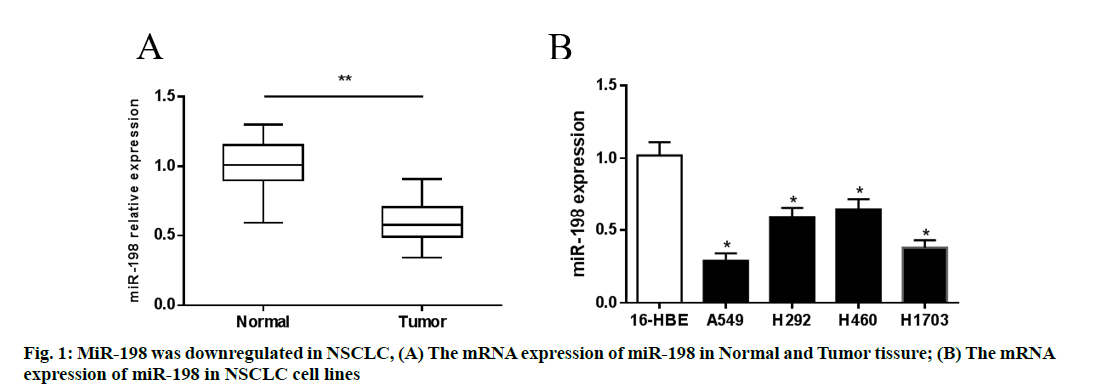
- *Corresponding Author:
- H. Shi
Medical College of Yangzhou University,Yangzhou 225001, China
E-mail: shihongcan@yzu.edu.cn
| Date of Received | 03 June 2020 |
| Date of Revision | 30 December 2020 |
| Date of Acceptance | 31 July 2021 |
| Indian J Pharm Sci 2021;83(4):758-764 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
The expression of microRNA 198 which is localized in human chromosome 13q13.3 is downregulated in many malignant tumors. The purpose of this study is to investigate the regulatory effects of microRNA 198 on non-small cell lung cancer and the associated regulatory mechanism. The expression levels of microRNA 198 in non-small cell lung cancer cell lines and tumor tissues were determined using the reverse transcription polymerase chain reaction method. Overexpression microRNA in adenocarcinomic human alveolar basal epithelial cells, non-small cell lung cancer cells and cell proliferation in each group were measured with the cell counting kit-8 method. Cell migration was determined with a cell scratch test, while transwell assay was used for the determination of cell migration and invasion capacities. Bioinformatics website was used to predict microRNA 198 target genes and verified with double luciferase assay. Western blot method was used to assay the protein expression level of toll-like receptor 4 in the cells and the effects of overexpression of microRNA 198 on the regulation of cell proliferation and migration by microRNA 198. The expressions of microRNA 198 in non-small cell lung cancer cell lines and tissues were significantly downregulated. However, overexpression of microRNA 198 through transfection significantly inhibited the proliferation, migration and invasion of lung cancer cell lines adenocarcinomic human alveolar basal epithelial cells and non-small cell lung cancer cells and directly targeted and inhibited the expression of toll-like receptor 4. Overexpression of toll-like receptor 4 reversed the regulatory effect of microRNA 198 to some extent. MicroRNA 198 inhibits the proliferation, migration and invasion of non-small cell lung cancer cells by targeting toll-like receptor 4, thereby playing the role of tumor suppressor gene in the development of non-small cell lung cancer.
Keywords
MicroRNA 198, non-small cell lung cancer, proliferation, toll-like receptor 4
Lung cancer is a common clinical malignancy with high morbidity and mortality worldwide. Non-small cell lung cancer (NSCLC) accounts for more than 80 % of all lung cancer cases. At present, the traditional clinical treatment for lung cancer is surgical resection combined with chemoradiotherapy. However, because most NSCLC cases are diagnosed at an advanced stage, the tumor cells have already metastasized. Moreover, since some patients have low sensitivity to chemotherapy, the overall therapeutic outcome for NSCLC is poor, resulting in low 5 y survival of patients [1]. Therefore, an in-depth study of the molecular mechanism involved in the pathogenesis of NSCLC and its metastasis and the development of new molecular markers for diagnosis and treatment of lung cancer are of great significance for improving the clinical treatment efficacy of NSCLC patients and for prolonging their survival.
