- *Corresponding Author:
- S. Sharma
Department of Pharmaceutical Sciences, Guru Nanak Dev University, Amritsar-143 005, India
E-mail: ss.gq2009@gmail.com
| Date of Submission | 09 August 2016 |
| Date of Revision | 19 February 2017 |
| Date of Acceptance | 01 August 2017 |
| Indian J Pharm Sci 2017;79(5):801-812 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A library of 3,4,6,7-tetrahydro-3,3,6,6-tetramethyl-9,10-diphenylacridine-1,8-dione derivatives were synthesized via microwave-assisted three-component reaction employing silica supported sulphuric acid under solvent free conditions and tested for in vitro cholesterol esterase inhibitory activity. All synthesized compounds exhibited moderate to good inhibition of cholesterol esterase enzyme. Among all the synthesized acridinedione derivatives, compound HSM-1a showed the most promising activity with IC50 value of 1.53 µM against cholesterol esterase enzyme. Enzyme kinetic studies carried out for HSM-1a, revealed its mixed-type inhibition approach. Molecular protein–ligand docking studies were also performed to figure out the key binding interactions of HSM-1a with the amino acid residues of the enzyme’s active site. Furthermore, acquiescence of some potent compounds to the Lipinski rule was also determined.
Keywords
Cholesterol esterase, acridinedione, Lipinski’s rule, docking, enzyme kinetics
Cholesterol is a vital component of cell membrane and possesses many physiological functions. The greatest percentage of cholesterol is used in cytoplasm for bile acid synthesis [1]. Hypercholesterolemia produced either by cholesterol feeding or by cholesterol-free, purified diets (“endogenous” hypercholesterolemia) results in the accumulation of cholesterol in adipose tissue. The reduction of cholesterol level in plasma, particularly in low density lipoprotein (LDL) lowers the risk of cardiovascular events [2,3]. There are various enzymes present in our body having contributory effect in diverse disease conditions [4-8]. Likewise, pancreatic cholesterol esterase (CEase) is an enzyme having prominent role in increasing the risk of heart disease. CEase is the member of α/β hydrolase family of proteins which catalyse the hydrolysis of cholesterol ester into free cholesterol in the lumen of small intestine [9,10]. It is also thought that the transport of cholesterol micelles to enterocytes is performed by this enzyme [11]. As the combined role of CEase in the absorption and transport of cholesterol, its inhibition is important by the development of novel moieties, which helps in treating hypercholesterolemia and associated diseases such as coronary heart disease [12].
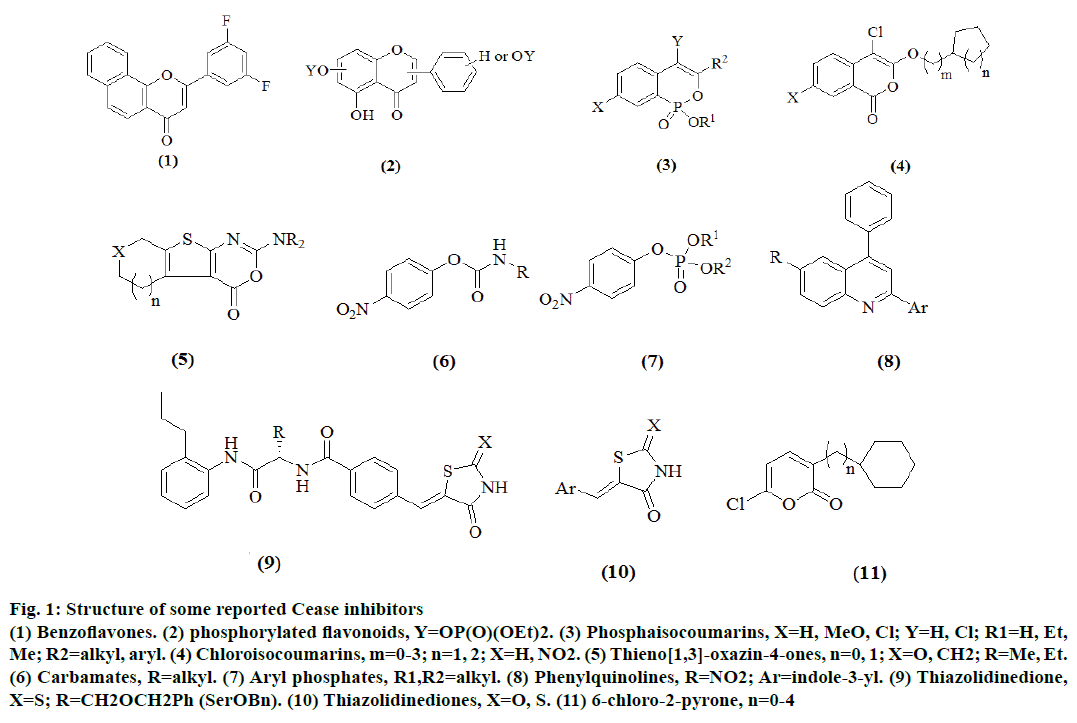
Since the last decade, there have been a number of scaffolds reported, which were biologically active against CEase and several classes of potent CEase inhibitors have been developed [13] thus far, including 6-chloro-2-pyrones [14,15], thieno [1,3]- oxazin-4-ones [16,17], carbamates [18-25], aryl phosphates and phosphonates [26,27], chloroisocoumarins [28], phosphaisocoumarins [29], thiazolidinediones [30] and benzoflavones [31] (Figure 1). However, most of these inhibitors were not highly selective and they could also inhibit other serine hydrolases such as acetylcholinesterase (AChE), butyrylcholinesterase (BuChE), Pseudomonas species lipase (PSL), chymotrpsin (CT) and trypsin [32]. Acridine is one of the prevalent biologically active heterocycle due to its presence in many medicinally important molecules, such as amsacrine (1, cytotoxic and antiviral agent), botiacrine (2, antiparkinsonian drug), clomacran (3, tranquilizer), monometacrine (4, antidepressant) and acridinecarboxamide (DACA, 5 antitumor) as shown in Figure 2 [33].
Figure 1: Structure of some reported Cease inhibitors
(1) Benzoflavones. (2) phosphorylated flavonoids, Y=OP(O)(OEt)2. (3) Phosphaisocoumarins, X=H, MeO, Cl; Y=H, Cl; R1=H, Et,
Me; R2=alkyl, aryl. (4) Chloroisocoumarins, m=0-3; n=1, 2; X=H, NO2. (5) Thieno[1,3]-oxazin-4-ones, n=0, 1; X=O, CH2; R=Me, Et.
(6) Carbamates, R=alkyl. (7) Aryl phosphates, R1,R2=alkyl. (8) Phenylquinolines, R=NO2; Ar=indole-3-yl. (9) Thiazolidinedione,
X=S; R=CH2OCH2Ph (SerOBn). (10) Thiazolidinediones, X=O, S. (11) 6-chloro-2-pyrone, n=0-4
In view of various medicinal attributes of acridines, we report here for the first time, acridinedione derivatives as potent CEase inhibitors. Thus, in the present study, various acridinedione analogues were synthesized via microwave-assisted three-component reaction employing silica supported sulphuric acid under solvent free conditions and evaluated for their inhibitory potential against CEase enzyme using spectrophotometric assay. The catalytic influence of HBF4-SiO2 was also investigated in detail to optimize the reaction conditions. The type of inhibition and the interactions of the most potent inhibitor with CEase enzyme had also been figured out.
