- *Corresponding Author:
- M. L. Chen
Department of Psychiatry
Fu Jen Catholic University
New Taipei City, Taiwan
E-mail: cater0656@hotmail.com
| Date of Accepted | 02 April 2020 |
| Date of Revised | 13 February 2020 |
| Date of Received | 06 August 2019 |
| Indian J Pharm Sci 2020;82(3):409-419 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In the present investigation antipsychotic drug-induced miRNA modulations in glial cells were analyzed through miRNA expression in antipsychotic drugs-treated C6 cells using a miRNA microarray. Seventeen miRNAs were upregulated by haloperidol and 10 miRNAs were upregulated by risperidone in C6 cells. Thirty-six miRNAs were downregulated by clozapine in C6 cells. Six miRNAs (rno-let-7a-5p, rno-miR-221-3p, rno-miR-152-3p, rno-miR-345-5p, rno-miR-3473 and rno-miR-551b-5p) were selected for validation by reverse transcription-quantitative polymerase chain reaction, the results of which showed almost the same expression trends as the microarray results. mRNA and protein expression of potential targets for some validated miRNAs was examined using reverse transcription-quantitative polymerase chain reaction, western blot analysis and immunofluorescence staining. Reduction of miRNA (rno-let-7a-5p, rno-miR-152-3p and rno-miR-221-3p) caused by clozapine treatment could further induce expressions of predicted target genes/proteins (atg12, cdkn1b, c-myc and K-ras) in C6 cells. miRNAs regulation induced by haloperidol or risperidone were not closely related to predicted target genes/proteins expressions. It was concluded that the regulatory trends between antipsychotic drug-induced miRNA and the mRNA/protein expression of potential targets of the examined miRNAs were not quite the same. Clozapine, but not haloperidol or risperidone, could modulate miRNA expression to further regulate protein expression in C6 cells.
Keywords
Haloperidol, Risperidone, Clozapine, MicroRNA, Microarray
MicroRNAs (miRNAs) constitute one of the two major classes of noncoding RNA molecules. miRNAs are 22-24 nucleotides in length and are found in eukaryotic cells. miRNAs function in transcriptional and posttranscriptional regulation of gene expression. miRNAs are encoded by nuclear deoxyribonucleic acid (DNA) and act by base-pairing with complementary sequences in mRNA molecules, resulting in gene silencing via translational repression or target mRNA degradation[1,2]. The human genome may encode over 1000 miRNAs, which target approximately 60 % of mammalian genes and are abundant in many human cell types[3,4]. Animal miRNAs typically present only partial complementarity to their mRNA targets[5]. A given miRNA may have multiple different mRNA targets and a given target might similarly be targeted by multiple miRNAs. miRNA studies have revealed multiple roles in negative regulation (through transcript degradation and translational suppression) and possible involvement in positive regulation (through transcriptional and translational activation). miRNAs are likely to participate in most biological processes by affecting gene regulation[6]. miRNAs have been revealed that can regulate the nervous system[7]. Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (which involves miR- 132, miR-134, and miR-124), synapse formation and synapse maturation (in which miR-134 and miR- 138 are thought to be involved)[8]. Some studies have found altered miRNA expression in schizophrenia. Accumulated evidence has revealed that miRNAs are potential targets for studying the pathogenesis of psychotic disorders. Recent studies have demonstrated that artificially synthesized miRNAs can affect the synaptic plasticity of oligodendrocytes and cognitive function by downregulating Nogo-A expression in transgenic rats[9]. Various genome-wide studies have mentioned correlations between miRNA-137 and early age of onset of psychosis[10]. Additionally, miRNA-382 expression has been found to be elevated in the olfactory neuroepithelia of schizophrenia patients[11]. Polymorphisms in the 3’ untranslated regions of COMT and RGS4, which are two schizophrenia-related genes, have been reported to affect miRNA binding on the genes[12]. These results suggest that miRNAs are highly correlated with psychotic disorders.
Antipsychotic drugs (APDs) can ease the psychotic symptoms of schizophrenic patients in the clinic. Various studies have reported that APDs might modulate the biological functions of cells by regulating miRNA expression. Perkins et al. found that haloperidol increased miR-199a, miR-128a and miR-128b expression in rats[13]. Other studies have also revealed that APDs can induce various miRNA regulations and further trigger signaling cascades in rat brains or neuronal cell lines[14-16]. MiR-22 and miR-29b involved in DNMT1-, DNMT3A-, DNMT3B- and MBD2- derived global methylation of DNA have been found to be induced by haloperidol[16]. A recent study also mentioned that chronic treatment of rats with clozapine induced miRNA 132 expression and further decreased enhancer of Zeste 1 (EZH1) expression, contributing to behavioral abnormalities and gene expression changes[17].
Clinical evidence indicated that the pathogenesis of schizophrenia might be closely related to disturbed functions and abnormalities of glial cells in schizophrenia[18-20]. Increasing numbers of pharmacological and animal studies have also shown the importance of glial cells in schizophrenia. In vivo evidence has also revealed the differential impacts of typical and atypical antipsychotics on intracortical myelin in adults with schizophrenia[21-24]. It is believed that dysfunction of glial cells in the central nervous system might play an important role in the development of psychiatric disease. Moreover, the complexity of cell types in brain tissue might mask the effects of APDs and further mislead researchers with regards to APDinduced gene/protein regulation in different cell types.
To investigate the effects of APDs on the modulation of miRNA expression and related target protein regulation in glial cells, the miRNA expression profiles of C6 cells treated with haloperidol, risperidone or clozapine were analyzed using an Agilent miRNA microarray chip. It was found that APDs could regulate various miRNAs. The expression of some miRNAs was validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and the results were found to be consistent with the microarray results. Additionally, western blot analysis and cell immunofluorescence staining was used to examine the relations between miRNA expression and miRNA target protein regulation in C6 cells.
Materials and Methods
Cell culture and APD treatment:
C6 cells obtained from the Bioresource Collection and Research Center (BCRC) of the Food Industry Research and Development Institute (FRDI), Taiwan, were maintained in high-glucose DMEM (Gibco, Inc., NY, USA) supplemented with 26 mM NaHCO3, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 10 % horse serum and 2 % FBS (Gibco, Inc.). Haloperidol, risperidone or clozapine was added to the medium daily for 5 d. The final concentrations of haloperidol (2 μg/ml), risperidone (4 μg/ml) and clozapine (25 μg/ml) were calculated based on therapeutic concentrations in the brain. Risperidone used in this study was purchased from Janssen Pharmaceutica (Beerse, Belgium) while clozapine and haloperidol were purchased from Sigma-Aldrich (St. Louis MO, USA).
