid Triterpenoids with Amino Acid Residues of S Glycoprotein
Compound 1 forms three hydrogen bonds. The C=O group at C-2 forms hydrogen bond with Asp467. The –OH group at C-3 forms two hydrogen bonds with Asn422 and Glu465. In compound 2 the –OH group at C-3 forms two hydrogen bonds with Ser349 and Asn450. The –COOMe group at C-20 also form another hydrogen bond with Asn354. Compound 3 forms two hydrogen bonds by Ser371 and Ser373 with –OH group at C-3 and two hydrogen bonds by Asp364 with –COOH group at C-20. Compound 4 forms no hydrogen bond with amino acid residues. Compound 5 forms five hydrogen bonds. Lys378 forms two hydrogen bonds with C=O group at C-2 and –OH group at C-3 simultaneously. The –OH group at C-3 also makes another hydrogen bond with Cys379. The –OH group at C-22 makes two hydrogen bonds with Ala372 and Phe374. Compound 6 forms two hydrogen bonds of C=O group at C-21 and –OH group at C-20 with Arg355 and Glu516 respectively.
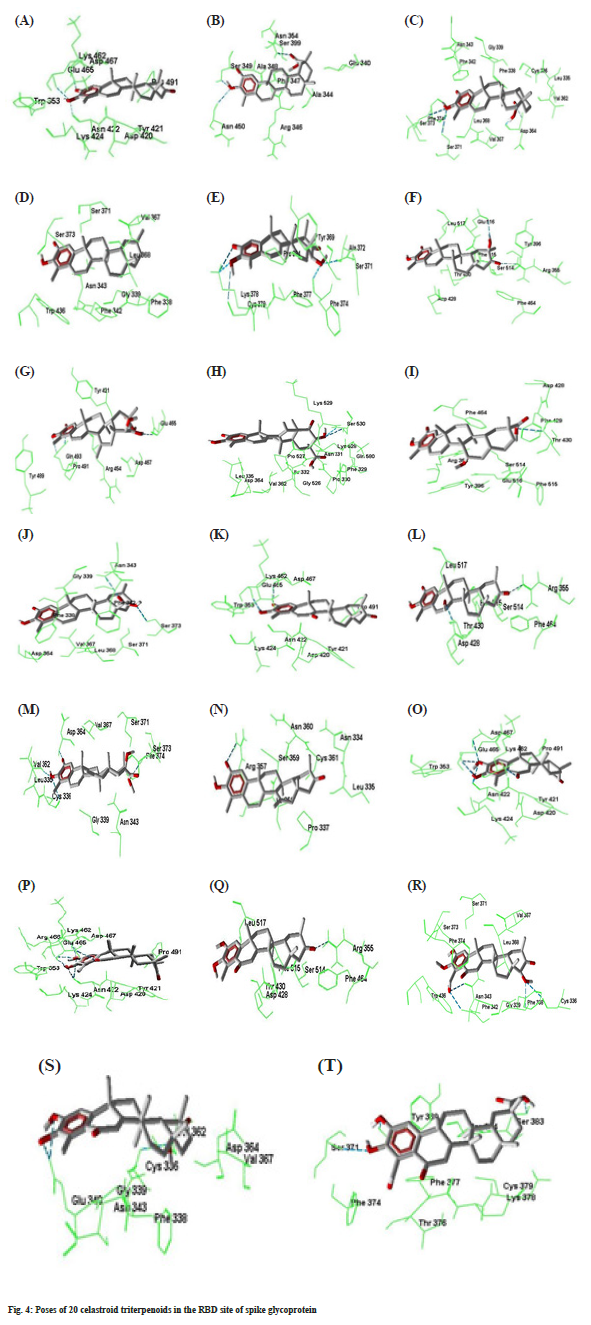
Compound 7 forms three hydrogen bonds. The C=O group at C-2 and –OH group at C-3 forms two hydrogen bonds simultaneously with Gln493. Again – OH group at C-21 makes hydrogen bond with Glu465. In compound 8, -OH group at C-21 forms two hydrogen bonds with Ser530 which is outside of the RBD site. Compounds number 9 forms two hydrogen bonds with Arg355 and Thr430. The -COOMe group at C-17 of compound 10 forms two hydrogen bonds with Asn343 and Ser373. Compounds 11 forms two hydrogen bonds by Asp467 and Glu465 with C=O at C-2 and –OH at C-3 respectively. Compound number 12 makes two hydrogen bonds by Asp428 and Arg355 with the –OH at C7 and C=O groups at C21 respectively. Compounds number 13 creates four hydrogen bonds with Cys336, Val362, Asp364 and Ser373 at the different parts of the molecule. Compound 14 forms one hydrogen bond by Arg357 with C=O group at C-2. Both compounds number 15 and 16 generates six hydrogen bonds. The –OH groups at C-2 and C-3 forms five hydrogen bonds with Asn422, Glu465 and Asp467. The C=O group at C-6 also makes another hydrogen bond with Lys462. Compounds 17 forms one hydrogen bond. The –OH groups at C-23 and C-22 forms four