Materials and Methods
Most reagents were purchased from Sigma-Aldrich, Loba and CDH, India and used without further purification. All yields refer to isolated products after purification. Products were characterized by comparison with authentic samples and by spectroscopic data (1H and 13C NMR). 1H NMR and 13C NMR spectra were recorded on Avance III HD 500 MHz Bruker Biospin Nuclear Magnetic Resonance Spectrometer. The spectra were measured in CDCl3 and Dimethyl sulfoxide (DMSO)-d6 relative to TMS (0.00 ppm). In 1H NMR chemical shifts were reported in δ values using tetramethylsilane as internal standard with number of protons, multiplicities (s-singlet, d-doublet, t-triplet, q-quartet, m-multiplet, dd-double doublet) and coupling constants (J) in Hz (Hertz) in the solvent indicated. Melting points were determined in open capillaries and were uncorrected.
Procedure for the preparation of SiO2-H2SO4
To a 250 ml dried round bottom flask (RBF), a suspension of silica gel (50 g, 230-400 mesh) in acetone (100 ml) was added. To this suspension, a solution of concentrated sulphuric acid in acetone (1 ml in 15 ml acetone) was added drop wise. The reaction mixture was stirred at room temperature for 8 h. After 8 h, the solvent was removed under reduced pressure and the yellowish brown powder obtained was stored in desiccator for longer period of time.
General procedure for the synthesis of acridinedione derivatives
To a 50 ml conical flask, dimedone (1 mmol), substituted benzaldehyde (1 mmol), substituted aniline (1 mmol) were added. In the mixture 1 mol% of silicated sulphuric acid was added as a catalyst. The reaction mixture was irradiated Biotage Microwave Synthesizer operating at 150° with the microwave power maximum level of 400 W for 10 min. After the completion of reaction (confirmed by TLC), the reaction mixture was dissolved in ethanol and then filtered to remove silica. The filtrate was dried under reduced pressure to get the crude product.
Characterization data
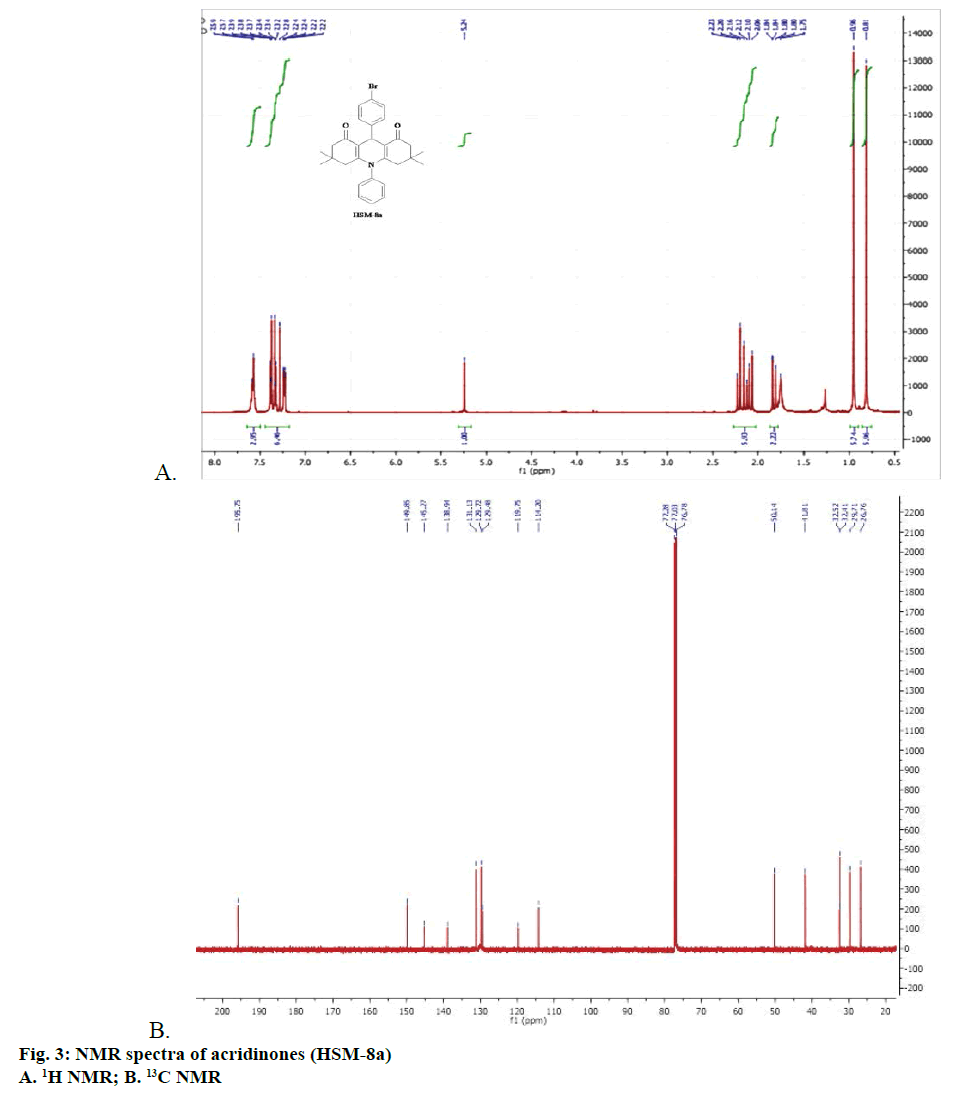
In the present investigation, 26 acridinedione derivatives viz.,3,4,6,7-tetrahydro-3,3,6,6-tetramethyl-9,10- diphenylacridine-1,8-diones have been synthesized in the presence of catalytic amount of silica-H2SO4. All the synthesized compounds (HSM1a-HSM13a and HSM1b-HSM13b) with diversification in chemical structures were characterized by melting point, 1H NMR, 13C NMR and IR spectroscopy.
3 , 4 , 6 , 7 - t e t r a h y d r o - 3 , 3 , 6 , 6 - t e t r a m e t h y l - 9 , 10-diphenylacridine-1,8(2H,5H,9H,10H)-dione (HSM-1a): Yellowish solid; yield 85%; MP: 182-184°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.18-7.20 (5H, m), 7.04-7.14 (5H, m), 5.46 (1H, s), 2.56-2.67 (4H, m), 1.88 (4H, m), 1.20 (6H, s), 0.99 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 154.29, 141.89, 134.12, 131.84, 129.23, 128.57, 118.72, 116.20, 113.29, 111.35, 52.31, 41.32, 40.13, 38.25, 37.73. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203.
9-(4-fluorophenyl)-3,4,6,7-tetrahydro-3,3,6,6- tetramethyl-10-phenylacridine-1,8-(2H,5H,9H,10H)- dione (HSM-2a): Brown powder; yield 77%; MP: 180-182°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.08 (2H, d, J=8.0 Hz), 7.01-7.05 (2H, m), 6.89 (2H, d, J=8.0 Hz), 6.63 (1H, m), 6.46 (2H, m), 5.43 (1H, s), 2.87-2.90 (4H, m), 1.89-1.97 (4H, m), 1.20 (6H, s), 1.00 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 157.29, 152.87, 141.89, 132.12, 130.84, 129.23, 128.57, 124.32, 117.72, 111.32, 51.31, 40.32, 32.25, 27.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1398, 1478, 1221, 1203.