Total RNA preparation and Agilent Rat R19 miRNA array analysis:
Total RNA from APD-treated and untreated C6 cells was extracted using TRIzol RNA isolation reagent according to the manufacturer’s instructions (Invitrogen Life Technologies, CA, USA). In brief, C6 cells were lysed in 1 ml of TRIzol reagent, incubated for 5 min on ice, vortexed and centrifuged at 12 000 rpm at 4° for 10 min. The supernatant was removed to a fresh tube, mixed with 200 μl of chloroform, incubated for 5 min on ice and centrifuged at 12 000 rpm at 4º for 10 min. The upper layer of the solution was mixed with 2-propanol and the RNA was collected by centrifugation at 12 000 rpm at 4º for 20 min. The RNA pellet was washed twice with 70 % ethanol, airdried and dissolved in 1 mM Na citrate solution. Two independent batches of APD-induced miRNAs from APD-treated C6 cells were analyzed using an Agilent Rat R19 miRNA array (Agilent Technologies, CA, USA). RNA labeling, hybridization, image scanning and raw data collection were performed by the service department of Welgene Biotech Co., Ltd. (Taiwan, ROC.) (https://www.welgene.com.tw) according to the technical recommendations of Agilent Technologies. The APD-induced miRNA modulation profiles were also analyzed by Welgene Biotech Co., Ltd. using GeneSpring 11.5.
RT-qPCR:
Reverse transcription (RT) reactions were performed using a SuperScript III Reverse Transcription Kit (Invitrogen Life Technologies) according to the manufacturer’s instructions. For each of the tested miRNAs, specific stem-loop RT primers, forward primers and universal reverse primers were designed according to Chen et al.[25], and the primers are listed in Table 1. The sequences of the PCR primers for the tested target genes are listed in Table 2. U6 snRNA was used as an internal control to calibrate target miRNA expression and actin was used as an internal control to calibrate the expression of the tested genes. For each of the tested target genes, random hexamers were used to produce cDNA in the reverse transcription reaction. The cDNA was then diluted with an equal volume of ddH2O and stored for further analysis. The cDNA was diluted and added to a mixture containing SYBR Green PCR Master Mix (Applied Biosystems, CA, USA). The reactions were carried out using an ABI 7900 Real-time PCR detection system. The general PCR conditions included 10 min of polymerase activation at 95° followed by 50 cycles of denaturation at 95° for 15 s, primer annealing at 60° for 30 s, and elongation at 72° for 60 s. The relative quantification method was used to obtain ΔΔCt values to calculate changes in the expression of miRNAs or tested target genes. The data were collected from four independent batches of experiments for miRNAs or from 3 batches for tested target genes. The normalized expression ratios of the examined miRNAs or tested genes were assessed using one-way ANOVA followed by the Dunnett post hoc test. One-way ANOVA and the Dunnett post hoc test were performed with SPSS Statistics 17.0.
| miRNA | HAL | RIS | CLO |
|---|---|---|---|
| rno-miR-103-3p, rno-miR-107-3p, rno-miR-148b-3p, rno-miR-17-5p, rno-miR-185-5p, rno-miR-31a-5p, rno-miR-335, rno-miR-345-5p*, rno-miR-466b-5p, rno-miR-542-5p, rno-miR-9a-5p | + | ||
| rno-miR-324-5p, rno-miR-3473*, rno-miR-34a-5p, rno-miR-455-5p, rno-miR-551b-5p* | + | + | |
| rno-miR-152-3p* | + | - | |
| rno-miR-20a-3p, rno-miR-290, rno-miR-34b-5p, rno-miR-34c-5p, rno-miR-532-5p | + | ||
| rno-let-7a-5p*, rno-let-7f-5p, rno-miR-100-5p, rno-miR-125b-5p, rno-miR-128-3p, rno-miR-129-2-3p, rno-miR-146a-5p, rno-miR-146b-5p, rno-miR-151-5p, rno-miR-15b-5p, rno-miR-196b-5p, rno-miR-199a-3p, rno-miR-218a-5p, rno-miR-221-3p*, rno-miR-22-3p, rno-miR-22-5p, rno-miR-25-3p, rno-miR-26b-5p, rno-miR-28-5p, rno-miR-322-3p, rno-miR-325-3p, rno-miR-342-3p, rno-miR-352, rno-miR-361-5p, rno-miR-365-3p, rno-miR-374-5p, rno-miR-384-5p, rno-miR-421-3p, rno-miR-425-5p, rno-miR-450a-5p, rno-miR-598-3p, rno-miR-92a-3p, rno-miR-93-5p, rno-miR-98-5p, rno-miR-99a-5p | - |
Table 1: Treatment with Haloperidol, Risperidone or Clozapine on Mirna Expression in C6 Cells
| Validated miRNA | Group | Mean±SD | df | F | ANOVA-p | MD | p valuea |
|---|---|---|---|---|---|---|---|
| rno-let-7a-5p | CTRL | 1.032±0.319 | 12 | 4.7999 | 0.020000 | ||
| HAL | 0.856±0.074 | -0.1753 | 0.7720 | ||||
| RIS | 0.968±0.526 | -0.0633 | 0.9840 | ||||
| CLO | 0.281±0.094 | -0.7510* | 0.0140 | ||||
| rno-miR-152-3p | CTRL | 1.002±0.065 | 12 | 42.9092 | 0.000001 | ||
| HAL | 1.739±0.177 | 0.7376* | 0.0030 | ||||
| RIS | 2.472±0.449 | 1.4701* | 0.0000 | ||||
| CLO | 0.667±0.058 | -0.3350 | 0.1807 | ||||
