- *Corresponding Author:
- Beibei Zhou
Department of Traditional Chinese Medicine,
The Fourth Hospital of Hebei Medical University,
Shijiazhuang,
Hebei, 050011,
China
E-mail: zhoubeibei2020@126.com
| Date of Received | 09 July 2020 |
| Date of Revision | 15 May 2021 |
| Date of Acceptance | 21 July 2021 |
| Indian J Pharm Sci 2021;83(4):742-749 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Lung cancer is one of the most deadly tumor disease in the world and most lung cancer is non-small cell lung cancer. MicroRNAs are considered as a variety of endogenous non-coding RNA, acting as interference during the process after transcription of gene. Recently, microRNA-142-5p, one of the microRNAs, was reported to enhance anti-tumor immune of non-small cell lung cancer cells by targeting programmed cell death receptor ligand 1. Programmed cell death receptor ligand 1 was also believed to be a regulator of cell proliferation and migration. However, the effect of microRNA-142-5p on proliferation and invasion of non-small cell lung cancer cells via regulating programmed cell death receptor ligand 1 was still unknown. Thus, this study demonstrated that there was low expression level of microRNA-142-5p in different nonsmall cell lung cancer cell lines. Programmed cell death receptor ligand 1 was proved to be the target gene of microRNA-142-5p and microRNA-142-5p also had negative correlation with programmed cell death receptor ligand 1. The high expression of microRNA-142-5p lead to suppression of proliferation and invasion in non-small cell lung cancer cells by down regulation of programmed cell death receptor ligand 1. The mechanism of microRNA-142-5p affecting proliferation and invasion of non-small cell lung cancer cells via targeting programmed cell death receptor ligand 1 might give a new thought of tumor therapy.
Keywords
Non-small cell lung cancer, microRNA-142-5p, programmed cell death receptor ligand 1, proliferation, immunity
Lung cancer is of the highest mortality rate in all tumor related disease and nearly 80 %~85 % lung cancer is non-small cell lung cancer (NSCLC), one type of lung cancer, with delayed accurate diagnosis, high degree of malignity and high rate of death, so early diagnosis and effective therapy need to be developed [1-4]. In recent years, microRNAs (miRNAs) become a new research focus of tumor targeted therapy [5]. Many studies have showed that miRNAs, such as miRNA-221, miRNA-223, miRNA-615-3p and etc., have the potential to be used in NSCLC therapy, for working as biomarkers or therapeutic targets [6-8]. The application of miRNAs in NSCLC is involved with expression of multiple genes, because miRNAs are a type of small non-coding RNA working as interference of the gene expression process after transcription, according to the strict complementary nature of the base pairs [9-11]. Programmed cell death receptor ligand 1 (PD-L1) is target gene of many miRNAs, which reduces the immune response induced by T cell and regulates biological behavior of tumor cell to affect genesis and development of NSCLC [12,13]. Jia et al. [14] have found that miR-142-5p regulated expression of PD-L1 enhancing the antitumor immunity of NSCLC cell. However, there is no research about the effect of miR-142-5p on proliferation and invasion of NSCLC cell through targeting PD-L1. Thus, our study aims to investigate the effect of miR-142-5p on proliferation and invasion of NSCLC cell through targeting PD-L1, in the hope of providing a new biological marker and a novel therapy for clinical diagnosis and treatment of NSCLC.
Materials and Methods
Cell culture:
Human normal bronchial epithelium cell line BEAS-2B, and human NSCLC cell lines A549, NCI-H1299 and NCI-H1650, were purchased from KeyGEN BioTECH (Nanjing, China) and cultured in Dulbecco's modified eagle medium (DMEM) added with 10 % fetal bovine serum (FBS) at 37° in a humidified 5 % CO2 incubator.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) for objective cell for transfection selection:
Total RNA of BEAS-2B, A549, NCI-H1299 and NCI-H1650 cells was extracted with TRIzol reagent (Invitrogen, USA). To execute qRT-PCR, total RNA was reverse transcribed to complementary DNAs (cDNA) by using PrimeScript™ RT reagent Kit (Takara, Japen). qRT-PCR analyses were conducted with TB Green® Premix Ex Taq™ kit (Takara, Japen). The prime sequence is shown in Table 1. The PCR system was 20 μl in total as follows: 2X TB Green Premix Ex Taq 10 μl, PCR forward primer 0.4 μl, PCR reverse primer 0.4 μl, 6-carboxy-X-rhodamine (ROX) reference dye 0.4 μl, cDNA 2 μl, double distilled water (ddH2O) 6.8 μl. PCR reaction conditions: pre-denaturation at 95° for 30 s, denaturation at 95° for 5 s, annealing at 60° for 30 s, totaling 40 cycles. All the protocols were carried out according to the instruction of manufacture. U6 was set as the internal reference for miR-142-5p and BEAS-2B cell was as control group. Comparative threshold cycle (Ct) values were analyzed by 2-ΔΔct. Each detection was performed in triplicate and the results were averaged.