9-(2-chlorophenyl)-3,4,6,7-tetrahydro-3,3,6,6- tetramethyl-10-phenylacridine-1,8(2H,5H,9H,10H)- dione (HSM-3a): Yellow solid; yield 82%; MP: 184-186°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.15 (1H, m), 7.01-7.05 (5H, m), 6.66 (1H, m), 6.56 (2H, m), 5.59 (1H, s), 2.35 (4H, m), 1.69 (4H, m), 1.25 (6H, s), 1.10 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.93, 160.69, 153.67, 143.89, 133.12, 127.84, 126.23, 121.57, 120.30, 118.24, 116.02, 114.5, 113.75, 52.31, 43.32, 41.33, 31.35, 27.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 748.
9-(3-chlorophenyl)-3,4,6,7-tetrahydro-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H,9 H,10H)-dione (HSM-4a): Brown powder; yield 84%; MP: 168-170°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.07 (3H, m), 7.01 (2H, m), 6.94 (1H, m), 6.60 (1H, m), 6.46 (2H, m), 4.79 (1H, s), 2.35-2.40 (4H, m), 1.69 (4H, m), 1.20 (6H, s), 1.00 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.93, 158.29, 153.87, 141.89, 130.12, 129.84, 128.23, 126.57, 118.54, 116.32, 114.15, 113.75, 51.31, 42.32, 41.33, 31.65, 26.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 743.
9 - ( 4 - c h l o r o p h e n y l ) - 3 , 4 , 6 , 7 - t etrahydro-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H,9H,10H)– dione (HSM-5a): Reddish brown solid; yield 80%; MP: 172-174°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.19 (2H, d, J=8.5 Hz), 7.16 (2H, m), 7.01 (2H, d, J=8.5 Hz), 6.46- 6.62 (3H, m), 4.43 (1H, s), 2.86 (4H, s), 1.88 (12H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.91, 153.67, 131.32, 130.64, 129.63, 128.87, 118.92, 116.36, 51.51, 40.92, 40.53, 32.05, 27.46. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 750.
9-(2-bromophenyl)-3,4,6,7-tetrahydro-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H,9 H,10H)-dione (HSM-6a): Yellow powder; yield 80%; MP: 160-162°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.31 (1H, m), 7.10-7.15 (3H, m), 6.90 (2H, m), 6.60-6.70 (1H, m), 6.40-6.50 (2H, m), 5.56 (1H, s), 2.46-2.57 (4H, m), 1.99 (4H, m), 1.49 (6H, s), 1.19 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 158.29, 153.87, 143.89, 131.12, 130.84, 120.30, 118.32, 114.25, 112.75, 55.92, 51.31, 42.32, 41.33, 31.65, 26.89. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 598.
9-(3-bromophenyl)-3,4,6,7-tetrahydro-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H,9 H,10H)-dione (HSM-7a): Brown powder; yield 67%; MP: 182-184°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.24 (2H, m), 7.01-7.04 (4H, m), 6.60 (1H, m), 6.46 (2H, m), 5.56 (1H, s), 2.56-2.57 (4H, m), 1.59 (4H, m), 1.19 (6H, s), 1.02 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 156.29, 133.12, 131.28, 130.84, 129.23, 128.57, 120.30, 118.32, 116.23, 113.75, 111.75, 52.31, 42.32, 41.13, 31.25, 25.83. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 600.
9-(4-bromophenyl)-3,4,6,7-tetrahydro-3,3,6," 6-tetramethyl-10-phenylacridine-1,8(2H,5H, 9H,10H)-dione (HSM-8a): Pale yellow powder; yield 83%; MP: 190-192°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.56-7.59 (3H, m), 7.38 (2H, d, J=10 Hz), 7.33 (2H, d, J=10 Hz), 7.22-7.24 (2H, d, J=10 Hz), 5.24 (1H, s), 2.06-2.23 (6H, m), 1.80-1.84 (2H, m), 0.96 (6H, m) , 0.81 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 195.75, 149.85, 145.27, 138.94, 131.13, 129.72, 129.48, 119.75, 114.20, 50.14, 41.81, 32.52, 32.41, 29.71, 26.76. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 597.
3,4,6,7-tetrahydro-9-(2-methoxyphenyl)-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H, 9H,10H)-dione (HSM-9a): Pale yellow powder; yield 84%; MP: 191-193°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.31 (1H, m), 7.10-7.15 (3H, m), 6.90 (2H, m), 6.60-6.70 (1H, m), 6.40-6.50 (2H, m), 5.56 (1H, s), 2.46-2.57 (4H, m), 1.99 (4H, m), 1.49 (6H, s), 1.19 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 158.29, 153.87, 143.89, 131.12, 130.84, 120.30, 118.32, 114.25, 112.75, 55.92, 51.31, 42.32, 41.33, 31.65, 26.89. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1101.
3,4,6,7-tetrahydro-9-(3-methoxyphenyl)-3,3,6, 6-tetramethyl-10-phenylacridine-1,8-(2H,5H, 9H,10H)-dione (HSM-10a): Yellowish solid; yield 85%; MP: 168-170°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.07 (3H, m), 7.01 (2H, m), 6.94 (1H, m), 6.60 (1H, m), 6.46 (2H, m), 5.79 (1H, s), 3.35 (3H, s), 2.35-2.40 (4H, m), 1.69 (4H, m), 1.20 (6H, s), 1.00 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.93, 158.29, 153.87, 141.89, 130.12, 129.84, 128.23, 126.57, 118.54, 116.32, 113.75, 57.57, 51.31, 42.32, 41.33, 31.65, 26.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1098.
3,4,6,7-tetrahydro-9-(4-methoxyphenyl)-3,3,6, 6 - t e t r a m e t h y l - 1 0 - p h e n y l a n t h r a c e n e - 1 , 8(2H,5H,9H,10H)-dione (HSM-11a): Yellowish powder; yield 77%; MP: 190-192°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.01 (2H, m), 6.96 (2H, d, J=8.5 Hz), 6.69 (2H, d, J=8.5 Hz), 6.46-6.50 (3H, m), 4.47 (1H, s), 3.35 (3H, s), 2.84 (4H, s), 1.88 (4H, s), 0.89 (6H, s), 0.85 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.93, 157.79, 152.67, 141.81, 131.12, 130.24, 129.63, 128.77, 118.72, 116.26, 114.29, 55.93, 51.51, 40.82, 40.43, 32.65, 27.43. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100.
3,4,6,7-tetrahydro-9-(3,4-dimethoxyphenyl)-3,3,6, 6-tetramethyl-10-phenylacridine-1,8(2H,5H,9H,10H)- dione (HSM-12a): Yellowish powder; yield 87%; MP: 170-174°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.01-7.05 (2H, m), 6.60 (2H, d, J=9.0 Hz), 6.55-6.57 (2H, m), 6.53 (2H, d, J=9.0 Hz), 5.46 (1H, s), 3.5 (6H, s), 2.56-2.67 (4H, m), 1.88 (4H, m), 1.20 (6H, s), 1.06 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 157.29, 152.87, 149.89, 146.85, 132.12, 130.84, 129.23, 128.57, 113.29, 111.35, 56.65, 51.31, 40.32, 40.13, 32.25, 27.73. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1103.