| rno-miR-221-3p | CTRL | 1.003±0.085 | 12 | 17.1193 | 0.000124 | ||
| HAL | 1.091±0.166 | 0.0884 | 0.5166 | ||||
| RIS | 0.951±0.081 | -0.0513 | 0.8315 | ||||
| CLO | 0.596±0.051 | -0.4068* | 0.0004 | ||||
| rno-miR-345-5p | CTRL | 1.012±0.188 | 12 | 15.6787 | 0.000188 | ||
| HAL | 1.596±0.417 | 0.5831* | 0.0177 | ||||
| RIS | 1.045±0.209 | 0.0326 | 0.9955 | ||||
| CLO | 0.372±0.053 | -0.6408* | 0.0099 | ||||
| rno-miR-3473 | CTRL | 1.01±0.174 | 12 | 33.6548 | 0.000004 | ||
| HAL | 1.724±0.101 | 0.7133* | 0.0001 | ||||
| RIS | 1.684±0.213 | 0.6732* | 0.0001 | ||||
| CLO | 0.856±0.103 | -0.1543 | 0.3919 | ||||
| rno-miR-551b-5p | CTRL | 1.021±0.257 | 12 | 62.9991 | 0.000000 | ||
| HAL | 2.413±0.472 | 1.3914* | 0.0048 | ||||
| RIS | 5.164±0.401 | 4.1429* | 0.0000 | ||||
| CLO | 1.006±0.723 | -0.0154 | 0.9999 |
Table 2: Rt-Qpcr Validation of Expression of Selected Mirnas
Preparation of total cellular extracts and western blot analysis:
APD-treated C6 cells at 80 % confluence were washed 3 times with PBS and then harvested by scraping. Total cytosol extracts were prepared by lysing cells in mammalian protein extraction buffer (GE Healthcare Life Sciences, NJ, USA). Proteins (30-60 μg) purified from C6 cells were separated on 10-15 % polyacrylamide gels containing sodium dodecyl sulfate and were then transferred to polyvinylidene difluoride membranes (Amersham Biosciences, Uppsala, Sweden). After blocking with Pierce protein-free blocking buffer (Thermo Fisher Scientific, Rockford, IL, USA), the membranes were incubated with 1:1000-1:2000 dilutions of antiactin and other targeted antibodies, antiautophagy-related protein 12 (ATG12), GTX24181, GeneTex; anticyclindependent kinase inhibitor 1B (CDKN1B), GTX34930, GeneTex; antiB-cell lymphoma 2 (Bcl2), sc-492, Santa Cruz Biotechnology; antiBcl-2-like protein 11 (Bcl2l11), GTX84980, GeneTex; anticMyc, GTX103436, GeneTex; and antiK-Ras, 415700, Thermo Fisher Scientific) at 4° for 12 h. Then, the membranes were incubated with horseradish peroxidase-conjugated goat antimouse or antirabbit antibodies at room temperature. The target protein signals were developed using an Amersham ECL kit (Amersham, Bucks, UK) and quantitated after normalization to actin expression. The normalized expression ratios of the examined proteins were assessed using one-way ANOVA followed by the Dunnett post hoc test. All the normalized expression ratios of control group will be designated as one to eliminate differences caused by the uneven loading quantities of total proteins, antibody binding efficiency and developing of chemiluminescent signals between biological repeats.
Immunofluorescence staining of C6 and B35 cells:
C6 cells were seeded onto a 6-well plate at a density of 5×103 cells per well with coverslips and cultured in medium for 5 d with APDs added daily as described in the drug treatment section. After APD treatment, the coverslips with C6 cells were washed with PBS and the cells were fixed by incubating the coverslips in paraformaldehyde for 15 min. The cells were permeabilized with methanol and further incubated with a specific antibody for 1 h at room temperature. The coverslips were washed with 0.1 % PBST and then incubated with specific secondary antibodies for 1 h at room temperature. The coverslips were then mounted on slides with SlowFade Gold antifade reagent with DAPI (Life Technologies, Eugene, OR, USA). ImageJ 1.50i downloaded from the NIH website (http://imagej.nih.gov/ij/) was used to quantify the fluorescence intensity.
Results and Discussion
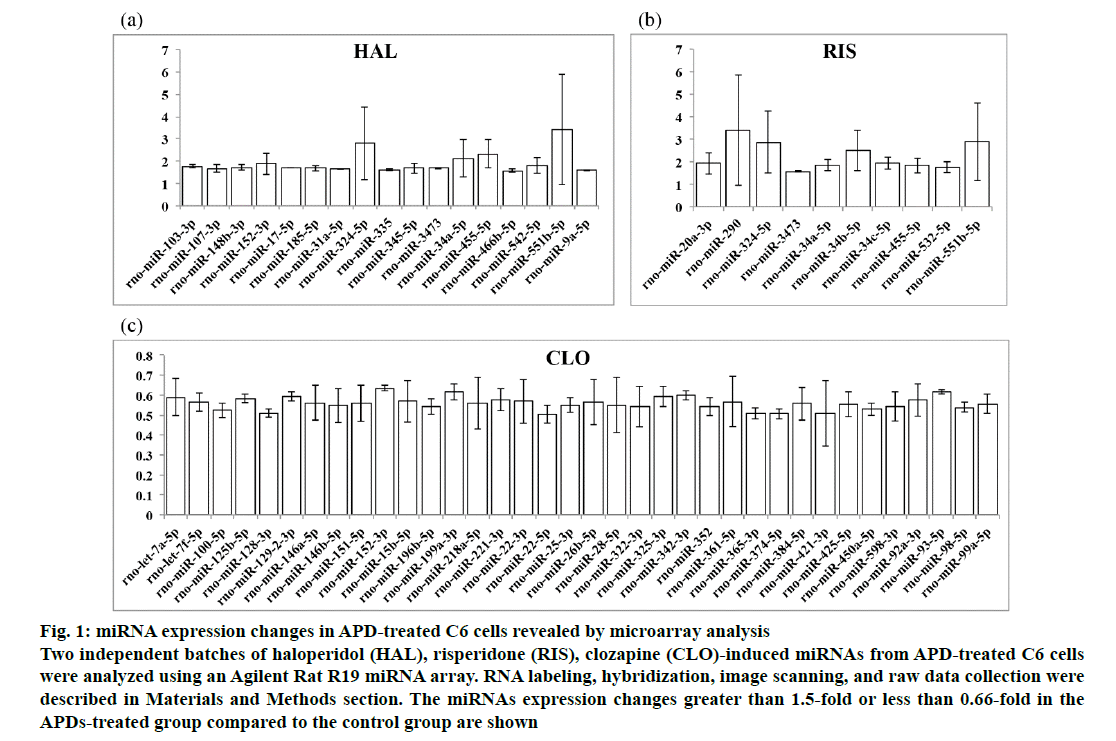
The miRNA expression levels in APD-treated C6 cells as revealed by the Agilent Rat R19 miRNA array were normalized to control C6 cell miRNA expression levels. The microarray data are available from the NCBI Gene Expression Omnibus (GEO: GSE 129908; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129908). The miRNAs with fold changes greater than 1.5-fold or less than 0.66-fold in the treated group compared to the control group are shown in fig. 1. Haloperidol was found to upregulate 17 (fig. 1a), risperidone upregulated 10 (fig. 1b) and clozapine downregulated 36 (fig. 1c) miRNAs in the APD-treated C6 cells. As shown in Table 1, 11 miRNAs were found to be upregulated by only haloperidol treatment in C6 cells. Five miRNAs were upregulated by only risperidone treatment in C6 cells. Five upregulated miRNAs were found in both haloperidol- and risperidone-treated C6 cells. miR-152- 3p was upregulated by haloperidol but downregulated by clozapine in C6 cells. Thirty-five miRNAs were downregulated by only clozapine in C6 cells.