Transfection of cells:
According to the qRT-PCR result, A549 cell was selected for transfection. MiR-negative control (miRNC), miR-142-5p mimic and miR-142-5p inhibitor were purchased from Applied Biosystems, USA. A549 cells were seeded in 24-well plates which added with DMEM containing 10 % FBS. When the cells reached to 80 % confluence, A549 was transfected with miR-NC, miR-142-5p mimic and miR-142-5p inhibitor by using Lipofectamine 2000 (Invitrogen, USA), respectively. After transfection for 6 h, the original medium was replaced with fresh medium.
Dual luciferase assay:
A biological prediction of miRNA targets website, TargetScan (http://www.targetscan.org/vert_72/), was used to predict the target sites of miR-142-5p on PDL1 (fig. 2A), and Luciferase Reporter Gene Detection Kit (Sigma, USA) was used to verify whether miR-142- 5p targeted PD-L1. As shown in fig. 2B, the wild type PD-L1 (WT-PD-L1) 3’-UTR luciferase reporter vector was constructed, accompanied with the mutated type of the putative miR-142-5p-binding site in the 3’-UTR region of PD-L1 designed. WT-PD-L1 3’-UTR plasmid was co-transfected with miR-NC or miR-142-5p mimic into A549 cell, while mutant type PD-L1 (MUTPD- L1) 3’-UTR plasmid was co-transfected with miR-NC or miR-142-5p mimic into A549 cell. After transfection for 6 h, the original medium was replaced with fresh medium. After transfection for 24 h, the cells were lysed for dual luciferase reporter gene detection. The fluorescence value of renilla plasmid was used as the internal reference. Relative luciferase activity was obtained from the ratio of firefly to renilla luciferase. Each detection was performed in triplicate and the results were averaged.
qRT-PCR of miR-142-5p expression in transfected cells:
Total RNA of transfected cells was extracted with TRIzol reagent (Invitrogen, USA). To execute qRT-PCR, total RNA was reverse transcribed to cDNA by using PrimeScript™ RT reagent Kit (Takara, Japen). qRT-PCR analyses were conducted with TB Green® Premix Ex Taq™ kit (Takara, Japen). The prime sequence is shown in Table 1. All the protocols were carried out according to the instruction of manufacture. U6 was set as the internal reference for miR-142-5p, and miR-NC was as control group. Comparative Ct values were analyzed by 2-ΔΔct. Each detection was performed in triplicate and the results were averaged.
| Primer sequence | |
|---|---|
| miR-142-5p | forward primer(5′-3′):CAUAAAGUAGAAAGCACUACU |
| reverse primer(5′-3′):UAGUGCUUUCUACUUUAUGUU | |
| U6 | forward primer(5′-3′):CTCGCTTCGGCAGAC |
| reverse primer(5′-3′):AACGCTTACGAATTT |
Table 1: Primer Sequences Of MIR-142-5P and U6 PCR
Western blotting of PD-L1 expression in transfected cells:
After transfection for 48 h, the transfected A549 cells were lysed and total proteins were collected by using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, China). Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate proteins, and then transferred the proteins to a polyvinylidene fluoride (PVDF) membrane. After the membrane was incubated with 5 % non-fat dried milk block for 90 min, PD-L1 (1:600) and beta (β)-Actin (1:2000, Abcam, UK) antibodies were used to incubate with PVDF membrane at 4° overnight. The next day added with anti-rabbit Immunoglobulin G (IgG) secondary antibody conjugated to horseradish peroxidase (1:1000, Abcam, UK). After incubation for 1 h, the electrochemiluminescence (ECL) detection system (Thermo fisher scientific, USA) was used for detection. β-Actin was probed as a loading control, and Image-Pro plus 6.0 (Media Cyber-netics, USA) was used to analyze the gray value of the bands. Each detection was performed in triplicate and the results were averaged.
3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) assay:
The transfected A549 cells were seeded in 96-well plates at 5×103/well, and placed in an incubator at 37° and 5 % CO2. 20 μl MTT solution (5 g/l) was added to each well, after cells were cultured for 12 h, 24 h, 36 h and 48 h, respectively. For each time for addition of MTT solution, with further incubation for 4 h, the supernatant was carefully discarded and 150 μl dimethyl sulfoxide (DMSO) solution was added to each hole, and then used for detection. The 96-well plate was placed on a shaker shaking for 15 min in the dark, and then absorbance values of each well were detected at the wavelength of 490 nm using PR 4100 microplate reader (Bio-Rad, USA). Each detection was performed in triplicate, and the results were averaged.