3,4,6,7-tetrahydro-9-(3,4,5-trimethoxyphenyl)- 3,3,6,6- tet ramethyl -10 -phenylacr idine- 1 , 8-(2H,5H,9H,10H)-dione (HSM-13a): Reddish brown powder; yield 72%; MP: 158-160°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.01-7.05 (2H, m), 6.60 (1H, m), 6.46 (2H, m), 6.08-6.10 (2H, m), 4.79 (1H, s), 3.65 (3H, s), 3.58 (3H, s), 3.50 (3H, s), 2.46 (4H, s), 1.90 (4H, m), 1.29 (6H, s) , 1.10 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 197.63, 157.29, 153.87, 140.89, 131.12, 130.84, 129.23, 128.57, 120.30, 118.32, 111.75, 52.31, 42.32, 41.13, 31.25, 28.73. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1101.
3,4,6,7-tetrahydro-10-(4-methoxyphenyl)-3,3,6, 6-tetramethyl-9-phenylacridine-1,8(2H,5H,9 H,10H)- dione (HSM-1b): black powder; yield 80%; MP: 172-174°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.06 -7.14 (5H, m), 6.50 (2H, d, J=8.0 Hz), 6.38 (2H, d, J=8.0 Hz), 5.40 (1H, s), 3.56 (3H, s), 2.36- 2.47 (4H, m), 1.79 (4H, m), 1.19 (6H, s), 1.10 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 159.29, 143.89, 131.12, 130.84, 129.23, 127.57, 120.01, 124.30, 117.32, 111.75, 56.65, 52.31, 43.32, 41.33, 32.25, 27.73. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100.
9 - ( 4 - f l u o r o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine-1,8 (2H,5H,9H,10H)-dione (HSM-2b): Yellowish powder; yield 79%; MP: 191-193°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.02-7.04 (2H, m), 6.90-7.00 (2H, m), 6.52-6.55 (2H, m), 6.36-6.39 (2H, m), 5.37 (1H, s), 3.85 (3H, s), 2.53-2.64 (2H, m), 2.06-2.48 (2H, m), 1.76-2.03 (4H, m), 1.50 (6H, s), 1.09 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 157.29, 152.87, 141.89, 132.12, 130.84, 129.23, 128.57, 124.32, 117.72, 111.32, 57.65, 51.31, 40.32, 32.25, 27.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1399, 1478, 1221, 1203, 1100.
9 - ( 2 - c h l o r o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine-1, 8(2H,5H,9H,10H)-dione (HSM-3b): Black brown powder; yield 83%; MP: 184-186°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.05-7.15 (4H, m), 6.52 (2H, d, J=8.0 Hz), 6.38 (2H, d, J=8.0 Hz), 6.79-6.92 (2H, m), 5.46 (1H, s), 3.89 (3H, s), 2.55-2.59 (2H, m), 2.2-2.33 (4H, m), 1.75 (6H, s), 1.55 (6H, s); 13C NMR (CDCl3, 100 MHz, δ, TMS=0): 196.80, 158.21, 138.05, 130.14, 129.04, 127.81, 127.44, 120.63, 120.45, 115.23, 114.99, 111.66, 56.47, 51.02, 37.01, 29.27, 20.42. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 749.
9-(3-chlorophenyl)-3,4,6,7-tetrahydro-3,3,6, 6-tetramethyl-10-(4-methoxyphenyl)-1,8(2H,5 H,9H,10H)-dione (HSM-4b): Yellow solid; yield 74%; MP: 174-176°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 6.95-7.05 (4H, m), 6.55-6.56 (2H, m), 6.35 (2H, m), 5.35 (1H, s), 3.88 (3H, s), 2.55-2.67 (4H, m), 1.97 (4H, m), 1.28 (6H, s), 1.04 (6H, s); 13C NMR (CDCl3, 100 MHz, δ, TMS=0): 196.69, 157.86, 160.62, 137.64, 136.76, 128.33, 123.21, 117.03, 114.34, 113.52, 55.61, 51.32, 36.78, 35.74, 27.32. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 748.
9 - ( 4 - c h l o r o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine- 1,8(2H,5H,9H,10H)-dione (HSM-5b): Yellow powder; yield 70%; MP: 182-184°; 1H NMR (CDCl3, 500 MHz, δ, TMS= 0): 6.90-7.0 (4H, m), 6.40-6.50 (4H, m), 4.52 (1H, s), 3.53 (3H, s) 2.84-2.94 (4H, m), 1.97 (4H, m), 1.82 (12H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.73, 155.29, 153.87, 143.89, 131.12, 130.84, 129.23, 128.57, 124.30, 112.75, 56.92, 51.31, 42.22, 41.03, 31.15, 25.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 750.
9 - ( 2 - b r o m o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine-1,8 (2H,5H,9H,10H)-dione (HSM-6b): Yellow solid; yield 80%; MP: 184-186°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.30 (1H, m), 6.90-7.00 (3H, m), 6.45-6.55 (2H, m), 6.30-6.35 (2H, m), 5.26 (1H, s), 3.52 (3H, s), 2.23-2.33 (4H, m), 1.67 (4H, m), 1.33 (6H, s), 1.14 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.43, 160.63, 152.67, 141.89, 134.12, 125.84, 124.23, 123.57, 118.04, 116.07, 57.12, 52.21, 43.30, 41.31, 31.34, 27.43. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 599.
9 - ( 3 - b r o m o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine- 1,8(2H,5H,9H,10H)-dione (HSM-7b): Reddish brown powder; yield 72%; MP: 180-182°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.05-7.15 (4H, m), 6.85-6.95 (1H, m), 6.45-6.55 (1H, m), 6.25-6.30 (1H, m), 4.52 (1H, s), 3.53 (3H, s), 2.87-2.97 (4H, m), 1.96 (4H, m), 1.84 1.19 (6H, s), 1.10 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.63, 155.69, 153.37, 143.87, 134.82, 130.83, 129.83, 123.30, 112.75, 56.92, 52.31, 42.32, 41.33, 31.35, 27.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 598.
9 - ( 4 - b r o m o p h e n y l ) - 3 , 4 , 6 , 7 - t e t r a h y d r o - 1 0 - (4-methoxyphenyl)-3,3,6,6-tetramethylacridine-1, 8(2H,5H,9H,10H)-dione (HSM-8b): Brownish solid; yield 75%; MP: 164-166°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.30 (2H, m), 6.90-7.00 (2H, m), 6.55 (2H, m) , 6.35 (2H, m), 4.36 (1H, s), 3.50 (3H, s), 2.37-2.47 (4H, m), 1.59 (4H, m), 1.12 (6H, s), 1.02 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 197.63, 156.87, 143.89, 132.12, 130.84, 129.23, 128.87, 124.42, 118.32, 116.23, 57.31, 52.31, 41.13, 31.25, 26.73. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 600.
3,4,6,7-tetrahydro-9-(2-methoxyphenyl)-10- (4-methoxyphenyl)-3,3,6,6-tetramethylacridine- 1,8(2H,5H,9H,10H)-dione (HSM-9b): Yellowish brown solid; yield 75%; MP: 186-188°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 6.95 (2H, m), 6.65 (2H, m), 6.50 (2H, m), 6.35 (2H, m), 4.29 (1H, s), 3.52 (3H, s), 3.32 (3H, s), 2.23-2.33 (4H, m), 1.67 (4H, m ), 0.98 (6H, s), 0.89 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 198.43, 160.63, 152.67, 141.89, 131.12, 125.84, 124.23, 114.54, 113.75, 57.12, 52.21, 43.30, 41.31, 31.34, 27.43. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 1094.