hydrogen bonds with Phe342, Asn343, Phe338 and Cys336 respectively in compound number 18. Both compound number 19 and 20 forms three hydrogen bonds. The best docking conformation of 20 celastroid triterpenoids along with the important amino acid residues of RBD of spike glycoprotein are presented in fig. 4. The binding energies of poses of 20 ligands along with hydrogen bond interacting amino acids along with MM/PB(GB) SA scores are presented in Table 2.
| Docked Pose | Hydrogen bond residues | Number of hydrogen bond | Free energy of binding kcal/mol | MM/PB(GB)SA score kcal/mol |
|---|---|---|---|---|
| A | Glu465, Asp467, Asn422 | 3 | -8.8 | -13.61 |
| B | Ser349, Asn354, Asn450 | 3 | -7.9 | -6.01 |
| C | Asp364, Ser371, Ser373 | 4 | -7.85 | -13.99 |
| D | - | - | -8.06 | -10.12 |
| E | Lys378, Cys379, Ala372, Phe374 | 5 | -8.98 | -6.61 |
| F | Arg355, Glu516 | 2 | -8.18 | -13.89 |
| G | Glu465, Gln493 | 3 | -7.52 | -7.86 |
| H | Ser530 | 2 | -8.11 | -13.46 |
| I | Arg355, Thr430 | 2 | -7.54 | -7.89 |
| J | Asn343, Ser373 | 2 | -8.38 | -6.45 |
| K | Glu465, Asp467 | 2 | -8.72 | -8.29 |
| L | Arg355, Asp428 | 2 | -8.11 | -14.13 |
| M | Cys336, Val362, Asp364, Ser373 | 4 | -6.69 | -10.09 |
| N | Arg357 | 1 | -7.98 | -8.15 |
| O | Asn422, Glu465, Lys462, Asp467 | 6 | -9.45 | -8.85 |
| P | Asn422, Glu465, Lys462, Asp467 | 6 | -10.01 | -16.31 |
| Q | Arg355 | 1 | -8.18 | -10.98 |
| R | Cys336, Phe338, Phe342, Asn343 | 4 | -7.74 | -9.2 |
| S | Cys336, Glu340 | 3 | -7.96 | -8.46 |
| T | Tyr369, Ser371, Ser383 | 3 | -7.86 | -15.84 |
Table 2: Free Energy of Binding and MM/PB(GB)SA Score of Different Compounds Along With Hydrogen Bond Interacting Amino Acids
Though intermolecular hydrogen bonds may play as an actor determining the 3D position of the ligand in the RBD site, there are some other energy also contribute to the free energy induced by bonding. In the output dlg file of AutoDock’s calculation, van der Walls interaction and electrostatic interaction energies are presented separately. Although the electrostatic energy variation from one another is too small, but the van deer Waals energy contributes some part to the total energy.