Cell invasion assay:
The transfected A549 cells after transfection for 48 h were collected, adjusted to 5×104/well, and inoculated on 6-well plates. Rinse with phosphate buffered saline (PBS) twice, cells were inoculated in the upper chamber where 200 μl DMEM culture medium was added, and 500 ml DMEM containing 10 % FBS was added to the lower chamber. After incubation at 37° for 48 h, the substrates and cells that did not pass through the membrane surface in the upper chamber were wiped, washed with PBS for 3 times, fixed with 4 % paraformaldehyde for 20 min, and washed with ddH2O for 3 times. After the process of drying, the cells were stained with 0.5 % crystal violet, and the cell invasion was observed. 5 sights under the microscope were randomly selected. Each detection was performed in triplicate and the results were averaged.
Statistical analysis:
Statistical package for the social sciences (SPSS) v22.0 (IBM, USA) was used to perform statistics and analysis of experimental data. Data were expressed as the mean±standard deviation (SD) (x?±s). Statistical significance of the means between two independent groups was analyzed by Student’s t-test (two-tailed) or one-way analysis of variance (ANOVA), while two-way ANOVA was used for comparisons among multiple groups, with p<0.05 considered statistically significant.
Results and Discussion
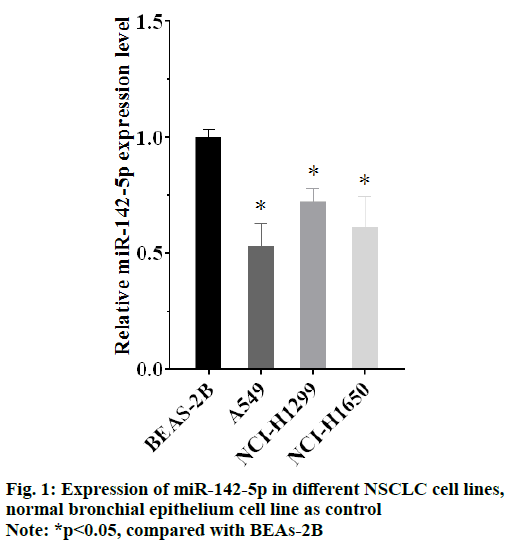
