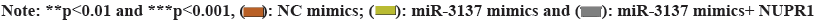- *Corresponding Author:
- Chunyan Cai
Department of Gynaecology ,The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, Huaian, Jiangsu 223300, China
E-mail: sisicai@126.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “185-192” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cervical cancer belongs to the second most frequent kind of cancer in female as well as a major cause of mortality in female all over the world. MicroRNAs are crucial modulators of multiple diseases. Based on GSE86100 database, microRNA-3137 expression was low in cervical cancer tissues and the difference was most significant, thus we further probed the potential along with mechanism of microRNA-3137 in cervical cancer cells. Reverse transcription-quantitative polymerase chain reaction together with Western blot was implemented for examining gene expression. 5-ethynyl-2'-deoxyuridine and flow cytometry analyses were adopted for assessing cervical cancer cells proliferation and apoptosis. Immunofluorescence was implemented to measure the fluorescence intensity of autophagy marker LC3II. Target scan together with luciferase reporter assay were implemented for confirming the direct binding of microRNA-3137 and nuclear protein transcriptional regulator 1. The results revealed that, microRNA-3137 was under expressed in cervical cancer cells. Overexpressed microRNA-3137 repressed cervical cancer cells proliferation while elevated cells apoptosis. Besides, nuclear protein transcriptional regulator 1 was directly targeted and negatively regulated by microRNA-3137 and was highly expressed in cervical cancer cells. Moreover, nuclear protein transcriptional regulator 1 silence promoted autophagy in cervical cancer cells and rescue assays further validated that the microRNA-3137/nuclear protein transcriptional regulator 1 axis suppressed cervical cancer cells proliferation while promoted cells apoptosis and autophagy through inactivation of the phosphatidylinositol-3-kinase/ protein kinase B/mammalian target of rapamycin pathway. Collectively, our study demonstrated that microRNA-3137/nuclear protein transcriptional regulator 1 axis affected cervical cancer cells proliferation, apoptosis, along with autophagy by means of inhibiting the phosphatidylinositol-3-kinase/protein kinase B/ mammalian target of rapamycin pathway, which may offer novel insights into the mechanism of cervical cancer progression as well as a novel therapy targeting for cervical cancer therapy.
Keywords
Cervical cancer, microRNA-3137, nuclear protein transcriptional regulator 1, autophagy, phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin pathway
Cervical Cancer (CC) belongs to the second most frequent cancer among female as well as a major cause of mortality in female in the world[1]. Infection of high-risk carcinogenic Human Papilloma Viruses (HPVs) is the most critical risk factor for CC[2]. Early CC often has no obvious symptoms and signs. As the lesion progresses, the following manifestations including vaginal bleeding and discharge may occur[3]. Developments in the diagnosis together with therapy in CC have been observed, the current main treatments for CC are surgery, radiotherapy, chemotherapy and immunotherapy[4]. However, the long-time survival rate of CC patients was poor[5]. Pathological studies have certified that the molecular mechanism of CC progression is very complicated, which includes various gene mutations together with epigenetic changes[6]. Hence, further exploration of the internal mechanism underlying CC progression is of significant value for CC therapy.
MicroRNAs (miRNAs) belong to small non-coding RNAs (sncRNAs) which harbors 18-25 nucleotides and regulate messenger RNA (mRNA) expression through targeting the 3-Untranslated Region (3’-UTR) of mRNA[7]. Increasing data has proved that miRNAs act as a crucial part in the development of a variety of cancers, CC included[8,9]. In CC, numerous miRNAs have been unveiled to serve as tumor suppressors in CC progression. MiR-195-3p is down-regulated in CC and hinders cell proliferation by modulating BCDIN3D[10]. MiR-3156-3p is low-expressed in HPV-positive CC, as well as functions to be a tumor-repressive miRNA[11]. MiR-449a expression is reduced in CC and suppresses cell malignant behaviors[12]. Moreover, miR-3137 is a potential biomarkers for diabetic kidney disease[13] and its expression and role in cancers are still unclear.
Autophagy is identified to be a cytoprotective process, which promotes cell survival[14]. It is also a highly conserved multistep lysosomal degradation process, where cellular components are separated in autophagosomes and then fused with lysosomes to degrade the isolate[15]. Besides, autophagy can stimulate non-reversible autophagic cell death together with autophagic dependent apoptosis. An excess of autophagy may result in cell death, which implies that autophagy may be a mode of cell death or only a failed attempt for rescuing stressed cells from death[16]. Thus, appropriate autophagy can prevent cell death whereas immoderate autophagy may bring out autophagic cell death[17]. Nevertheless, given the complicacy of autophagy, further studies are required to probe its function in cell survival as well as death.
Autophagy can be promoted or inhibited by many different signaling pathways and the Phosphatidylinositol-3-Kinase (PI3K)/Protein Kinase B (AKT)/mammalian Target of Rapamycin (mTOR) pathway was the most classic[18,19]. mTOR is a crucial modulator in the autophagy initiation phase. Stimulation of mTOR can repress cell autophagy, resulting in unlimited tumor cells proliferation[20]. Additionally, the PI3K/AKT/mTOR pathway is widely-implicated in cellular processes, containing cell proliferation, apoptosis, autophagy, as well as necroptosis[21]. It is often abnormally stimulated in the development of cancers, CC included[22,23].