The quinone-methides (compounds 1-14) are electrophilic and can form covalent bonds with appropriate nucleophiles. So we should closely examine the proximity of nucleophilic amino acids to the electrophilic sites of the quinone-methide triterpenoids (C6). It can be seen that C6 is surrounded with Asp364, Asp428, Asp420, Arg346, Arg355, Arg357, Cys336, Lys374, Tyr369, Tyr396 and Tyr421, which are important nucleophilic amino acids for the interaction of the C6 with the protein.
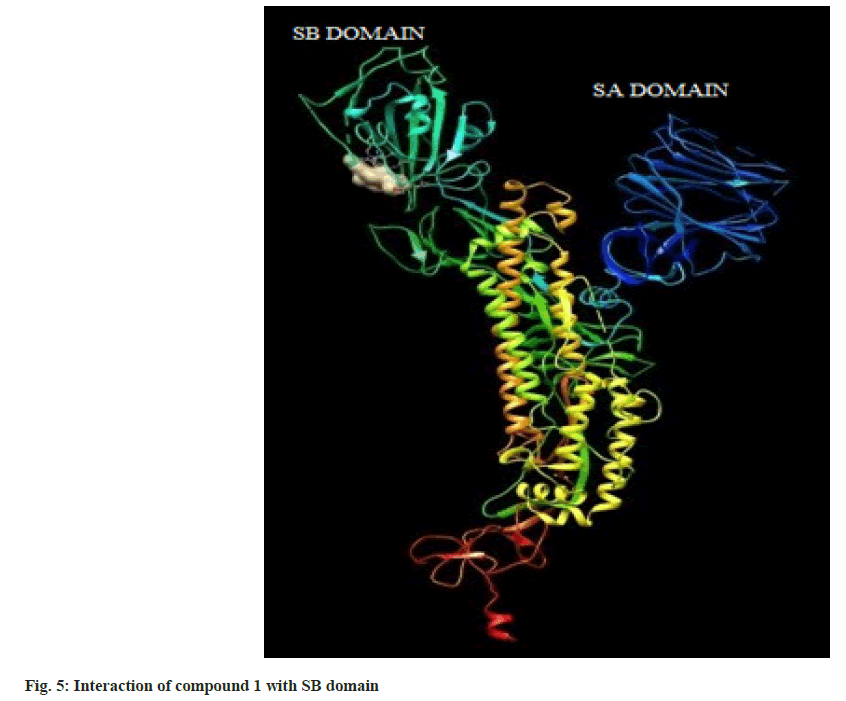
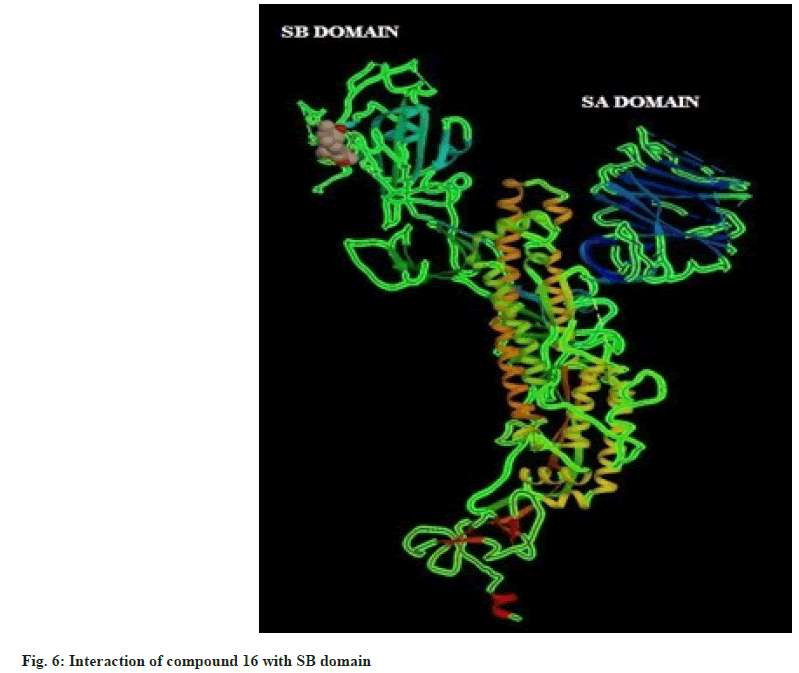
Hence on the basis of the binding free energy and MM/ PB(GB)SA score, two compounds, tingenone and 7,8-dihydro-6-oxo-tingenol i.e., ligand 1 and 16 may be promising candidates for further modification and structure-based drug design. The interactions of ligand 1 and 16 with SB domain of S1 unit are presented in fig. 5 and fig. 6 respectively.