The results of qRT-PCR showed that there was lower expression of miR-142-5p in all the NSCLC cells (p<0.05), compared with BEAS-2B cell, the human normal bronchial epithelium cell line. It indicated that NSCLC was connected with abnormality expression of miR-142-5p. Moreover, expression of miR-142-5p in A549 cell line was lower than other NSCLC cells, as shown in fig. 1. A549 cell was selected as object for the following experiment, for its lowest expression of miR-142-5p.
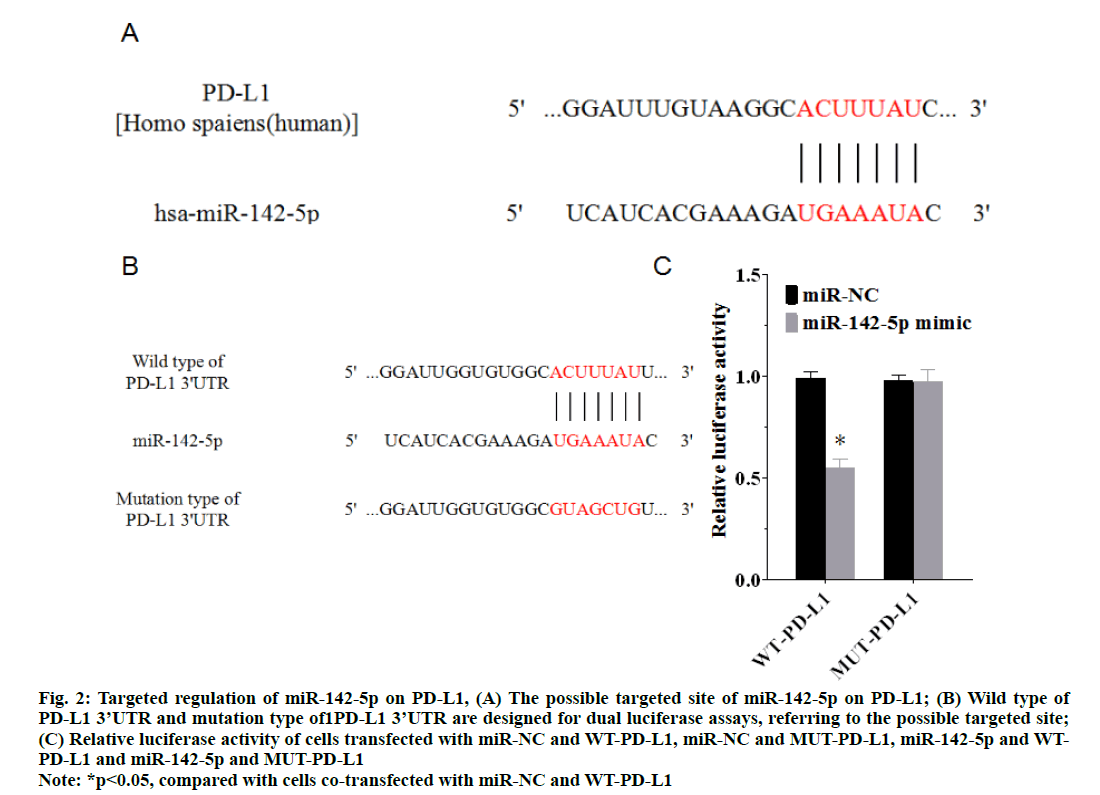
According to TargetScan, a biological prediction of miRNA targets website, there is a possible target site of miR-142-5p on the PD-L1 3’UTR, as shown in fig. 2A. A luciferase reporter assay was carried out to prove that PD-L1 was the target gene of miR-142-5p. The result of luciferase reporter assay demonstrated that the luciferase activity in cells co-transfected with miR-142-5p mimic and WT-PD-L1 was significantly reduced (p<0.05), in comparison with cells co-transfected with miR-NC and WT-PD-L1, whereas no significant difference was identified in cells co-transfected with miR-142-5p mimic and MUT-PD-L1 (p>0.05) (fig. 2C). These results showed that PD-L1 was a target gene of miR-142-5p, because miR-142-5p mimic directly regulate the expression of WT-PD-L1 differing from other groups.
Figure 2: Targeted regulation of miR-142-5p on PD-L1, (A) The possible targeted site of miR-142-5p on PD-L1; (B) Wild type of PD-L1 3’UTR and mutation type of1PD-L1 3’UTR are designed for dual luciferase assays, referring to the possible targeted site; (C) Relative luciferase activity of cells transfected with miR-NC and WT-PD-L1, miR-NC and MUT-PD-L1, miR-142-5p and WTPD- L1 and miR-142-5p and MUT-PD-L1 *p<0.05, compared with cells co-transfected with miR-NC and WT-PD-L1
Note: *p<0.05, compared with cells co-transfected with miR-NC and WT-PD-L1