In our study, the gene expression profiles of miRNA in CC patients (GSE86100) were downloaded from the Gene Expression Omnibus (GEO) database and discovered that miR-3137 expression was the most significantly declined in CC tissues, thus we further probed the expression, potential together with mechanism of miR-3137 in CC cells.
Materials and Methods
Cell culture:
acquired from Pro cell (Wuhan, China), as well as cultured in Roswell Park Memorial Institute (RPMI)-1640 medium, Minimal Essential Medium (MEM) and McCoy’s 5A medium, respectively. Human normal Cervical Epithelial cell line (HCerEpic) was acquired from Shenzhen Haodi Huatuo Biotechnology Co., Ltd (Shenzhen, China), as well as cultured in Dulbecco's Modified Eagle's Medium (DMEM). All cell mediums were added with 10 % Fetal Bovine Serum (FBS) and cultivated in an incubator with 5 % Carbon dioxide (CO2) at 37°.
Cell transfection:
MiR-3137 mimics together with the Negative Control (NC) mimics, specific short-hairpin RNA (shRNA) against Nuclear Protein Transcriptional Regulator 1 (NUPR1) (sh-DANCR) along with the sh-NC, NUPR1 overexpression plasmid together with the empty vector, which were obtained from RiboBio (Guangzhou, China). The above plasmids were transfected into CC cells by means of the Lipofectamine 3000 reagent.
Cell transfection:
Total RNA was obtained from cells by use of TRIzol (Takara) and then reverse-transcribed into complementary Deoxyribonucleic acid (cDNA) based on the reverse transcription kit (Gima). qPCR was implemented using SYBR Premix Ex Taq II (Takara). U6 or Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) served as an internal control for miRNA or mRNA. The 2−ΔΔCT method was implemented to analyze relative gene expression. The primers used for analysis were indicated as follows: miR-3137 forward: 5’-GGTCCTCTGTAGCCTGGGAGCAATG-3’, reverse: 5’-CCAGTGCAGGGTCCGAGGT-3’; NUPR1: forward: 5’-GACAGAGCTGGAGATGAGG-3’, reverse: 5’-GCGGGAATAAGTCCTAGGG-3’; GAPDH: forward: 5’-AACGTGTCAGTGGTGGACCTG-3’, reverse: 5’-AGTGGGTGTCGCTGTTGAAGT-3’; U6 forward: 5’-CTCGCTTCGGCAGCACA-3’, reverse: 5’-AACGCTTCACGAATTTGCGT-3’.
5-Ethynyl-2'-deoxyuridine (EdU):
EdU detection kit (RiboBio, Guangzhou, China) was adopted to test cell proliferation. Cells (5×103 cells/well) were cultured in 96-well plates, followed by adding 10 µl of EdU labeling media for incubation for 2 h. Cells were stained with the anti-EdU working solution as well as 4',6-Diamidino-2-Phenylindole Dihydrochloride (DAPI) after treating 4 % paraformaldehyde together with 0.5 % Triton X-100. Next, cells were visualized based on a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry:
CC cells after transfection were harvested, collected and suspended, and then stained with Annexin V-Fluorescein Isothiocyanate (FITC) and Propidium Iodide (PI) in the darkness. Afterwards, the apoptosis rate was measured by flow cytometry (FACS Calibur, BD Biosciences).
Luciferase reporter assay:
The pmiR-GLO reporter vector (GenePharma) was utilized to synthesize the NUPR1 3’-UTR reporter plasmid according to the binding sites between NUPR1 3’-UTR and miR-3137. Then, CC cells were transfected with reporter plasmids and miR-3137 mimics/NC mimics. After 48 h, the luciferase activity was examined using dual-luciferase reporter assay system (Promega). Renilla luciferase served as the internal transfection control.
Immunofluorescence:
Cells after transfection were grown on cover slips and then washed with Phosphate Buffered Saline (PBS). Upon fixing with 4 % paraformaldehyde as well as permeabilizing with 0.1 % Triton X-100, cells were sealed by 1 % Bovine Serum Albumin (BSA) for 30 min. Afterwards, cells were treated with the primary rabbit antibody against LC3-II (Abcam; 1:1000) overnight at 4° and then treated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Beyotime, 1:500) for 1 h. The nuclei were stained with DAPI. The samples were observed on a fluorescence microscope (Olympus, Tokyo, Japan).
Western blot:
Total proteins were lysed from cells using Radioimmunoprecipitation Assay (RIPA) reagent. Protein lysates were electrophoresed through Sodium Dodecyl-Sulfate/Polyacrylamide Gel Electrophoresis (SDS/PAGE) and transferred to polyvinylidene fluoride membranes. Followed by being blocked, the membranes were then cultivated with primary antibodies overnight and then incubated with a corresponding horseradish peroxidase-conjugated secondary antibody (Abcam, 1:2000). The protein bands were visualized using an Electrochemiluminescent (ECL) substrate. The primary antibodies used were p62 (Abcam, 1:10 000), Beclin1 (Abcam, 1:1000), Autophagy Related-3 (ATG3) (Abcam, 1:10 000), ATG5 (Abcam, 1:1000), p-AKT (Abcam, 1:500), AKT (Abcam, 1:500), p-mTOR (Abcam, 1:1000), mTOR (Abcam, 1: 10 000) and GAPDH (Abcam, 1:1000).