3,4,6,7-tetrahydro-9-(3-methoxyphenyl)-10- (4-methoxyphenyl)-3,3,6,6-tetramethylacridine- 1,8(2H,5H,9H,10H)-dione (HSM-10b): Brown powder; yield 79%; MP: 172-174°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 7.03 (1H, m), 6.50-6.60 (5H, m), 6.32-6.35 (1H, m), 5.33 (1H, s), 3.74 (3H, s), 3.71 (3H, s), 2.76-2.86 (4H, m), 1.70-1.80 (4H, m), 1.20 (12H, s); 13C NMR (CDCl3, 125 MHz, δ, TMT=0): 197.63, 156.29, 151.87, 141.89, 129.23, 128.57, 123.32, 117.72, 111.32, 57.25, 51.31, 40.32, 32.25, 27.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1098, 1092.
3,4,6,7-tetrahydro-9,10-bis(4-methoxyphenyl)- 3,3,6,6-tetramethylacridine-1,8(2H,5H,9H,10H)-dione (HSM-11b): Brown black solid; yield 82%; MP: 182- 184°; 1H NMR (CDCl3, 300 MHz, δ, TMS=0): 7.33 (2H, d, J=8.1 Hz), 7.11 (2H, m), 7.02 (2H, m), 6.78 (2H, d, J=8.1 Hz), 5.20 (1H, s), 3.74 (3H, s), 3.71 (3H, s), 2.03-2.16 (4H, m), 1.65-1.87 (4H, m), 0.94 (6H, s), 0.80 (6H, s); 13C NMR (CDCl3, 100 MHz, δ, TMS=0): 194.93, 157.29, 152.87, 142.89, 130.14, 129.87, 128.23, 126.55, 124.34, 113.75, 56.22, 51.31, 42.32, 41.33, 31.65, 26.53. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1100, 1098.
3,4,6,7-tetrahydro-9-(3,4-dimethoxyphenyl)-10- (4-methoxyphenyl)-3,3,6,6-tetramethyl acridine- 1,8(2H,5H,9H,10H)-dione (HSM-12b): Brown solid; yield 73%; MP: 188-190°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 6.50-6.55 (4H, m), 6.35-6.40 (3H, m), 5.417 (1H, s), 2.57-2.74 (4H, m), 3.54 (3H, s), 3.51 (3H, s), 3.48 (3H, s), 2.24-2.37 (4H, m), 1.90-2.11 (4H, m), 0.104 (6H, s), 0.90 (6H, s); 13C NMR (CDCl3, 100 MHz, δ, TMS=0): 196.49, 146.44, 146.24, 137.64, 133.31, 132.39, 129.43, 123.58, 123.41, 115.72, 114.71, 36.81, 36.57, 20.22. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1101, 1098, 1095.
3,4,6,7-tetrahydro-9-(3,4,5-trimethoxyphenyl)-10- (4-methoxyphenyl)-3,3,6,6-tetramethyl acridine- 1,8(2H,5H,9H,10H)-dione (HSM-13b): Yellowish brown powder; yield 69%; MP: 170-172°; 1H NMR (CDCl3, 500 MHz, δ, TMS=0): 6.50 (2H, m), 6.35 (2H, m), 6.02-6.05 (2H, m), 4.56 (1H, s), 3.64 (3H, s), 3.59 (3H, s), 3.49 (6H, s), 2.85-2.95 (4H, m), 1.49 (4H, m), 1.59 (6H, s), 1.30 (6H, s); 13C NMR (CDCl3, 125 MHz, δ, TMS=0): 196.63, 152.87, 144.89, 134.51, 133.12, 132.74, 129.43, 127.77, 124.62, 118.62, 116.33, 57.37, 52.35, 41.73, 31.55, 26.33. IR (KBr, cm-1): 2903, 2874, 1710, 1720, 1600, 1478, 1221, 1203, 1102, 1099.
In vitro CEase assay
Porcine CEase inhibition was assayed spectrophotometrically at 405 nm at 25°. Assay buffer was 100 mM sodium phosphate, 100 mM NaCl, pH 7.0. A stock solution of CEase (200 μg/ml) was prepared in 100 mM sodium phosphate buffer, pH 7.0 and kept at 25°. A 1:200 dilution was done with the same buffer immediately before starting the measurement. Sodium taurocholate (12 mm) was dissolved in assay buffer and kept at 25°. A stock solution of para-nitrophenyl butyrate (20 mm) was prepared in acetonitrile. The final concentration of acetonitrile was 3%, of the substrate para-nitrophenyl butyrate 20 μM, and of sodium taurocholate 6 mM. Assays were performed with a final concentration of 10 ng/ml of CEase. Into a cuvette containing 430 μl assay buffer, 500 μl of the sodium taurocholate solution, 20 μl acetonitrile, 10 μl of the para-nitrophenyl butyrate solution, and 30 μl of an inhibitor solution in DMSO were added and thoroughly mixed. After incubation for 5 min at 25°, the reaction was initiated by adding 10 μl of the enzyme solution 1 μg/ml [34].
Enzyme kinetics study
Synthesized compounds were further investigated for the type of inhibition and enzyme kinetics studies were carried out. The Lineweaver-Burk plot will be established from which we can calculate the Km, Vmax of the slope of inhibitor and the value of α (a constant that defines the degree to which inhibitor binding affects the affinity of the enzyme for substrate) [35].
Molecular properties
Molecular properties of the synthesized compounds were calculated by Lipinski’s rule of five. Briefly, this simple rule is based upon the observation that most biological active drugs have a molecular weight (MW) of 500 or less, a log P not higher than 5, five or fewer hydrogen bond donor sites and ten or fewer hydrogen bond acceptor sites (N or O atoms).
Molecular modelling study
The 3D X-ray coordinates of human cholesterol esterase (hCEase) was obtained from protein data bank (PDB code 1F6W) [36]. Structure of compound will be draw in Chem Draw Ultra (2010) and subjected to energy minimization using the MM2 force field as implemented in Chem 3D Ultra software. The compound will be docked in to the active site of CEase using the GOLD 5.3.0 software [37]. GOLD performs genetic algorithm based ligand docking to optimize the conformation of ligand at the receptor binding site. Gold score scoring function would be employed to calculate the binding score. It utilizes Gold Score fitness function to evaluate the various conformations of ligand at the binding site and comprises of four components: protein-ligand hydrogen bond energy, protein-ligand van der Waals (vdw) energy, ligand internal vdw energy, and ligand torsional strain energy.
Results and Discussion
In order to investigate the catalytic efficiency of various Bronsted acids adsorbed on silica (BA-SiO2), a model reaction was performed for the synthesis of target compound with 1 mol% of various catalysts with the reaction time of 10 min under microwave irradiation. Table 1 revealed that sulphuric acid adsorbed on silica most efficiently catalysed the synthesis of the target compound. In an attempt to improve the yield and optimize the reaction conditions, the model reaction was carried out in the presence of different amounts of silicated sulphuric acid for various time intervals under similar conditions. The conditions were optimized for 100% conversion. A significant improvement in the yield was observed with 1 mol% of the catalyst with the reaction time of 10 min under microwave irradiation. Larger amount of catalyst did not improve the results (Table 2).