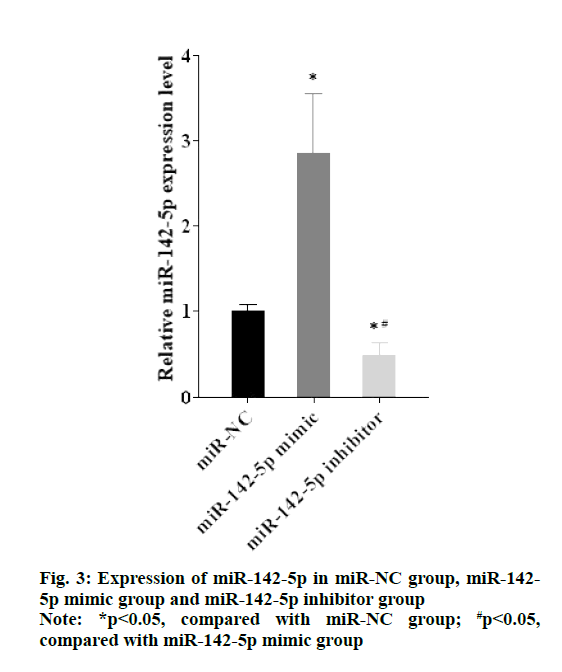
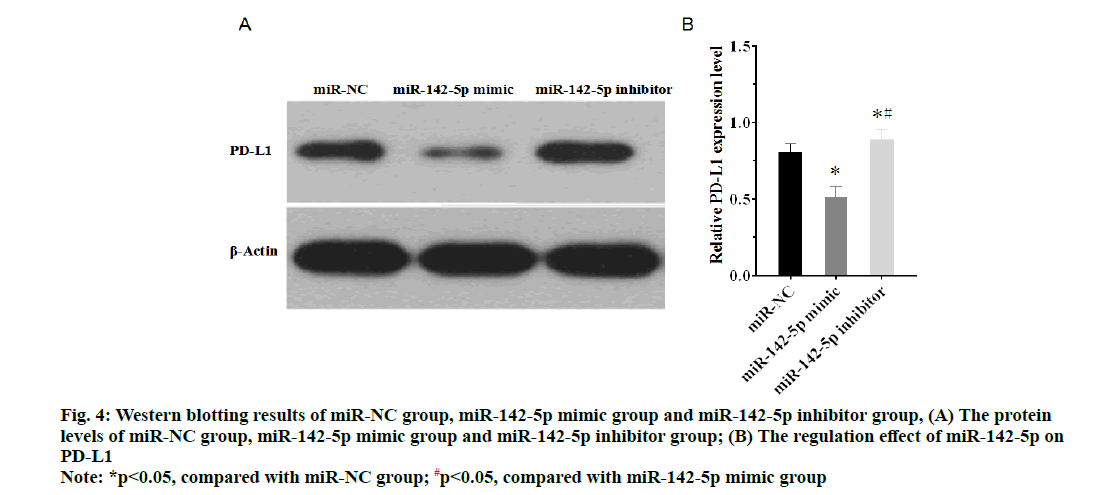
As shown in the fig. 3, expression of miR-142-5p in miR-142-5p mimic group was higher than miR-NC group, while expression of miR-142-5p in miR-142-5p inhibitor group was lower than miR-NC group (p<0.05). The results of western blotting manifested that PD-L1 highly expressed in miR-142-5p inhibitor group compared with miR-NC group, but in the miR-142-5p mimic group, expression of PD-L1 was obviously suppressed in comparison with miR-NC group (fig. 4A and fig. 4B). The findings suggested that miR-142-5p had a negative correlation with PD-L1, as higher the expression of miR-142-5p, the lower the expression of PD-L1.
Figure 4: Western blotting results of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group, (A) The protein levels of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group; (B) The regulation effect of miR-142-5p on PD-L1
Note: *p<0.05, compared with miR-NC group; #p<0.05, compared with miR-142-5p mimic group
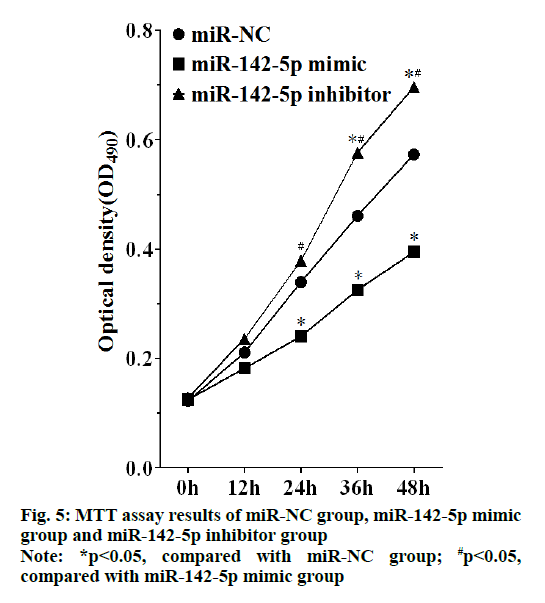
MTT assay showed that cell proliferation was promoted in miR-142-5p inhibitor group, which had statistically significant differences with miR-NC group and miR-142-5p mimic after culture for 24 h (p<0.05), and cell proliferation of miR-142-5p mimic group was highly suppressed, as there was statistically significant differences between miR-142-5p mimic group and miR-NC group after incubation for 36 h (p<0.05), as shown in the fig. 5. Therefore, miR-142-5p could restrain cell proliferation of NSCLC by suppressing PD-L1.