Statistical analysis:
All experiments were implemented at least for 3 times. All data were exhibited as mean±standard deviation. GraphPad 6.0 software (GraphPad, San Diego, California, USA) was adopted for statistical analysis. Differences were compared using the student’s t-test or one-way analysis of variance. p<0.05 was deemed as statistical significance.
Results and Discussion
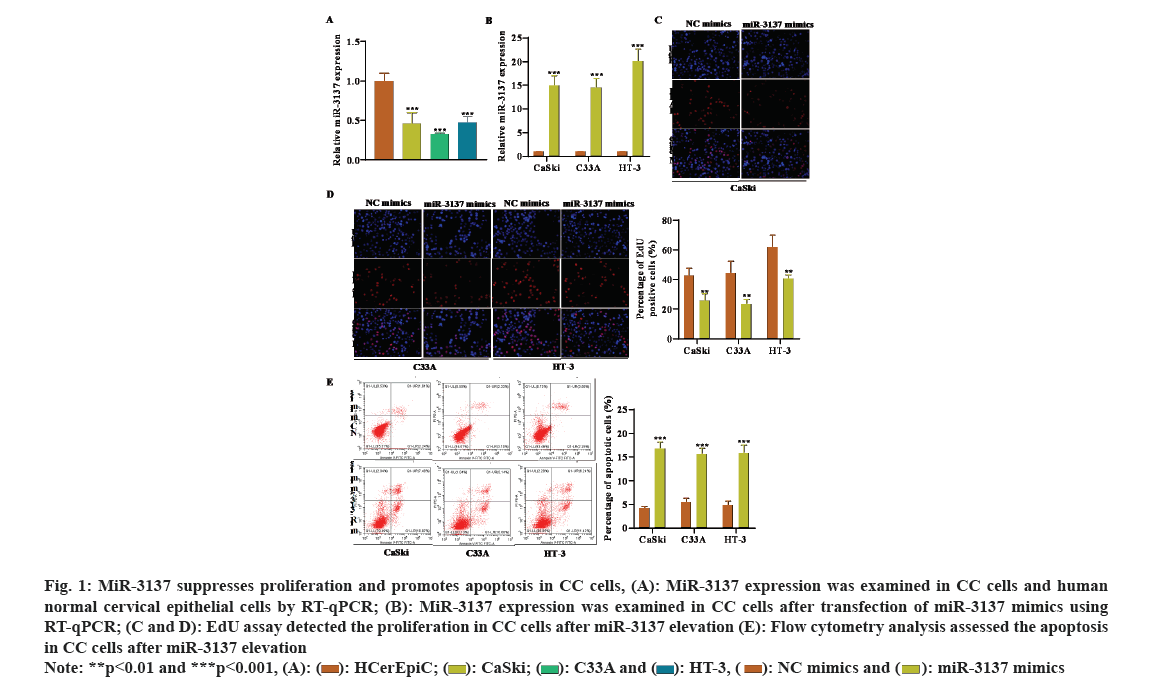
The gene expression profiles of miRNA in CC patients (GSE86100) were downloaded from the GEO database. MiR-3137 expression was low in CC tissues, with the most significant difference, thus we selected miR-3137 for the objective of our study. Subsequently, miR-3137 expression in CC cells was detected by RT-qPCR analysis. The outcome displayed that miR-3137 was down-regulated in CC cells (CaSki, C33A and HT-3) compared to human normal cervical epithelial cells (HCerEpic) (fig. 1A). To probe the impacts of miR-3137 on CC cell proliferation as well as apoptosis, miR-3137 mimics was transfected into CC cells to elevate miR-3137 expression. RT-qPCR analysis further verified the successful transfection, which showed in fig. 1B that, miR-3137 expression in CC cells was apparently enhanced followed by miR-3137 mimics transfection. Gain-of-function assays further demonstrated that miR-3137 overexpression distinctly lessened CC cells proliferation, while increased cell apoptosis (fig. 1C-fig. 1E). Collectively, miR-3137 was low-expressed in CC cells and inhibited CC cell proliferation as well as elevated cell apoptosis.