Figure 1: miRNA expression changes in APD-treated C6 cells revealed by microarray analysis
Two independent batches of haloperidol (HAL), risperidone (RIS), clozapine (CLO)-induced miRNAs from APD-treated C6 cells
were analyzed using an Agilent Rat R19 miRNA array. RNA labeling, hybridization, image scanning, and raw data collection were
described in Materials and Methods section. The miRNAs expression changes greater than 1.5-fold or less than 0.66-fold in the
APDs-treated group compared to the control group are shown
To confirm the results obtained by microarray analysis, miRNAs were randomly selected and validated by using real-time quantitative PCR (RT-qPCR). As shown in Table 2, rno-let-7a-5p and rno-miR-221-3p expression was downregulated by clozapine in C6 cells. rno-miR- 152-3p expression was shown to be increased by treating C6 cells with haloperidol or risperidone using one-way ANOVA. Clozapine downregulated rno-miR-152-3p expression in the treated group compared to the control group, as determined using Student’s t-test. Rno-miR- 345-5p expression was increased by haloperidol and decreased by clozapine in C6 cells. Haloperidol and risperidone also increased rno-miR-3473 and rno-miR- 551b-5p expression in C6 cells.
The target proteins of the miRNAs regulated by APD treatment in C6 cells was predicted with miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). Table 3 shows the matched potential targets related to haloperidol-, risperidone- or clozapine-regulated miRNAs. Some target genes such as K-Ras and c-Myc for rno-let-7a-5p, Atg12 for rno-miR-152-3p, Bcl2l11 and Cdkn1b for rno-miR-221-3p and Bcl2 for rnomiR- 34a-5p and rno-miR-15b-5p related to the APDregulated miRNAs identified in this study were selected to further examine the relations between target gene expression and miRNA expression using RT-qPCR. As shown in Table 4, it was investigated whether any of the 3 APDs increased expression of Atg12, Cdkn1b and K-Ras genes. c-Myc expression was increased by haloperidol and clozapine in C6 cells, as determined by one-way ANOVA and the Dunnett post hoc test. Student’s t-test indicated increased c-Myc expression in risperidone-treated C6 cells. Risperidone was also found to reduce Bcl2 mRNA expression in C6 cells. None of the 3 APDs affected Bcl2l11 mRNA expression in C6 cells.
| APD | miRNA | Target(s) | Validation Method* |
|---|---|---|---|
| Haloperidol | rno-miR-107-3p | Arnt, Cpt1a, Pdcd10, Srebf1 | 2 |
| rno-miR-17-5p | Canx, Golga2, Hspa4, Hspa8, Il6st, Stat3, Txnip, Wif1 | 2 | |
| rno-miR-152-3p | Atg12 | 2 | |
| rno-miR-185-5p | Vim | 1 | |
| rno-miR-31a-5p | Isl1, Klf5, Satb2 | 2 | |
| rno-miR-324-5p | Sept3 | 1 | |
| rno-miR-335 | Rasa1 | 2 | |
| rno-miR-34a-5p | Capn8, Tagln | 1 | |
| Acsl1, Bcl2, Ccnd1, Ccne2, E2f3, Grm7, Met, Mgst1, Mycn, Notch1, Sirt1, Sp1, Tp53 | 2 | ||
| rno-miR-455-5p | Scarb1 | 2 | |
| rno-miR-9a-5p | Fgf16 | 1 | |
| Brd4, Myocd, Nfkb1, Npy2r, Onecut2, Pmp22, Sirt1, Syt17, TLN1, Zeb2 | 2 | ||
| Risperidone | rno-miR-290 | Sept3, Acvr1, Adora1, Akap6, Arc, Capn8, Cntn4, Dclk1, Fgf16, Gmfb, Gnb1, Golph3, Klf15, Mapk1, Mapre1, Mgst1, Mmp9, Nr4a1, Ogt, Ptges, Ptp4a2, Rcan1, Ssr3, Stx1a, Syne1, Tagln, Vsnl1 | 1 |
| rno-miR-324-5p | Sept3 | 1 | |
| rno-miR-34a-5p | Capn8, Tagln | 1 | |
| Acsl1, Bcl2, Ccnd1, Ccne2, E2f3, Grm7, Met, Mgst1, Mycn, Notch1, Sirt1, Sp1, Tp53 | 2 | ||