Conflict of interest:
The authors declared no conflicts of interest.
References
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497-506.
[Crossref] [Google Scholar] [PubMed]
- Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol 2003;77(16):8801-11.
[Crossref] [Google Scholar] [PubMed]
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503(7477):535-8.
[Crossref] [Google Scholar] [PubMed]
- Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 2006;350(1):15-25.
[Crossref] [Google Scholar] [PubMed]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426(6965):450-4.
[Crossref] [Google Scholar] [PubMed]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):281-92.
[Crossref] [Google Scholar] [PubMed]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 2020;10(5):766-88.
[Crossref] [Google Scholar] [PubMed]
- Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020;17(6):613-20.
[Crossref] [Google Scholar] [PubMed]
- Day CJ, Bailly B, Guillon P, Dirr L, Jen FE, Spillings BL, et al. Multidisciplinary approaches identify compounds that bind to human ACE2 or SARS-CoV-2 spike protein as candidates to block SARS-CoV-2-ACE2 receptor interactions. mBio 2021;12(2):e03681-20.
[Crossref] [Google Scholar] [PubMed]
- Ramezani Z, Dayer MR, Noorizadeh S, Thompson M. Deactivation of SARS-CoV-2 via shielding of spike glycoprotein using carbon quantum dots: Bioinformatic perspective. COVID 2021;1(1):120-9.
- Sinha SK, Shakya A, Prasad SK, Singh S, Gurav NS, Prasad RS, et al. An in silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn 2021;39(9):3244-55.
[Crossref] [Google Scholar] [PubMed]
- Mengist HM, Dilnessa T, Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front Chem 2021;9:622898.
[Crossref] [Google Scholar] [PubMed]
- Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol Sin 2020;41(9):1141-9.
- Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep 2020;10(1):17699.
[Crossref] [Google Scholar] [PubMed]
- Sayed AM, Khattab AR, AboulMagd AM, Hassan HM, Rateb ME, Zaid H, et al. Nature as a treasure trove of potential anti-SARS-CoV drug leads: A structural/mechanistic rationale. RSC Adv 2020;10(34):19790-802.
- Setzer WN. A theoretical investigation of cytotoxic activity of celastroid triterpenoids. J Mol Model 2009;15:197-201.
[Crossref] [Google Scholar] [PubMed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30(16):2785-91.
[Crossref] [Google Scholar] [PubMed]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with chargeâ€based desolvation. J Comput Chem 2007;28(6):1145-52.
[Crossref] [Google Scholar] [PubMed]
- Huey R, Goodsell DS, Morris GM, Olson AJ. Grid-based hydrogen bond potentials with improved directionality. Lett Drug Des Discov 2004;1(2):178-83.
- Thomsen R, Christensen MH. MolDock: A new technique for high-accuracy molecular docking. J Med Chem 2006;49(11):3315-21.
[Crossref] [Google Scholar] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605-12.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Wang X, Li Y, Lei T, Wang E, Li D, et al. farPPI: A webserver for accurate prediction of protein-ligand binding structures for small-molecule PPI inhibitors by MM/PB (GB) SA methods. Bioinformatics 2019;35(10):1777-9.
[Crossref] [Google Scholar] [PubMed]