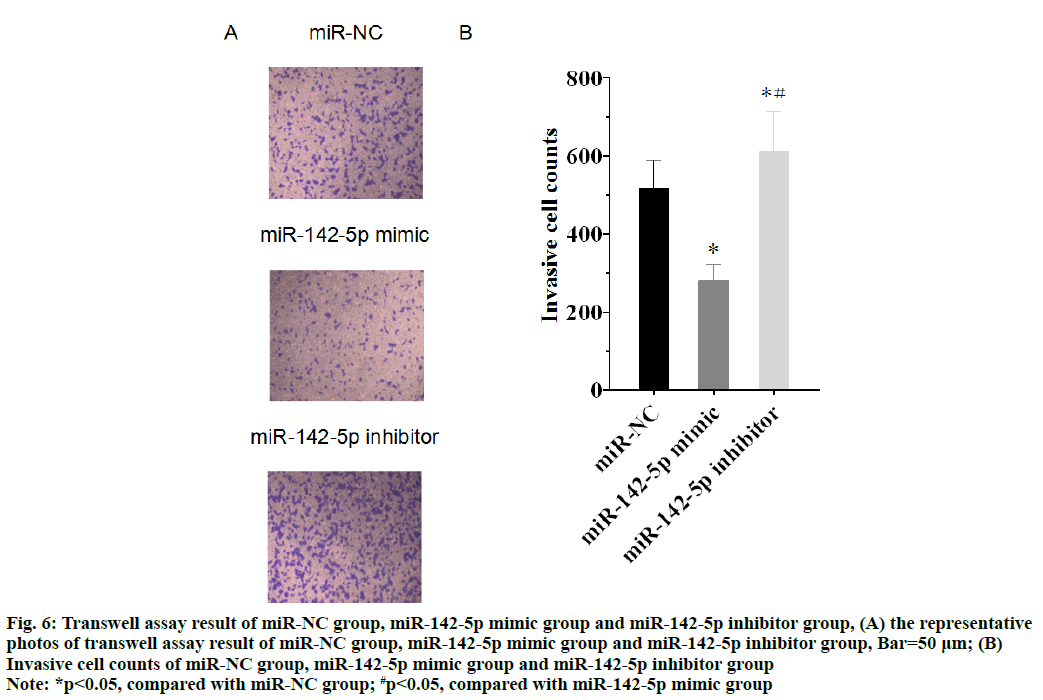
The invasive cell counts of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group were respectively 516.61±72.87, 283.52±38.99 and 613.41±100.48. As shown in the fig. 6A and fig. 6B, the cell invasion was inhibited in miR-142-5p mimic group, whereas the invasive cell counts increased in miR-142-5p inhibitor group, compared with miR-NC group (p<0.05). The result proved that miR-142-5p had the ability to inhibit NSCLC cell invasion by suppression of PD-L1.
Figure 6: Transwell assay result of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group, (A) the representative photos of transwell assay result of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group, Bar=50 μm; (B) Invasive cell counts of miR-NC group, miR-142-5p mimic group and miR-142-5p inhibitor group
Note: *p<0.05, compared with miR-NC group; #p<0.05, compared with miR-142-5p mimic group
Lung cancer is the most deadly cancer in the worldwide, and NSCLC is the main type of different lung cancers [15]. Increasing researches of NSCLC help with the understanding of the molecular changes affecting the occurrence and development of NSCLC, which has revolutionized the medicine and therapy applied to treating NSCLC [16-18]. As we all know, gene is related to genesis and progression of multiple diseases, which especially plays an important role in a variety of cancers [19,20]. The mechanisms of cancer affected by gene is different, as genes are divided into oncogenes and antioncogenes which promote or suppress the cancer, respectively [21-23]. These genes always connected with expressions of cytokines and receptors that regulate diverse cell activities, such as proliferation, differentiation, migration and apotosis [24,25]. What’s more, some cytokines and its receptors affect various types of cancers, according to the similar mechanism, so it’s crucial for tumor therapy to figure out the mechanism between gene and cancer. For a instance, PD-1 and PD-L1 are now the most eye-catching targets of cancer therapy which have been used in treatment of lung cancer, gastric cancer, breast cancer and etc., and many blockades of PD-1 and PD-L1 are already developed or during experiment, and all of them aim to inhibit PD-1/PD-L1 pathway, to activate the immune system to eliminate tumor cells [26-29]. To regulate the cytokines and receptors needs to affect the expression of gene in transcription or translation. In these years, researchers suggest that miRNAs are a bunch of non-coding RNAs (ncRNA) and considered as crucial regulators of mRNA after transcription [30,31]. MiRNAs result in the interference of mRNA translation by binding with mRNA, according to the principle of complementary base pairing, and miR-142-5p is one of those miRNAs, which is reported to be related to several cancers [32-34]. Besides, Wan et al. [35] found that miR-142-5p could enhance the anti-tumor immunity of NSCLC by regulating PD-L1. Thus, we hypothesized that miR-142-5p might have effects on proliferation and invasion of NSCLC cell via targeting PD-L1, and designed miR-NC, miR-142-5p mimic and miR-142-5p inhibitor for following experiments.
As different NSCLC cell lines usually have different gene expression levels, the comparison of miR-142-5p expression in different NSCLC cells is used to select the suitable NSCLC cell for following experiments, and human normal bronchial epithelium cell as control to see whether the expression level of PD-L1changes and how much it has changed. All the NSCLC cells show lower expression of miR-142-5p than normal bronchial epithelium cell which means that there is down-regulation of miR-142-5p in NSCLC cells. A549 cell is selected for transfection, because it has the lowest miR-142-50 level in NSCLC cells.