Fig. 1: MiR-3137 suppresses proliferation and promotes apoptosis in CC cells, (A): MiR-3137 expression was examined in CC cells and human normal cervical epithelial cells by RT-qPCR; (B): MiR-3137 expression was examined in CC cells after transfection of miR-3137 mimics using RT-qPCR; (C and D): EdU assay detected the proliferation in CC cells after miR-3137 elevation (E): Flow cytometry analysis assessed the apoptosis in CC cells after miR-3137 elevation 
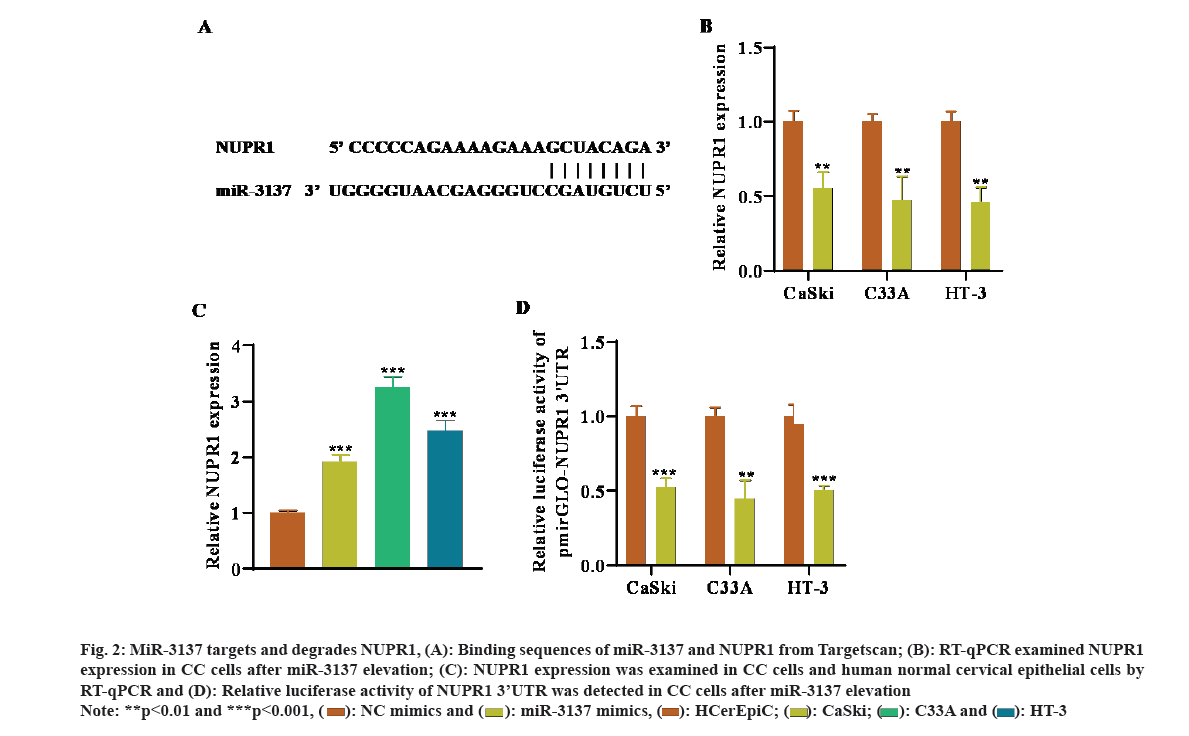
Next, the latent mechanism of miR-3137 in CC cells was probed. By means of Target scan (https://www.targetscan.org/vert_72/) website, the target mRNAs of miR-3137 were predicted. NUPR1 was chosen for the downstream mRNA of miR-3137 due to the largest absolute value of the total context score with miR-3137 and their predicted binding sites were indicated in fig. 2A. RT-qPCR analysis further detected that NUPR1 expression was remarkably declined in CC cells after miR-3137 elevation (fig. 2B). Besides, NUPR1 was high-expressed in CC cells in comparison with HCerEpic (fig. 2C). In addition, the outcome of luciferase reporter assays revealed that, transfecting miR-3137 mimics overtly lessened the relative luciferase activity of NUPR1 3’UTR, further verifying the direct binding of miR-3137 and NUPR1 (fig. 2D). Taken together, NUPR1 was directly targeted by miR-3137 in CC cells.
Fig. 2: MiR-3137 targets and degrades NUPR1, (A): Binding sequences of miR-3137 and NUPR1 from Targetscan; (B): RT-qPCR examined NUPR1 expression in CC cells after miR-3137 elevation; (C): NUPR1 expression was examined in CC cells and human normal cervical epithelial cells by RT-qPCR and (D): Relative luciferase activity of NUPR1 3’UTR was detected in CC cells after miR-3137 elevation