| Catalysts (BA-SiO2) | %Yield |
|---|---|
| HBF4-SiO2 | 43 |
| HNO3-SiO2 | 40 |
| HCl-SiO2 | 35 |
| H2SO4- SiO2 | 85 |
| HClO4-SiO2 | 37 |
| SiO2 | - |
Table 1: Percentage yield of acridinedione with various catalysts
| Catalyst | Amount of catalyst (mol%) | Time (min) | Yield (%) |
|---|---|---|---|
| Silicated sulphuric acid | 0.1 | 5 | 15 |
| 10 | 55 | ||
| 15 | 30 | ||
| 20 | 25 | ||
| Silicated sulphuric acid | 0.5 | 5 | 20 |
| 10 | 68 | ||
| 15 | 48 | ||
| 20 | 35 | ||
| Silicated sulphuric acid | 1.0 | 5 | 70 |
| 10 | 85 | ||
| 15 | 80 | ||
| 20 | 74 | ||
| Silicated sulphuric acid | 1.5 | 5 | 70 |
| 10 | 77 | ||
| 15 | 75 | ||
| 20 | 73 |
Table 2: Percent yield with varying mol% of the catalyst and time of exposure to microwave irradiation
The recyclability of the catalyst was investigated using the model reaction. After the completion of the reaction, ethanol was added to the mixture and filtered for separation of the catalyst. It was washed thrice with acetone, dried in oven at 100° for 30 min prior to use and tested for its activity in the subsequent run. The catalyst was tested for four runs. It was observed that the recovered catalyst was recycling in subsequent runs without significant decrease in activity even after four runs (Table 3).
| Cycle | Time (min) | Yield (%) |
|---|---|---|
| 0 | 10 | 85 |
| 1 | 10 | 83 |
| 2 | 10 | 82 |
| 3 | 10 | 80 |
The catalyst was tested for four runs. It was observed that the recovered catalyst was recycling in subsequent runs without significant decrease in activity even after four runs
Table 3: Effect of recyclability of catalyst on product yield
All the synthesized compounds (HSM-1a–HSM-13a and HSM-1b–HSM-13b) with diversification in chemical structures were characterized by melting point, 1H NMR, 13C NMR and IR spectroscopy. NMR spectra of one of the representative compound from the series (HSM-8a) is given in Figure 3.
All the synthesized compounds (HSM1a–HSM13a and HSM1b–HSM13b) were evaluated for their inhibitory potential against CEase enzyme using spectrophotometric assay. Compounds with CEase enzyme inhibition of more than 60% at 50 μM were further evaluated at 1 and 10 μM in order to calculate their IC50 values, Table 4 revealed an interesting structure activity relationship for acridinedione derivatives as CEase inhibitors. Any substitution on both the phenyl rings (at 9th and 10th position of acridinedione nucleus) significantly decreased the activity (compare HSM-1a with all other synthetics).
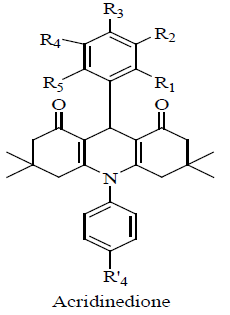 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Code | Substitutions | %Inhibition | IC50 (µM)±SD | |||||||
| R1 | R2 | R3 | R4 | R5 | R’4 | 1 µM | 10 µM | 50 µM | ||
| HSM-1a | H | H | H | H | H | H | 41 | 67 | 93 | 1.53±0.4 |
| HSM-2a | H | H | F | H | H | H | 31 | 55 | 79 | 5.12±0.9 |
| HSM-3a | Cl | H | H | H | H | H | 28 | 49.9 | 73.3 | 10±1.4 |
| HSM-4a | H | Cl | H | H | H | H | 25 | 42 | 68.1 | 10.59±1.3 |
| HSM-5a | H | H | Cl | H | H | H | 26 | 40.1 | 62.5 | 10.98±1.5 |
| HSM-6a | Br | H | H | H | H | H | - | - | 58.4 | - |
| HSM-7a | H | Br | H | H | H | H | - | - | 57.5 | - |
| HSM-8a | H | H | Br | H | H | H | - | - | 52 | - |
| HSM-9a | OCH3 | H | H | H | H | H | - | - | 49.1 | - |
| HSM-10a | H | OCH3 | H | H | H | H | - | - | 42.1 | - |
| HSM-11a | H | H | OCH3 | H | H | H | - | - | 41.5 | - |
| HSM-12a | H | OCH3 | OCH3 | H | H | H | - | - | 40.3 | - |
| HSM-13a | H | OCH3 | OCH3 | OCH3 | H | H | - | - | 39 | - |
| HSM-1b | H | H | H | H | H | OCH3 | - | - | 40 | - |
| HSM-2b | H | H | F | H | H | OCH3 | - | - | 35.7 | - |
| HSM-3b | Cl | H | H | H | H | OCH3 | - | - | 31.1 | - |
| HSM-4b | H | Cl | H | H | H | OCH3 | 28.9 | - | ||
| HSM-5b | H | H | Cl | H | H | OCH3 | - | - | 25.2 | - |
| HSM-6b | Br | H | H | H | H | OCH3 | - | - | 23.3 | - |
| HSM-7b | H | Br | H | H | H | OCH3 | - | - | 22.4 | - |
| HSM-8b | H | H | Br | H | H | OCH3 | - | - | 20.6 | - |
| HSM-9b | OCH3 | H | H | H | H | OCH3 | - | - | 19.9 | - |
| HSM-10b | H | OCH3 | H | H | H | OCH3 | - | - | N.A | - |
| HSM-11b | H | H | OCH3 | H | H | OCH3 | - | - | N.A | - |
| HSM-12b | H | OCH3 | OCH3 | H | H | OCH3 | - | - | N.A | - |
| HSM-13b | H | OCH3 | OCH3 | OCH3 | H | OCH3 | - | - | N.A | - |
| PIC-a* | 1.9 | |||||||||
| PIC-b* | 4.8 | |||||||||
SD-Standard deviation, *PIC-phosphoisocoumarins
Table 4: Results of in vitro cholesterol esterase assay
However substitution of halogen on ring D showed some potency against CEase enzyme in the decreasing order with an increase in the size of the halogen (Figure 4). Compounds with deactivating groups on both D and E rings have significant CEase inhibitory potential in comparison to compounds with activating groups on both D and E rings. Therefore, overall preference order for substituents on both D and E rings could be described as: H>4-F>2-Cl>3-Cl>4-Cl>2-Br>3-Br>4- Br>2-OCH3>3-OCH3>4-OCH3>3,4-OCH3>3,4,5- OCH3. This pattern of activity is in full agreement with the molecular modelling studies, which suggest that HSM-1a fits well in the binding pocket of CEase enzyme and any substitution owing to stearic hindrance may decrease the binding efficiency, thereby, declined inhibitory potential against the CEase enzyme.
The Lineweaver-Burk plot (Figure 5) revealed that compounds HSM-1a was mixed-type CEase inhibitor. The pattern of graph shows that it is a form of mixed inhibition scenario. The Km, Vmax and slope are all affected by the inhibitor. The pattern is different from those characteristics of competitive, non-competitive and uncompetitive inhibition as the inhibitors have increased the Km and slope (Km/Vmax) while decreasing the Vmax. From Figure 5, it was observed that intersecting lines on the graph converge to the left of the y-axis and above the x-axis, which indicates that the value of α (a constant that defines the degree to which inhibitor binding affects the affinity of the enzyme for substrate) is greater than 1. This confirms that the inhibitor preferentially binds to the free enzyme and not the enzyme substrate complex. Therefore, the mode of inhibition of HSM-1a is mixed-type.