| rno-miR-34c-5p | p53-ps, Sipa1 | 2 | |
| rno-miR-455-5p | Scarb1 | 2 | |
| Clozapine | rno-let-7a-5p | Hmox1, Vim | 1 |
| Calm1, Kras, Myc, Tagln | 2 | ||
| rno-let-7f-5p | Kcnj16 | 1 | |
| rno-miR-100-5p | Aqp4 | 1 | |
| rno-miR-125b-5p | Sema4d, Cyp24a1 | 2 | |
| rno-miR-128-3p | Bmi1, Snap25 | 2 | |
| rno-miR-146a-5p | Irak1, Klf4, Nfkb1, Smad4, Snap25, STAT1, Traf6 | 2 | |
| rno-miR-146b-5p | Klf4 | 2 | |
| rno-miR-15b-5p | Arl2, Bcl2, Cdc25a | 2 | |
| rno-miR-151-5p | Fndc1, Fxyd1, Tnfsf10 | 2 | |
| rno-miR-152-3p | Atg12 | 2 | |
| rno-miR-218a-5p | Cntn4, Kcnj16 | 1 | |
| Tgm2, REST | 2 | ||
| rno-miR-221-3p | Bcl2l11, Cdkn1b, Cdkn1c | 2 | |
| rno-miR-22-5p | Pag1 | 2 | |
| rno-miR-25-3p | Vsnl1 | 1 | |
| Nox4, Kat2b | 2 | ||
| rno-miR-26b-5p | Akap6 | 1 | |
| Ccnd1, Ctgf, GATA4, Pten, Trpc3 | 2 | ||
| rno-miR-342-3p | Adora1, Kcnj16 | 1 | |
| rno-miR-374-5p | Vsnl1 | 1 | |
| rno-miR-384-5p | Pik3cd | 2 | |
| rno-miR-450a-5p | Hnrnpk, Wisp2 | 2 | |
| rno-miR-92a-3p | Vim, Vsnl1 | 1 | |
| Slc12a5 | 2 | ||
| rno-miR-93-5p | KCNJ14, Mgst1, Sirt1, Sp1, Epha4, Rb1 | 2 | |
| rno-miR-98-5p | Tagln | 1 | |
| Bcl2l1, Ccnd2, Stat3 | 2 | ||
| rno-miR-99a-5p | Aqp4, Fgf16 | 1 | |
| Bmpr2 | 2 |
Table 3: Mirnas and Their Potential Target(S) Affected by Haloperidol, Risperidone or Clozapine in C6 Cells
| Target gene | Group | Mean±SD | df | F | ANOVA-p | MD | p valuea |
|---|---|---|---|---|---|---|---|
| Atg12 | CTRL | 1.007±0.146 | 11 | 20.0669 | 0.000 | ||
| HAL | 1.705±0.182 | 0.69733* | 0.0026 | ||||
| RIS | 1.790±0.141 | 0.78280* | 0.0012 | ||||
| CLO | 2.025±0.201 | 1.01727* | 0.0002 | ||||
| Cdkn1b | CTRL | 1.003±0.091 | 11 | 105.1039 | 0.000 | ||
| HAL | 1.774±0.167 | 0.77127* | 0.0005 | ||||
| RIS | 3.075±0.105 | 2.07173* | 0.0000 | ||||
| CLO | 1.968±0.190 | 0.96473* | 0.0001 | ||||
| Bcl2l11 | CTRL | 1.003±0.092 | 11 | 0.3671 | 0.779 | ||
| HAL | 0.959±0.052 | -0.04403 | 0.9620 | ||||
| RIS | 0.929±0.065 | -0.07403 | 0.8569 | ||||
| CLO | 1.041±0.253 | 0.03833 | 0.9741 | ||||
| Bcl2 | CTRL | 1.010±0.168 | 11 | 6.1389 | 0.018 | ||
| HAL | 0.951±0.059 | -0.05877 | 0.9131 | ||||
| RIS | 0.569±0.137 | -0.44053* | 0.0111 | ||||
| CLO | 0.801±0.158 | -0.20977 | 0.2199 | ||||
| c-Myc | CTRL | 1.011±0.192 | 11 | 8.8099 | 0.006 | ||
| HAL | 1.783±0.231 | 0.77143* | 0.0390 | ||||
| RIS | 1.720±0.146 | 0.708833 | 0.0562 | ||||
| CLO | 2.304±0.522 | 1.29263* | 0.0024 | ||||
| K-Ras | CTRL | 1.003±0.086 | 11 | 13.4039 | 0.002 | ||
| HAL | 1.931±0.277 | 0.92897* | 0.0072 | ||||
| RIS | 2.009±0.338 | 1.00683* | 0.0045 | ||||
| CLO | 2.318±0.299 | 1.31513* | 0.0008 |
Table 4: Rt-Qpcr Validation of the Expression of Mirna-Targeted Genes
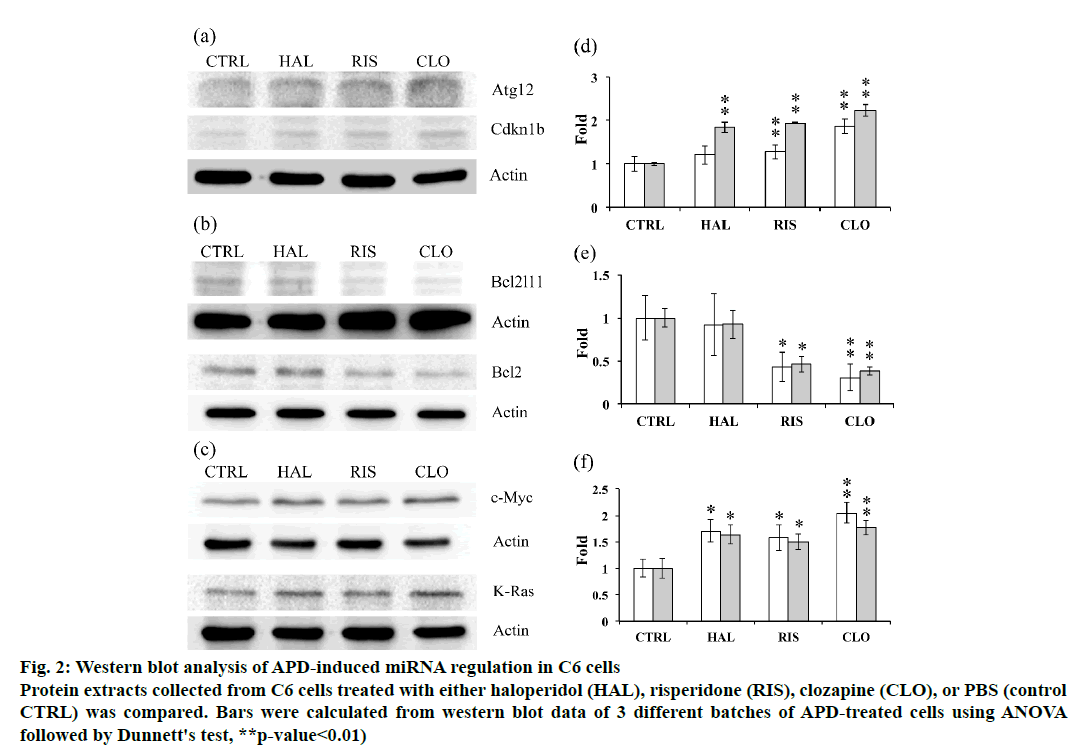
The expression of potential targets of the tested miRNAs was further examined using western blot analysis to unravel the relations between miRNA regulation and target protein expression. Atg12 expression was found to be increased by clozapine treatment, while haloperidol and risperidone did not significantly affect Atg12 expression in C6 cells. The expression of Cdkna1b was upregulated by all 3 APDs (fig. 2). Bcl2l11 and Bcl2 expression in C6 cells was downregulated by risperidone and clozapine but not by haloperidol (fig. 2). Induction of c-Myc and K-Ras expression was observed in C6 cells treated with any of the 3 APDs tested (fig. 2).