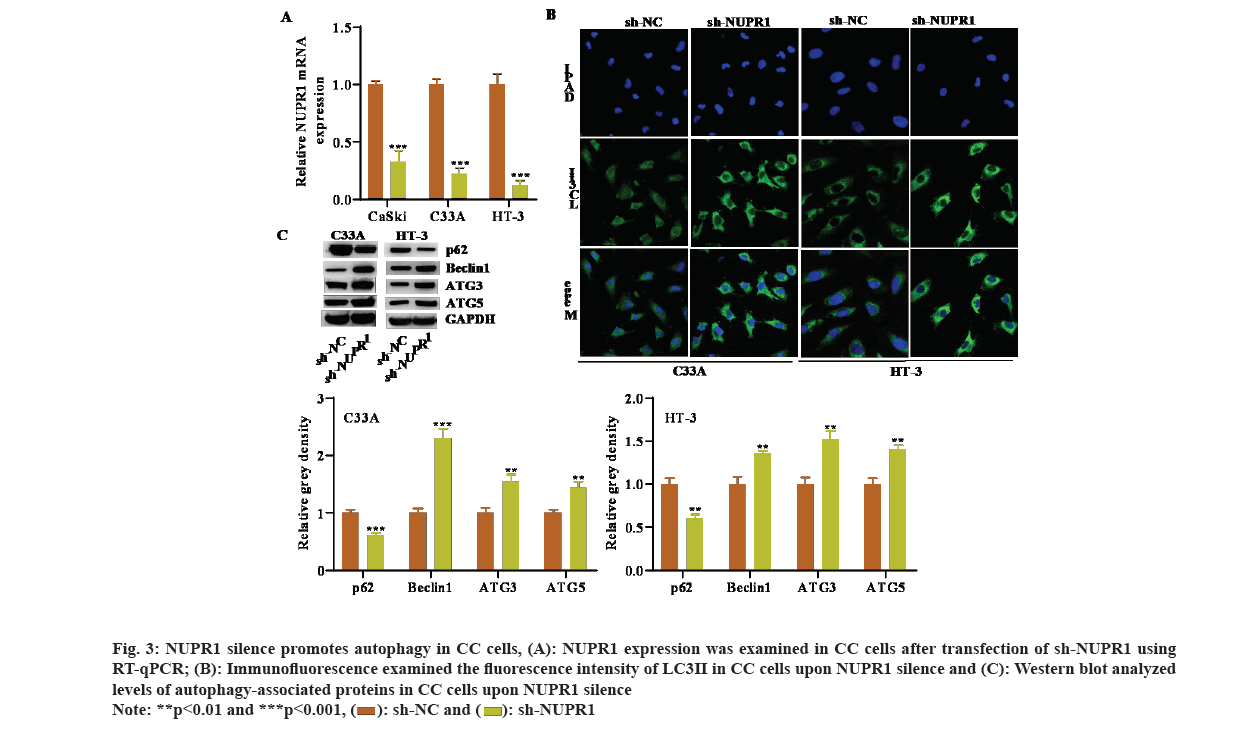
Several studies have reported that NUPR1 is associated with autophagy[24]. Hence, we speculated that NUPR1 may be also implicated in modulating autophagy in CC cells. We first silenced NUPR1 expression in CC cells by transfecting specific shRNA against NUPR1 (fig. 3A). We then observed an obvious increase in the immunofluorescence intensity of LC3 after NUPR1 knockdown (fig. 3B). Moreover, levels of autophagy-associated proteins were examined by Western blot and outcomes uncovered that reduced NUPR1 expression decreased p62 protein level, while increased the protein levels of Beclin1, ATG3, as well as ATG5 (fig. 3C). Collectively, NUPR1 silence promoted autophagy in CC cells.
Fig. 3: NUPR1 silence promotes autophagy in CC cells, (A): NUPR1 expression was examined in CC cells after transfection of sh-NUPR1 using RT-qPCR; (B): Immunofluorescence examined the fluorescence intensity of LC3II in CC cells upon NUPR1 silence and (C): Western blot analyzed levels of autophagy-associated proteins in CC cells upon NUPR1 silence 
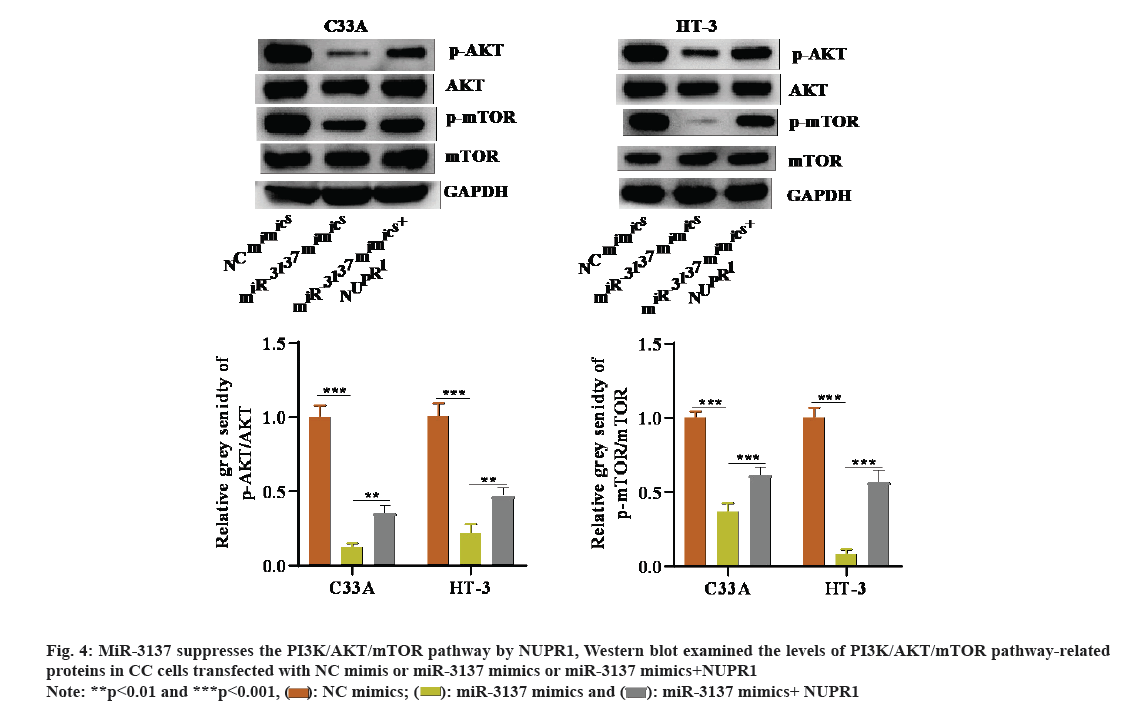
It is well-known that one of the classic pathways regulating autophagy is the PI3K/AKT/mTOR pathway[25]. Hence, it was assumed that the miR-3137/NUPR1 axis might modulate the PI3K/AKT/mTOR pathway in CC cells. Through Western blot, we discovered that p-AKT and p-mTOR protein levels were weakened in CC cells in presence of miR-3137 overexpression. However, co-transfection of NUPR1 overexpression plasmid could reverse this inhibitory effect on p-AKT and p-mTOR levels caused by miR-3137 elevation (fig. 4A). In a word, miR-3137 suppressed the PI3K/AKT/mTOR pathway via targeting NUPR1.