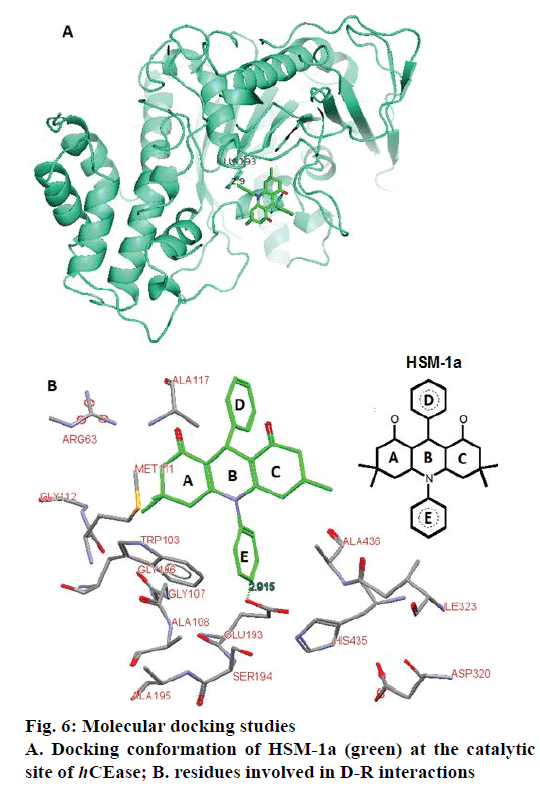
Molecular properties of the potent acridinedione synthetics have been calculated. Each potent derivative has only one violation from Lipinski’s rule of five (Table 5). A docking study was performed to investigate the recognition pattern between CEase and most potent CEase HSM-1a, (Figure 6). The active site of hCEase is a large groove and consists of a catalytic triad and an oxyanion hole. Here, the catalytic triad “Ser194-His435-Asp320’ resides at the bottom of a deep surface depression (groove). The oxyanion hole in hCEase is formed by backbone amide groups of conserved residues Gly107, Ala108, and Ala195 and play important role in ester hydrolysis reaction.
| Properties | Compounds | ||||
|---|---|---|---|---|---|
| HSM-1a | HSM-2a | HSM-3a | HSM-4a | HSM-5a | |
| TPSA | 37.38 | 37.38 | 37.38 | 46.61 | 37.38 |
| Mol. wt. | 425.57 | 443.56 | 460.02 | 455.60 | 460.02 |
| cLogP | 6.56 | 6.72 | 6.72 | 7.21 | 7.24 |
| nOHNH | 0 | 0 | 0 | 0 | 0 |
| nON | 3 | 3 | 3 | 3 | 3 |
| nrotb | 2 | 2 | 2 | 2 | 2 |
| Volume | 411.80 | 416.73 | 425.33 | 425.33 | 425.33 |
| No. of violations | 1 | 1 | 1 | 1 | 1 |
TPSA: topological polar surface area; nOHNH: number of hydrogen bonds donors and nON: acceptors; nrotb: number of rotatable bonds
Table 5: Molecular properties
The HSM-1a fits well at the hCEase active site and established by a hydrogen bond, electrostatic and hydrophobic interactions. The binding site residues and overall binding mode of HSM-1a is shown in Figure 6. The aromatic ring E is placed near the catalytic triad and an oxyanion hole. The C=O group of Glu193 is involved in H-bond interaction with aromatic hydrogen of ring E (d=2.4 Å). The ring E is also surrounded by various nonpolar residues such as Trp103, Gly106, Gly107, Ala108, and Ala463, which are suggested to be involved in van der Waals interaction with HSM- 1a. The ring A is involved on hydrophobic interactions with Met111 and Gly112. The aromatic ring D placed parallel to the side chain of Ala117. This was observed that the docked conformation of HSM-1a completely cover the catalytic triad and an oxyanion hole of hCEase. As discussed previously that catalytic triad and an oxyanion hole are vital for the catalytic action of hCEase. Therefore, this can be suggested that the binding conformation of HSM-1a blocks the approach of the normal substrates of hCEase to its catalytic assembly and thus inhibits its activity.
Present study provides an efficient methodology for the synthesis of acridinedione derivatives employing H2SO4-SiO2 at 1 mol% for 10 min under microwave irradiation. All the synthesized compounds were evaluated for in vitro CEase inhibition. Results of biological evaluation revealed the promising inhibitory profile of acridinediones, as the most potent derivative, HSM-1a, displayed strong enzyme inhibition with IC50 value of 1.53 μM. Enzyme kinetic studies performed on HSM-1a confirmed that the inhibitor preferentially binds to the free enzyme and not the enzyme substrate complex and was mixed type CEase inhibitor. Molecular modelling study figured out the various interesting facts about the binding interactions of HSM- 1a and amino acids residues of CEase further supported the mixed type aspect of enzyme inhibition. In view of potency against CEase enzyme and calculated molecular properties, HSM-1a appears to be the good hit among the series of acridinedione derivatives and displays promising attributes for detailed investigation.
Acknowledgements
Authors thank Professor Ajaib Singh Brar, Vice Chancellor, Guru Nanak Dev University, Amritsar for their constant support and encouragement.
Financial assistance
Nill.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sota O. The Interleukin-6 inflammation pathway from cholesterol to aging – Role of statins, bisphosphonates and plant polyphenols in aging and age-related diseases. Immun Ageing 2007;4:1-22.
- Maron DJ, Fazio S, Linton MF. Current perspectives on statins. Circulation 2000;101:207-13.
- Auer J, Eber B. Current aspects of statins. J Clin Basic Cardiol 1999;2:203-8.
- Zhong HJ, Liu LJ, Chong CM, Lu L, Wang M, Chan DS, et al. Discovery of a Natural Product-Like iNOS Inhibitor by Molecular Docking with Potential Neuroprotective Effects In Vivo. PLoS One 2014;9:0092905.
- Liu LJ, Leung KH, Chan DS, Wang YT, Ma DL, Leung CH. Identification of a natural product-like STAT3 dimerization inhibitor by structure-based virtual screening. Cell Death Dis 2014;5:e1293-302.
- Ma DL, Chan DSH, Wei G, Zhong HJ, Yang H, Leung LT, et al. Virtual screening and optimization of Type II inhibitors of JAK2 from a natural product library. Chem Comm 2014;50;13885-88.
- Leung CH, Liu LJ, Lu L, He B, Kwong DW, Wong CY, et al. A metal-based tumour necrosis factor-alpha converting enzyme inhibitor. Chem Comm 2015;51:3973-76.
- Zhong HJ, Lu L, Leung KH, Wong CCL, Peng C, Yan SC, et al. An iridium(III)-based irreversible protein–protein interaction inhibitor of BRD4 as a potent anticancer agent. Chem Sci 2015;6:5400-08.
- Myers-Payne SC, Hui DY, Brockman HL, Schroeder F. Cholesterol esterase: a cholesterol transfer protein. Biochemistry 1995;34:3942-47.
- Gallo LL, Clark SB, Myers S, Vahouny GV. Cholesterol absorption in rat intestine: role of cholesterol esterase and acyl coenzyme A: cholesterol acyltransferase. J Lipid Res 1984;25:604-12.
- Heng S, Tieu W, Hautmann S, Kuan K, Pedersen DS, Pietsch M, et al. New cholesterol esterase inhibitors based on rhodanine and thiazolidinedione scaffolds. Bioorg Med Chem 2011;19:7453-63.
- Heidrich JE, Contos LM, Hunsaker LA, Deck LM, Vander Jagt DL. Inhibition of pancreatic cholesterol esterase reduces cholesterol absorption in the hamster. BMC Pharmacol 2004;4:5.
- Feaster SR, Quinn DM. Mechanism-Based Inhibitors of Mammalian Cholesterol Esterase. Methods Enzymol 1997;286:231-52.