Figure 2: Western blot analysis of APD-induced miRNA regulation in C6 cells
Protein extracts collected from C6 cells treated with either haloperidol (HAL), risperidone (RIS), clozapine (CLO), or PBS (control
CTRL) was compared. Bars were calculated from western blot data of 3 different batches of APD-treated cells using ANOVA
followed by Dunnett's test, **p-value<0.01)
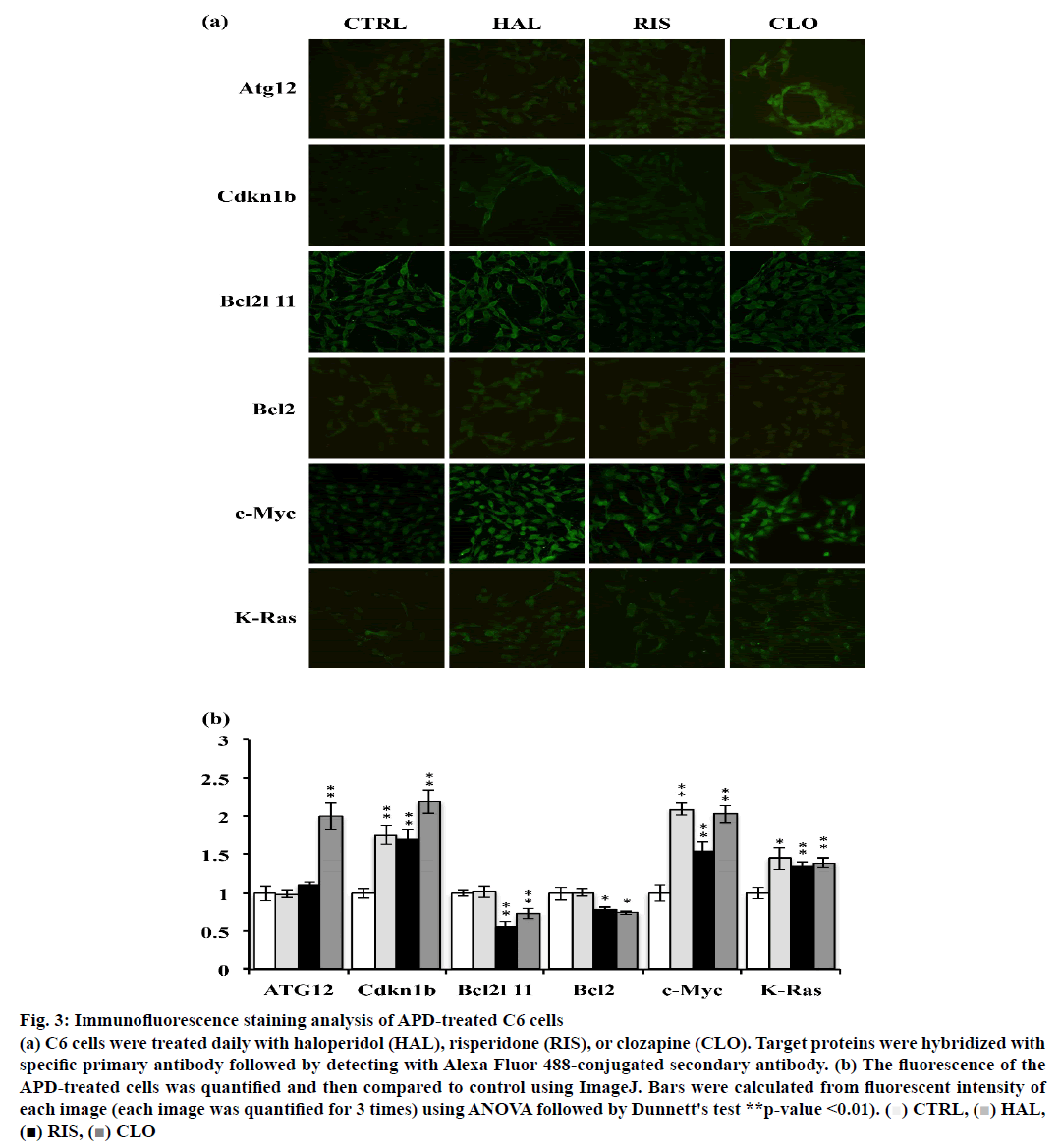
Atg12, Cdkn1b, Bcl2l11, Bcl2, c-Myc and K-Ras expression was examined in APD-treated C6 cells using immunofluorescence staining. As shown in fig. 3a and b, clozapine increased Atg12 expression. Cdkn1b expression was increased by all 3 APDs in C6 cells. Haloperidol did not significantly modulate Bcl2l11 expression, but risperidone and clozapine reduced Bcl2l11 expression in C6 cells. It was also observed that c-Myc expression was upregulated by these 3 APDs and accumulated in the cell nuclei of C6 cells.
Figure 3: Immunofluorescence staining analysis of APD-treated C6 cells
(a) C6 cells were treated daily with haloperidol (HAL), risperidone (RIS), or clozapine (CLO). Target proteins were hybridized with
specific primary antibody followed by detecting with Alexa Fluor 488-conjugated secondary antibody. (b) The fluorescence of the
APD-treated cells was quantified and then compared to control using ImageJ. Bars were calculated from fluorescent intensity of
each image (each image was quantified for 3 times) using ANOVA followed by Dunnett's test **p-value <0.01). ( )CTRL,(
)CTRL,( ) HAL,
(■) RIS, (
) HAL,
(■) RIS, ( ) CLO
) CLO
In this study, whether various APDs could regulate the expression of different miRNAs in C6 cells was investigated through microarray profiling (fig. 1 and Table 1). The expression of selected miRNAs was validated using RT-qPCR. miRNA-target interactions (MTIs) were searched to predict miRNA targets in miRTarBase. Western blot analysis and immunofluorescence staining were used to examine the potential targets of the selected APD-regulated miRNAs. Table 5 shows the regulation of miRNAs and their potential targets induced by haloperidol, risperidone and clozapine. We found that rno-miR- 152-3p expression was downregulated by clozapine according to microarray profiling and RT-qPCR analysis. The potential target, Atg12, was increased by clozapine according to western blot analysis (fig. 2a) and immunofluorescence staining (fig. 3a). Interestingly, some recent studies have revealed that miR-152 is found upregulated in the postmortem brain of schizophrenic patients[26]. Other studies have also shown that clozapine and olanzapine can induce Atg12 expression in the rat prefrontal cortex or in SH-SY5Y cells[27,28]. A previous study mentioned that olanzapine in combination with chloroquine can increase Bcl2l11 and Cdkn1b expressions to further inhibit autophagyinduced protective effects in cells[28]. Present investigation showed that rno-miR-221-3p expression was decreased by clozapine (Table 2), which might suggest that rno-miR-221-3p can increase Cdkn1b and Bcl2l11 expression and inhibit autophagy-induced protective effects in C6 cells. Subsequent validation also revealed that Cdkn1b mRNA or protein expression could be upregulated by any of the 3 APDs (Table 4). In contrast, risperidone and clozapine downregulated Bcl2l11 expression in C6 cells (Table 4).