Rescue experiments were further implemented to validate the influence of the miR-3137/NUPR1 axis on influencing CC cell proliferation, apoptosis and autophagy. It was exhibited in fig. 5A-fig. 5C that, the lessened cell proliferation as well as the elevated cell apoptosis caused by miR-3137 overexpression was rescued by simultaneous NUPR1 elevation. Besides, NUPR1 up-regulation offset the reduced p62 protein level and enhanced Beclin1, ATG3 as well as ATG5 protein levels in miR-3137 mimic’s transfected CC cells (fig. 5D). Collectively, miR-3137 suppressed proliferation and promoted apoptosis and autophagy by NUPR1 in CC cells.
Fig. 5: MiR-3137 suppresses proliferation and promotes apoptosis and autophagy by NUPR1. C33A cells were transfected with NC mimis or miR-3137 mimics or miR-3137 mimics+NUPR1, followed by rescue assays, (A): EdU examined cell proliferation; (B and C) Flow cytometry assessed cell apoptosis and (D): Western blot examined autophagy-associated protein levels

The use of miRNAs as potential biomarkers and therapeutic targets has been extensively studied in a variety of cancers[26]. Similarly, there are also several miRNAs functioning as therapeutic and prognostic biomarkers in CC, such as miR-3154 and miR-600[27]. In this research, we elucidated the capability of the miR-3137/NUPR1 axis to suppress CC cells proliferation while promote cells apoptosis as well as autophagy via repressing the PI3K/AKT/mTOR pathway.
As documented before, miR-3137 is up-regulated in hepatocellular carcinoma cells and significantly associated with an unfavorable prognosis in hepatocellular carcinoma patients[28]. Unlike with the above report, our research clarified that miR-3137 was under expressed in CC cells. Besides, overexpressed miR-3137 could hinder cell proliferation whereas elevate cell apoptosis in CC, which was consistent with many previous studies on miRNA in CC[29,30].
MiRNAs act on post-transcriptionally through binding to mRNA 3’UTR and thereby modulating mRNA expression and this model has been widely applied in exploring the molecular mechanism of miRNAs in cancers, CC included[31]. For instance, miR-192-5p targets TRPM7 to hinder proliferation in CC cells[32]. MiR-499a accelerates the progression and chemo resistance of CC cells by binding to SOX6[33]. In line with this evidence, our study also predicted the potential mRNA that bound to miR-3137 using bioinformatics prediction. NUPR1 was proved to be directly targeted by miR-3137 as well as was negatively regulated by miR-3137 in CC cells. NUPR1 belongs to a transcription factor dependent on its C-terminal helix-loop-helix structure and its function may be mediated by a variety of kinases, factors or cellular stress factors[34]. Importantly, NUPR1 has been demonstrated to be up-regulated in many malignancies as well as facilitate their progression[35]. NUPR1 enhances breast cancer cells proliferation through stimulating Transcription Factor E3 (TFE3) transcription[36]. NUPR1 enhances pancreatic cancer development by inhibiting apoptosis[37]. Nevertheless, its potential in CC is elusive. In our research, we discovered that NUPR1 was high-expressed in CC cells, which may support the oncogenic role of NUPR1 in cancers.
Autophagy has a critical potential in maintaining intracellular homoeostasis in reaction to cellular stress[38]. NUPR1 works as a master modulator of cellular clearance by accelerating autophagy process[39]. NUPR1 drives auto lysosomal efflux via promoting SNAP25 transcription in cancer cells[40]. NUPR1 silence hinders autophagic flux in hepatocellular carcinoma cells[41]. NUPR1 silence reduces autophagy in multiple myeloma cells by the PI3K/AKT/mTOR pathway[42]. However, our study indicated that NUPR1 silence promoted autophagy in CC cells, which may be attributed to different NUPR1 potential in different cancer types.
The PI3K/AKT/mTOR pathway is drawn in various human cancers, as well as can affect apoptosis together with proliferation in cancer cells[43]. Hence, repressing this pathway is considered as a promising method for cancer therapy. Additionally, the PI3K/AKT/mTOR pathway was deemed to be the major pathway in modulating autophagy[44]. The repression of PI3K/AKT/mTOR pathway enhances cell autophagy[45]. In our research, we further demonstrated that the miR-3137/NUPR1 axis suppressed CC cells proliferation while promoted cells apoptosis as well as autophagy via repressing the PI3K/AKT/mTOR pathway.
In conclusion, our study presented that miR-3137 was low-expressed in CC cells and worked as a tumor repressor in CC. More importantly, our study validated that the miR-3137/NUPR1 axis suppressed CC cells proliferation while promoted cells apoptosis as well as autophagy via inhibiting the PI3K/AKT/mTOR pathway. Our research might provide a novel sight for CC therapy.