By using the miRNA targets website, TargetScan, a possible target site of miR-142-5p on the PD-L1 3’ UTR was determined and dual luciferase assays were executed to identify the targeting relation between miR-142-5p and PD-L1. The results of assays proved the target site was reliable, which also identified that PD-L1 was the target gene of miR-142-5p. Next, western blotting shows that inhibition of miR-142-5p causes higher expression of PD-L1, while up-regulation of miR-142-5p suppresses PD-L1 expression. The results manifest that miR-142-5p has a negative correlation with PD-L1 in NSCLC cell, which probably is the mechanism of miR-142-5p affects NSCLC cells, because the higher expression of miR-142-5p will cause lower expression level of PD-L1.
In this study, the results of MTT assays and transwell assays indicate that miR-142-5p suppresses the proliferation and invasion of NSCLC cells. NSCLC cells with high expression of miR-142-5p lose partial capability of proliferation, and high expression of miR-142-5p also makes the ability of migration weakened in transwell assays. As miR-142-5p could regulate expression of PD-L1, and Xie et al. [36] has found that PD-L1 expression induces cell proliferation in NSCLC cells. Therefore, we could conclude that miR-142-5p suppresses proliferation of NSCLC cells via targeting PD-L1. A previous study demonstrated that PD-L1 might drive the invasion and metastasis of NSCLC cells in vivo [37]. Combining this study result and our experiment results, the findings suggest that miR-142-5p down-regulates invasion and migration of NSCLC cells via targeting PD-L1. PD-1 and PD-L1 are believed to be important targets of immunotherapy, and PD-L1 is the main ligand of PD-1 which express on many types of cells, especially on tumor cells such as lung cancer [38]. PD-L1 plays an important role in escaping of tumor cells from the immune response, because its suppressing effect on the maturation of tumor-infiltrating dendritic cells [39-40]. Thus, blockade of interaction between PD-1 and PD-L1 is considered as a promising immunotherapy, which has been applied in clinic. In our study, miR-142-5p has the potential to be one of the PD-1/PD-L1 inhibitors for immune therapy, which needs further investigation to prove its availability for diagnosis or treatment.
In conclusion, PD-L1 is the target gene of miR-142-5p and there might be a negative correlation between them. Overexpression of miR-142-5p suppresses proliferation and invasion of NSCLC cell via targeting PD-L1. The findings of this study might provide promising novel targets of clinical treatment and pharmaceutical development, which could help with enhance the therapeutic effect and reduce the side effect of cancer treatment.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553(7689):446-54.
- Reck M, Rabe KF. Precision diagnosis and treatment for advanced non–small-cell lung cancer. New Eng J Med 2017;377(9):849-61.
- de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res 2018;7(3):220-3.
- Deng HY, Wang YC, Ni PZ, Li G, Yang XY, Lin YD, et al. Radiotherapy, lobectomy or sublobar resection? A meta-analysis of the choices for treating stage I non-small-cell lung cancer. Eur J Cardiothorac Surg 2017;51(2):203-10.
- Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem 2017;8(1):45-56.
- Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene 2017;620:46-53.
- Ma HP, Kong WX, Li XY, Li W, Zhang Y, Wu Y. miRNA-223 is an anticancer gene in human non-small cell lung cancer through the PI3K/AKT pathway by targeting EGFR. Oncol Rep 2019;41(3):1549-59.
- Liu J, Jia Y, Jia L, Li T, Yang L, Zhang G. MicroRNA 615-3p inhibits the tumor growth and metastasis of NSCLC via inhibiting IGF2. Oncol Res 2019;27(2):269-79.
- Arab A, Karimipoor M, Irani S, Kiani A, Zeinali S, Tafsiri E, et al. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet 2017;216:150-8.
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol 2017;15(1):1.
- Liao J, Shen J, Leng Q, Qin M, Zhan M, Jiang F. MicroRNA?based biomarkers for diagnosis of non?small cell lung cancer (NSCLC). Thorac Cancer 2020;11(3):762-8.
- Seifert AM, Zeng S, Zhang JQ, Kim TS, Cohen NA, Beckman MJ, et al. PD-1/PD-L1 blockade enhances T-cell activity and antitumor efficacy of imatinib in gastrointestinal stromal tumors. Clin Cancer Res 2017;23(2):454-65.
- Jiang L, Su X, Zhang T, Yin X, Zhang M, Fu H, et al. PD-L1 expression and its relationship with oncogenic drivers in non-small cell lung cancer (NSCLC). Oncotarget 2017;8(16):26845.
- Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun 2017;488(2):425-31.