| miRNA | APD | Microarray | RT-qPCR | Target | APD | RT-qPCR | Western Blot | IF |
|---|---|---|---|---|---|---|---|---|
| HAL | I | I | HAL | I | - | - | ||
| rno-miR-152-3p | RIS | - | I | Atg12 | RIS | I | I | - |
| CLO | D | D | CLO | I | I | I | ||
| HAL | I | I | I | |||||
| HAL | - | - | Cdkn1b | RIS | I | I | I | |
| rno-miR-221-3p | RIS | - | - | CLO | I | I | I | |
| CLO | D | D | HAL | - | D | - | ||
| Bcl2l11 | RIS | - | D | D | ||||
| CLO | - | D | D | |||||
| HAL | I | I | I | |||||
| HAL | - | - | c-Myc | RIS | - | I | I | |
| rno-let-7a-5p | RIS | - | - | CLO | I | I | I | |
| CLO | D | D | HAL | I | I | I | ||
| K-Ras | RIS | I | I | I | ||||
| CLO | I | I | I | |||||
| HAL | I | NA | HAL | - | - | - | ||
| rno-miR-34a-5p | RIS | I | NA | Bcl2 | RIS | D | D | D |
| CLO | - | NA | CLO | - | D | D |
Table 5: Regulation of Mirnas and Their Potential Targets Induced by Various Apds
Increased mRNA levels of c-myc, a protooncogene that is involved in cell proliferation, have been reported in rats receiving estrogen treatment followed by haloperidol injection[29]. Current data revealed that clozapine decreased rno-let-7a-5p expression and induced c-myc and K-Ras mRNA expression (Tables 2 and 4). Haloperidol and risperidone did not affect rno-let-7a-5p expression but induced c-myc mRNA and protein expression. These results suggested that clozapine might act through a different mechanism than haloperidol and risperidone to regulate c-myc and K-Ras expression and followed cell proliferation regulations. It was also found that rno-miR-34a-5p expression in C6 cells was increased by haloperidol and risperidone treatment (Table 1). The mRNA expression of Bcl2, a potential target of rno-miR-34a-5p, was also downregulated by risperidone (Table 4). In addition, risperidone and clozapine decreased Bcl2 protein expression (fig. 2b, Table 4). These results suggested that there are different regulatory mechanisms for Bcl2 expression among C6 cells treated with haloperidol, risperidone and clozapine.
In this study, it was found that various potential targets might be affected by the miRNAs that are modulated by the 3 APDs. As shown in Table 3, vimentin (Vim) expression might be regulated by both rno-let-7a-5p and rno-miR-92a-3p in clozapine-treated C6 cells. Synaptosomal nerve-associated protein 25 (SNAP- 25) expression in clozapine-treated C6 cells might be regulated by rno-miR-128-3p and rno-miR-146a-5p. Visinin-like protein 1 (Vsnl1) expression in clozapinetreated C6 cells might be regulated by rno-miR-25-3p and rno-miR-374-5p. Aquaporin-4 (Aqp4) expression in clozapine-treated C6 cells might be regulated by rno-miR-100-5p and rno-miR-99a-5p. Potassium inwardly-rectifying channel, subfamily J, member 16 (Kcnj16) expression in clozapine-treated C6 cells might be regulated by rno-let-7f-5p, rno-miR-152- 3p and rno-miR-342-3p. Neuronal-specific septin-3 (Sept3) expression in risperidone-treated C6 cells might be regulated by rno-miR-100-5p and rno-miR- 99a-5p. Different miRNAs might be involved in the expression regulation of one target gene. Additionally, APD-induced miRNA regulation might not cause predicted gene or protein expression modulation in C6 cells (Table 5). It can be concluded that APD-induced regulation of miRNAs is only part of the regulatory mechanism modulating the protein expression of the miRNA target genes.
The authors have previously described that Vim mRNA expression was induced by clozapine and that Hspa8 mRNA expression could also be downregulated by haloperidol, risperidone or clozapine in C6 cells[30]. In this study, it was observed that haloperidol induced rnomiR185- 5p expression (Table 1), which was proposed to downregulate Vim mRNA expression. In contrast, it was found that clozapine inhibited rno-let-7a-5p and rno-miR-92a-3p (Table 1 and 2), which might increase Vim mRNA expression. This result is consistent with previously reported gene expression findings. However, none of the 3 APDs studied changed Vim protein expression, according to the previous reports. Present data showed that haloperidol could induce rno-miR- 17-5p (Table 1), which might potentially reduce Hspa8 mRNA expression. As shown in a previous study, any of the 3 APDs could downregulate mRNA expression of Hspa8, but none of the APDs affect protein expression of Hspa8. These results suggested that APD-induced Vim or Hspa8 protein expression regulation could be modulated by various regulatory mechanisms other than miRNA-mediated protein modulation in C6 cells.
Haloperidol, risperidone and clozapine are commonly used APDs in clinic to ease the symptoms of schizophrenic patients. The receptor binding profile of haloperidol, a typical APD, is quite different from the atypical APDs, such as risperidone and clozapine. Haloperidol bound mainly to dopamine D2 receptor while risperidone bound to serotonin 2A (5-HT2A) receptor[31]. Clozapine bound various receptors such as dopamine D4, 5-HT2A, α1 and α2 adrenergic receptors, alpha-2 and histamine H1 receptor[32,33]. These suggested the molecular mechanisms of APDs might be varied according to the downstream signaling of receptors that APD bound and also the crosstalk between signaling pathways regulated by bound receptors. Previous study by the authors also mentioned that protein expression changes induced by APDs might be limited by cell itself to maintain the relative normal physiological function of cells[34]. In this study, it was found that clozapine did not induce miRNA expression according to the results of microarray analysis (Table 1). Also, it was found that clozapine treatment reduced miRNA rno-let-7a- 5p, rno-miR-152-3p and rno-miR-221-3p (Table 1 and 2), which could further induce expressions of predicted target genes/proteins, atg12, cdkn1b, c-myc and K-ras in C6 cells (Table 4, fig. 2a and c). It was also observed that miRNAs regulations induced by haloperidol or risperidone were not closely related to predicted target genes/proteins expressions. It was suggested that APDs might regulate protein expression not only through miRNA-mediated regulation but also through other regulatory mechanisms. Although some valuable information about APD-induced miRNA regulation was observed, detailed studies should still be conducted to clarify the relations between miRNA regulation and protein modulation in C6 cells.