Conflict of interests:
The authors declared no conflict of interests.
References
- Olusola P, Banerjee HN, Philley JV, Dasgupta S. Human papilloma virus-associated cervical cancer and health disparities. Cells 2019;8(6):622.
[Crossref] [Google Scholar] [PubMed]
- Wilailak S, Kengsakul M, Kehoe S. Worldwide initiatives to eliminate cervical cancer. Int J Gynecol Obstet 2021;155(1):102-6.
[Crossref] [Google Scholar] [PubMed]
- Downey K, deSouza NM. Imaging cervical cancer: Recent advances and future directions. Curr Opin Oncol 2011;23(5):519-25.
[Crossref] [Google Scholar] [PubMed]
- Kontostathi G, Zoidakis J, Anagnou NP, Pappa KI, Vlahou A, Makridakis M. Proteomics approaches in cervical cancer: Focus on the discovery of biomarkers for diagnosis and drug treatment monitoring. Expert Rev Proteomics 2016;13(8):731-45.
[Crossref] [Google Scholar] [PubMed]
- Deressa BT, Assefa M, Tafesse E, Kantelhardt EJ, Soldatovic I, Cihoric N, et al. Contemporary treatment patterns and survival of cervical cancer patients in Ethiopia. BMC Cancer 2021;21(1):1102.
[Crossref] [Google Scholar] [PubMed]
- Yu S, Li X, Zhang J, Wu S. Development of a novel immune infiltration-based gene signature to predict prognosis and immunotherapy response of patients with cervical cancer. Front Immunol 2021;12:3507.
- Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci 2019;20(24):6249.
[Crossref] [Google Scholar] [PubMed]
- He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, et al. miRNA-based biomarkers, therapies and resistance in cancer. Int J Biol Sci 2020;16(14):2628.
- Aftab M, Poojary SS, Seshan V, Kumar S, Agarwal P, Tandon S, et al. Urine miRNA signature as a potential non-invasive diagnostic and prognostic biomarker in cervical cancer. Sci Rep 2021;11(1):1-3.
- Jin M, Wang L, Zheng T, Yu J, Sheng R, Zhu H. MiR-195-3p inhibits cell proliferation in cervical cancer by targeting BCDIN3D. J Reprod Immunol 2021;143:103211.
[Crossref] [Google Scholar] [PubMed]
- Xia YF, Pei GH, Wang N, Che YC, Yu FS, Yin FF, et al. miR-3156-3p is down regulated in HPV-positive cervical cancer and performs as a tumor-suppressive miRNA. Virol J 2017;14(1):20.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Zhao Y, Xiong W, Ye W, Zhao W, Hua Y. MicroRNA-449a is down regulated in cervical cancer and inhibits proliferation, migration and invasion. Oncol Res Treat 2019;42(11):564-71.
[Crossref] [Google Scholar] [PubMed]
- Li X, Xu R, Liu X, Xu L, Xue M, Cheng Y, et al. Urinary miR-3137 and miR-4270 as potential biomarkers for diabetic kidney disease. J Clin Lab Anal 2020;34(12):e23549.
- Levy JM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17(9):528-42.
[Crossref] [Google Scholar] [PubMed]
- Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell 2011;147(4):728-41.
[Crossref] [Google Scholar] [PubMed]
- Glick D, Barth S, Macleod KF. Autophagy: Cellular and molecular mechanisms. J Pathol 2010;221(1):3-12.
[Crossref] [Google Scholar] [PubMed]
- Kocaturk NM, Akkoc Y, Kig C, Bayraktar O, Gozuacik D, Kutlu O. Autophagy as a molecular target for cancer treatment. Eur J Pharm Sci 2019;134:116-37.
[Crossref] [Google Scholar] [PubMed]
- Xu Z, Han X, Ou D, Liu T, Li Z, Jiang G, et al. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol 2020;104(2):575-87.
[Crossref] [Google Scholar] [PubMed]
- Zeng X, Ju D. Hedgehog signaling pathway and autophagy in cancer. Int J Mol Sci 2018;19(8):2279.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Zhang H. Regulation of autophagy by mTOR signaling pathway. Adv Exp Med Biol 2019;1206:67-83.
[Crossref] [Google Scholar] [PubMed]
- Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin Cancer Biol 2019;59:125-32.
[Crossref] [Google Scholar] [PubMed]
- Miricescu D, Totan A, Stanescu-Spinu II, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR signaling pathway in breast cancer: From molecular landscape to clinical aspects. Int J Mol Sci 2020;22(1):173.
[Crossref] [Google Scholar] [PubMed]
- Bossler F, Hoppe-Seyler K, Hoppe-Seyler F. PI3K/AKT/mTOR signaling regulates the virus/host cell crosstalk in HPV-positive cervical cancer cells. Int J Mol Sci 2019;20(9):2188.
[Crossref] [Google Scholar] [PubMed]
- Fan T, Wang X, Zhang S, Deng P, Jiang Y, Liang Y, et al. NUPR1 promotes the proliferation and metastasis of oral squamous cell carcinoma cells by activating TFE3-dependent autophagy. Signal Transduct Target Ther 2022;7(1):321.