- Deck LM, Baca ML, Salas SL, Hunsaker LA, Vander Jagt DL. 3-Alkyl-6-chloro-2-pyrones: selective inhibitors of pancreatic cholesterol esterase. J Med Chem 1999;42:4250-56.
- Stoddard HM, Brown WM, Deck JA, Hunsaker LA, Deck LM, Vander Jagt DL. Inhibition of yeast lipase (CRL1) and cholesterol esterase (CRL3) by 6-chloro-2-pyrones: comparison with porcine cholesterol esterase. Biochimica Biophysica Acta 2000;596:381-91.
- Pietsch M, Gutschow M. Synthesis of tricyclic 1,3-oxazin-4-ones and kinetic analysis of cholesterol esterase and acetylcholinesterase inhibition. J Med Chem 2005;48:8270-88.
- Pietsch M, Gutschow M. Alternate substrate inhibition of cholesterol esterase by thieno[2,3-d][1,3]oxazin-4-ones. J Biol Chem 2002;277:24006-13.
- Lin MC, Lin GZ, Hwang CI, Jian SY, Lin J, Shen YF, et al. Synthesis and evaluation of a new series of tri-, di-, and mono-N-alkylcarbamylphloroglucinols as conformationally constrained inhibitors of cholesterol esterase. Protein Sci 2012;21:1344-57.
- Chiou SY, Lai CY, Lin LY, Lin G. Probing stereoselective inhibition of the acyl binding site of cholesterol esterase with four diastereomers of 2'-N-alpha-methylbenzylcarbamyl-1, 1'-bi-2-naphthol. BMC Biochem 2005;6:17-25.
- Lin G, Chiou SY, Hwu BC, Hsieh CW. Probing structure-function relationships of serine hydrolases and proteases with carbamate and thiocarbamate inhibitors. Protein J 2006;5:33-43.
- Lin G, Liao WC, Chiou SY. Hammett analysis of the inhibition of pancreatic cholesterol esterase by substituted phenyl-N-butylcarbamate. Bioorg Med Chem 2000;8:2601-08.
- Lin G, Shieh CT, Ho HC, Chouhwang JY, Lin WY, Lu CP. Structure-reactivity relationships for the inhibition mechanism at the second alkyl chain binding site of cholesterol esterase and lipase. Biochemistry 1999;38:9971-81.
- Lin G, Shieh CT, Tsai YC, Hwang CI, Lu CP, Cheng GH. Structure-reactivity probes for active site shapes of cholesterol esterase by carbamate inhibitors. Biochim Biophys Acta 1999;1431:500-11.
- Lin G, Tsai YC, Liu HC, Liao WC, Chang CH. Enantiomeric inhibitors of cholesterol esterase and acetylcholinesterase. Biochim Biophys Acta 1998;1388:161-74.
- Feaster SR, Lee K, Baker N, Hui DY, Quinn DM. Molecular recognition by cholesterol esterase of active site ligands: structure-reactivity effects for inhibition by aryl carbamates and subsequent carbamylenzyme turnover. Biochemistry 1996;35:16723-34.
- Quistad GB, Liang SN, Fisher KJ, Nomura DK, Casida JE. Each lipase has a unique sensitivity profile for organophosphorus inhibitors. Toxicol Sci 2006;91:166-72.
- Schmidinger H, Birner-Gruenberger R, Riesenhuber G, Saf R, Susani-Etzerodt H, Hermetter A. Novel Fluorescent Phosphonic Acid Esters for Discrimination of Lipases and Esterases. Chembiochem 2005;6:1776-81.
- Heynekamp JJ, Hunsaker LA, Vander Jagt TA, Royer RE, Deck LM, Vander Jagt DL. Isocoumarin-based inhibitors of pancreatic cholesterol esterase. Bioorg Med Chem 2008;16:5285-94.
- Li B, Zhou B, Lu H, Ma L, Peng AY. Phosphaisocoumarins as a new class of potent inhibitors for pancreatic cholesterol esterase. Eur J Med Chem 2010;45:1955-63.
- Heng S, Tieu W, Hautmann S, Kuan K, Pedersen DS, Pietsch M, et al. New cholesterol esterase inhibitors based on rhodanine and thiazolidinedione scaffolds. Bioorg Med Chem 2011;19:7453-63.
- Singh H, Singh JV, Gupta MK, Singh P, Sharma S, Nepali K, et al. Benzoflavones as cholesterol esterase inhibitors: Synthesis, biological evaluation and docking studies. Bioorg Med Chem Lett 2017;27:850-54.
- Wei Y, Peng AY, Wang B, Ma L, Peng G, Du Y, et al. Synthesis and biological evaluation of phosphorylated flavonoids as potent and selective inhibitors of cholesterol esterase. Eur J Med Chem 2014;74:751-58.
- Ma Z, Saluta G, Kucera GL, Bierbach U. Effect of linkage geometry on biological activity in thiourea- and guanidine-substituted acridines and platinum–acridines. Bioorg Med Chem Lett 2008;18:3799-801.
- Muscia GC, Hautmann S, Buldain GY, Gutschow M. Synthesis and Evaluation of 2-(1H-Indol-3-yl)-4-phenylquinolines as Inhibitors of Cholesterol Esterase. Bioorg Med Chem Lett 2014;24:1545-49.
- Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. 2nd ed. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2005.
- Terzyan S, Wang CS, Downs D, Hunter B, Zhang XC. Crystal structure of the catalytic domain of human bile salt activated lipase. Protein Sci 2000;9:1783-90.
- https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/.
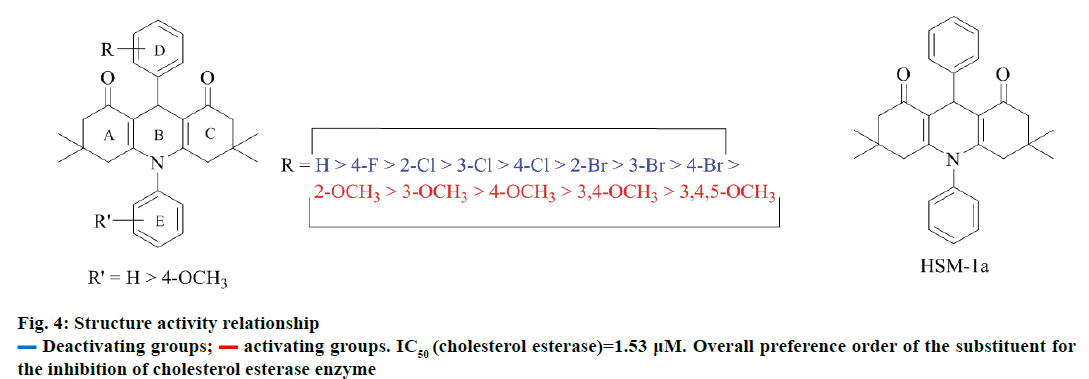
 Deactivating groups;
Deactivating groups;  activating groups. IC50 (cholesterol esterase)=1.53 µM. Overall preference order of the substituent for
the inhibition of cholesterol esterase enzyme
activating groups. IC50 (cholesterol esterase)=1.53 µM. Overall preference order of the substituent for
the inhibition of cholesterol esterase enzyme
 Control;
Control; 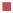 HSM-1a (1 µM);
HSM-1a (1 µM);  HSM-1a (10 µM);
HSM-1a (10 µM);  HSM-1a
(50 µM)
HSM-1a
(50 µM)