Acknowledgements
This work was supported by grants (TCRD-TPE-105-23 and TCRD-TPE-106-23) of the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, Taiwan, Republic of China. The author thanks the Core Laboratory of the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation for support.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33.
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005;37:766-70.
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105.
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 2007;17:118-26.
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 2007;91:209-17.
- Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genom 2009;10:154-68.
- Schratt G. microRNAs at the synapse. Nature reviews Neuroscience 2009;10:842-9.
- Tews B, Schonig K, Arzt ME, Clementi S, Rioult-Pedotti MS, Zemmar A, et al. Synthetic microRNA-mediated downregulation of Nogo-A in transgenic rats reveals its role as regulator of synaptic plasticity and cognitive function. Proc Natl Acad Sci U S A 2013;110:6583-8.
- Lett TA, Chakravarty MM, Felsky D, Brandl EJ, Tiwari AK, Goncalves VF, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry 2013;18:443-50.
- Mor E, Kano S, Colantuoni C, Sawa A, Navon R, Shomron N. MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol Dis 2013;55:1-10.
- Gong Y, Wu CN, Xu J, Feng G, Xing QH, Fu W, et al. Polymorphisms in microRNA target sites influence susceptibility to schizophrenia by altering the binding of miRNAs to their targets. Eur Neuropsychopharmacol 2013;23:1182-9.
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 2007;8:R27.
- Santarelli DM, Liu B, Duncan CE, Beveridge NJ, Tooney PA, Schofield PR, et al. Gene-microRNA interactions associated with antipsychotic mechanisms and the metabolic side effects of olanzapine. Psychopharmacol 2013;227:67-78.
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A 2009;106:3507-12.
- Swathy B, Banerjee M. Haloperidol induces pharmacoepigenetic response by modulating miRNA expression, global DNA methylation and expression profiles of methylation maintenance genes and genes involved in neurotransmission in neuronal cells. PloS One 2017;12:e0184209.
- Johnstone AL, O'Reilly JJ, Patel AJ, Guo Z, Andrade NS, Magistri M, et al. EZH1 is an antipsychotic-sensitive epigenetic modulator of social and motivational behavior that is dysregulated in schizophrenia. Neurobiol Dis 2018;119:149-58.
- Rothermundt M, Falkai P, Ponath G, Abel S, Burkle H, Diedrich M, et al. Glial cell dysfunction in schizophrenia indicated by increased S100B in the CSF. Mol Psychiatry 2004;9:897-9.
- Flynn SW, Lang DJ, Mackay AL, Goghari V, Vavasour IM, Whittall KP, et al. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol Psychiatry 2003;8:811-20.
- Pennington K, Dicker P, Hudson L, Cotter DR. Evidence for reduced neuronal somal size within the insular cortex in schizophrenia, but not in affective disorders. Schizophr Res 2008;106:164-71.
- Bartzokis G, Lu PH, Stewart SB, Oluwadara B, Lucas AJ, Pantages J, et al. In vivo evidence of differential impact of typical and atypical antipsychotics on intracortical myelin in adults with schizophrenia. Schizophr Res 2009;113:322-31.
- Peleg-Raibstein D, Knuesel I, Feldon J. Amphetamine sensitization in rats as an animal model of schizophrenia. Behav Brain Res 2008;191:190-201.
- Bartzokis G. Neuroglialpharmacology: white matter pathophysiologies and psychiatric treatments. Front Biosci 2011;16:2695-733.
- Nardin P, Tramontina AC, Quincozes-Santos A, Tortorelli LS, Lunardi P, Klein PR, et al. In vitro S100B secretion is reduced by apomorphine: effects of antipsychotics and antioxidants. Prog Neuropsychopharmacol Biol Psychiatry 2011;35:1291-6.
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33:e179.
- Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis 2012;46:263-71.
- Kim SH, Park S, Yu HS, Ko KH, Park HG, Kim YS. The antipsychotic agent clozapine induces autophagy via the AMPK-ULK1-Beclin1 signaling pathway in the rat frontal cortex. Prog Neuropsychopharmacol Biol Psychiatry 2018;81:96-104.
- Vucicevic L, Misirkic-Marjanovic M, Paunovic V, Kravic-Stevovic T, Martinovic T, Ciric D, et al. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy 2014;10:2362-78.
- Chernavsky AC, Valerani AV, Burdman JA. Haloperidol and oestrogens induce c-myc and c-fos expression in the anterior pituitary gland of the rat. Neurol Res 1993;15:339-43.
- Li T, Lee M, Tsai F, Chen Y, Lin Y, Chen M. Proteomic study revealed antipsychotics-induced nuclear protein regulations in B35 cells are similar to the regulations in C6 cells and rat cortex. BMC Pharmacol Toxicol 2018;19:9.
- Bymaster FP, Calligaro DO, Falcone JF, Marsh RD, Moore NA, Tye NC, et al. Radioreceptor binding profile of the atypical antipsychotic olanzapine. Neuropharmacology 1996;14:87-96.
- Schotte A, Janssen PF, Gommeren W, Luyten WH, Van Gompel P, Lesage AS, et al. Risperidone compared with new and reference antipsychotic drugs: in vitro and in vivo receptor binding. Psychopharmacology 1996;124:57-73.
- Schotte A, Janssen PF, Bonaventure P, Leysen JE. Endogenous dopamine limits the binding of antipsychotic drugs to D3 receptors in the rat brain: a quantitative autoradiographic study. Histochem J 1996;28:791-9.
- Chen ML. Two-dimensional gel electrophoresis revealed antipsychotic drugs induced protein expression modulations in C6 glioma cells. Prog Neuropsychopharmacol Biol Psychiatry 2013;40:1-11.