[Crossref] [Google Scholar] [PubMed]
- Ballesteros-Álvarez J, Andersen JK. mTORC2: The other mTOR in autophagy regulation. Aging Cell 2022;20(8):e13431.
[Crossref] [Google Scholar] [PubMed]
- Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J, et al. MiRNAs and cancer: Key link in diagnosis and therapy. Genes 2021;12(8):1289.
[Crossref] [Google Scholar] [PubMed]
- Zeng Y, Wang KX, Xu H, Hong Y. Integrative miRNA analysis identifies has-miR-3154, has-miR-7-3 and has-miR-600 as potential prognostic biomarker for cervical cancer. J Cell Biochem 2018;119(2):1558-66.
[Crossref] [Google Scholar] [PubMed]
- Enomoto H, Nakamura H, Nishikawa H, Nishimura T, Iwata Y, Nishiguchi S, et al. Hepatocellular carcinoma-associated microRNAs induced by hepatoma-derived growth factor stimulation. In Vivo 2020;34(5):2297-301.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Sun B, Zhao L, Liu Z, Xu Z, Tian Y, et al. Up-regulation of miRNA-148a inhibits proliferation, invasion and migration while promoting apoptosis of cervical cancer cells by down-regulating RRS1. Biosci Rep 2019;39(5).
[Crossref] [Google Scholar] [PubMed]
- Liao H, Pan Y, Pan Y, Shen J, Qi Q, Zhong L, et al. MicroRNA-874 is downregulated in cervical cancer and inhibits cancer progression by directly targeting ETS1. Oncol Rep 2018;40(4):2389-98.
[Crossref] [Google Scholar] [PubMed]
- Kabekkodu SP, Shukla V, Varghese VK, D'Souza J, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol Rev 2018;93(4):1955-86.
[Crossref] [Google Scholar] [PubMed]
- Dong RF, Zhuang YJ, Wang Y, Zhang ZY, Xu XZ, Mao YR, et al. Tumor suppressor miR-192-5p targets TRPM7 and inhibits proliferation and invasion in cervical cancer. Kaohsiung J Med Sci 2021;37(8):699-708.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Song Y, Mi Y, Jin H, Cao J, Li H, et al. microRNA-499a promotes the progression and chemoresistance of cervical cancer cells by targeting SOX6. Apoptosis 2020;25(3):205-16.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Song X, Kuang F, Zhang Q, Xie Y, Kang R, et al. NUPR1 is a critical repressor of ferroptosis. Nat Commun 2021;12(1):1-3.
[Crossref] [Google Scholar] [PubMed]
- Jiang L, Wang W, Li Z, Zhao Y, Qin Z. NUPR1 participates in YAP-mediate gastric cancer malignancy and drug resistance via AKT and p21 activation. J Pharm Pharmacol 2021;73(6):740-8.
[Crossref] [Google Scholar] [PubMed]
- Xiao H, Long J, Chen X, Tan MD. NUPR1 promotes the proliferation and migration of breast cancer cells by activating TFE3 transcription to induce autophagy. Exp Cell Res 2022:113234.
[Crossref] [Google Scholar] [PubMed]
- Hamidi T, Algül H, Cano CE, Sandi MJ, Molejon MI, Riemann M, et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest 2012;122(6):2092-103.
[Crossref] [Google Scholar] [PubMed]
- Li W, He P, Huang Y, Li YF, Lu J, Li M, et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 2021;11(1):222.
[Crossref] [Google Scholar] [PubMed]
- Fan T, Chen Y, He Z, Wang Q, Yang X, Ren Z, et al. Inhibition of ROS/NUPR1-dependent autophagy antagonises repeated cadmium exposure-induced oral squamous cell carcinoma cell migration and invasion. Toxicol Lett 2019;314:142-52.
[Crossref] [Google Scholar] [PubMed]
- Mu Y, Yan X, Li D, Zhao D, Wang L, Wang X, et al. NUPR1 maintains autolysosomal efflux by activating SNAP25 transcription in cancer cells. Autophagy 2018;14(4):654-70.
[Crossref] [Google Scholar] [PubMed]
- Augello G, Emma MR, Azzolina A, Puleio R, Condorelli L, Cusimano A, et al. The NUPR1/p73 axis contributes to sorafenib resistance in hepatocellular carcinoma. Cancer Lett 2021;519:250-62.
[Crossref] [Google Scholar] [PubMed]
- Li A, Li X, Chen X, Zeng C, Wang Z, Li Z, et al. NUPR1 silencing induces autophagy-mediated apoptosis in multiple myeloma cells through the PI3K/AKT/mTOR pathway. DNA Cell Biol 2020;39(3):368-78.
[Crossref] [Google Scholar] [PubMed]
- Nepstad I, Hatfield KJ, Grønningsæter IS, Reikvam H. The PI3K-Akt-mTOR signaling pathway in human acute myeloid leukemia (AML) cells. Int J Mol Sci 2020;21(8):2907.
[Crossref] [Google Scholar] [PubMed]
- Xue JF, Shi ZM, Zou J, Li XL. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed Pharmacother 2017;89:1252-61.
[Crossref] [Google Scholar] [PubMed]
- Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother 2018;103:699-707.
[Crossref] [Google Scholar] [PubMed]