- Bao SM, Hu QH, Yang WT, Wang Y, Tong YP, Bao WD. Targeting epidermal growth factor receptor in non-small-cell-lung cancer: current state and future perspective. Anticancer Agents Med Chem 2019;19(8):984-91.
- Barr Kumarakulasinghe N, Zanwijk NV, Soo RA. Molecular targeted therapy in the treatment of advanced stage non?small cell lung cancer (NSCLC). Respirology 2015;20(3):370-8.
- Tsoulos N, Papadopoulou E, Metaxa-Mariatou V, Tsaousis G, Efstathiadou C, Tounta G, et al. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncol Rep 2017;38(6):3419-29.
- Schrank Z, Chhabra G, Lin L, Iderzorig T, Osude C, Khan N, et al. Current molecular-targeted therapies in NSCLC and their mechanism of resistance. Cancers 2018;10(7):224.
- Lu CY, Loomer S, Ceccarelli R, Mazor KM, Sabin J, Clayton EW, et al. Insurance coverage policies for pharmacogenomic and multi-gene testing for cancer. J Pers Med 2018;8(2):19.
- Kikkawa A. Random matrix analysis for gene interaction networks in cancer cells. Sci Rep 2018;8(1):1-2.
- Biswas S, Rao CM. Epigenetics in cancer: fundamentals and beyond. Pharmacol Ther 2017;173:118-34.
- Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett 2017;396:130-7.
- Min HY, Lee HY. Oncogene-driven metabolic alterations in cancer. Biomol Ther 2018;26(1):45-56.
- Zhang E, Xu H. A new insight in chimeric antigen receptor-engineered T cells for cancer immunotherapy. J Hematol Oncol 2017;10(1):1.
- Moldawer LL, Rogy MA, Lowry SF. The role of cytokines in cancer cachexia. J Parenter Enter Nutr 1992;16:43S-9S.
- Li C, Zhang N, Zhou J, Ding C, Jin Y, Cui X, et al. Peptide blocking of PD-1/PD-L1 interaction for cancer immunotherapy. Cancer Immunol Res 2018;6(2):178-88.
- Jing W, Li M, Zhang Y, Teng F, Han A, Kong L, et al. PD-1/PD-L1 blockades in non-small-cell lung cancer therapy. Onco Targets Ther 2016;9:489-502.
- Saito H, Kono Y, Murakami Y, Shishido Y, Kuroda H, Matsunaga T, et al. Highly activated PD-1/PD-L1 pathway in gastric cancer with PD-L1 expression. Anticancer Res 2018;38(1):107-12.
- Schütz F, Stefanovic S, Mayer L, von Au A, Domschke C, Sohn C. PD-1/PD-L1 pathway in breast cancer. Oncol Res Treat 2017;40(5):294-7.
- Jiang L, Yu X, Ma X, Liu H, Zhou S, Zhou X, et al. Identification of transcription factor-miRNA-lncRNA feed-forward loops in breast cancer subtypes. Comput Biol Chem 2019;78:1-7.
- Pradhan AK, Emdad L, Das SK, Sarkar D, Fisher PB. The enigma of miRNA regulation in cancer. Adv Cancer Res 2017;135:25-52.
- Seok H, Lee H, Jang ES, Chi SW. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol Life Sci 2018;75(5):797-814.
- Wong JJ, Au AY, Gao D, Pinello N, Kwok CT, Thoeng A, et al. RBM3 regulates temperature sensitive miR-142–5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res 2016;44(6):2888-97.
- Xu R, Bi C, Song J, Wang L, Ge C, Liu X, et al. Upregulation of miR-142-5p in atherosclerotic plaques and regulation of oxidized low-density lipoprotein-induced apoptosis in macrophages. Mol Med Rep 2015;11(5):3229-34.
- Wan J, Ling X, Peng B, Ding G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol Rep 2018;40(1):272-82.
- Xie WB, Liang LH, Wu KG, Wang LX, He X, Song C, et al. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem 2018;46(2):654-63.
- Lin PL, Wu TC, Wu DW, Wang L, Chen CY, Lee H. An increase in BAG-1 by PD-L1 confers resistance to tyrosine kinase inhibitor in non–small cell lung cancer via persistent activation of ERK signalling. Eur J Cancer 2017;85:95-105.
- Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC). J Immunother Cancer 2018;6(1):1-27.
- Xu-Monette ZY, Zhang M, Li J, Young KH. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response?. Front Immunol 2017;8:1597.
- Cho YA, Yoon HJ, Lee JI, Hong SP, Hong SD. Relationship between the expressions of PD-L1 and tumor-infiltrating lymphocytes in oral squamous cell carcinoma. Oral Oncol 2011;47(12):1148-53.