Tumorigenesis is a process involving the abnormal expressions of various oncogenes and the inactivation of tumor suppressor genes. In recent years, studies on the regulatory effect of microRNA (miRNA) on tumors have attracted the interest of researchers. The miRNAs are endogenous single-stranded non-coding RNAs which are about 18-25 nucleotides in length with regulatory effects on about 30 % of gene expression in humans [2,3]. The expression of miRNAs is tissue-specific, conservative and spatiotemporal, i.e., different tissues have different miRNA expression profiles. Moreover, the expressions of miRNAs differ within the same tissues at different development stages and different physiological and pathological states [4,5]. Several studies have confirmed that miRNA is involved in the pathogenesis of NSCLC and that they play important regulatory roles in cell proliferation, differentiation, apoptosis and invasion.
Studies have shown that such as gastric cancer [6], thyroid cancer [7] and liver cancer [8]. Thus, miR-198 usually plays the role of the tumor suppressor gene. However, some studies have reported that the expression of miR-198 is upregulated in some malignant tumors, implying that it may play different regulatory roles in different tissues. There are limited reports about the effect of the miR-198 expression on NSCLC. In this study, reverse transcription polymerase chain reaction (RT-PCR) was used to discover that miR-198 expression was downregulated in NSCLC cell lines. Following the establishment of stable overexpression of miR-198 cell lines through cell transfection, it was observed that the proliferation, migration and invasion ability of cells were significantly reduced through a mechanism involving inhibition of toll-like receptor 4 (TLR-4) expression.
Materials and Methods
Cell culture:
The NSCLC cell lines adenocarcinomic human alveolar basal epithelial cells (A549), human pulmonary mucoepidermoid carcinoma cell line (H292), hypotriploid human cell line (H460) and non small cell lung cancer (H1703) and normal bronchial epithelial cell line (16-HBE) were purchased from American type culture collection (ATCC) in the United States of America. All tumor cells were cultured in roswell park memorial institute medium (RPMI) medium containing 10 % fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin in a 5 % CO2 incubator at 37°.
Reverse transcription polymerase chain reaction:
Total RNA was extracted with Trizol reagent and complementary DNA was obtained by reverse transcription. Then, the SYBR Green method was used for RT-PCR. The reaction conditions were 95° for 5 min and 40 cycles of 95° for 30 sec and 55° for 30 sec. The housekeeper gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference and each test was done in triplicate. A standard curve was drawn, and the expression level of the target gene was determined using the 2–∆∆Ct method. The sequences of the primers used were miR-198: Forward: 5'-GAATTTCGATGACGTCCATTGTCTTGG-3'; reverse: 5'-GGTACCGTAAAACTGCAGCGT AGTTGC-3', GAPDH: forward: 5'-AAGGTCGTACCCGGGTGCG-3'; reverse: 5'-TGAAGGCTGTTGAGCTAGC-3'.
Cell transfection:
Cells in the logarithmic growth stage were inoculated into a 6-well plate at a density of 1×105 cells/well. When the cells grew to about 80 % confluence, cell transfection was carried out. The operation process was strictly following the instructions of the Lipofectamine 3000 transfection kit. The target plasmid was dissolved in an appropriate amount of medium. Then, diluted Lipofectamine 3000 was added to each well, gently shaken and carefully mixed and then cultured in a 5 % CO2 incubator at 37°. The transfection efficiency was verified with RT-PCR. The cells were divided into blank groups and miRNAs-neighboring cells (miR-NC) combined the miR-198 mimics group.
Cell proliferation assays:
Cell growth in each group was determined with the cell counting kit-8 (CCK-8) method. Cells in the logarithmic growth stage were digested and made into a single-cell suspension. The cells were counted and seeded in 96-well plates at a density of 1×105 cells/well. Then, 10 μl CCK-8 solution was added to each well and the cells were further cultured. The optical density of cells was measured at 450 nm at the start and subsequently at 1, 2, 3 and 4 d. Three replicates were used for each well.
Clone experiment:
Cells in a good growth state were inoculated into a 6-well plate at a density of 5×103 cells/well, and then cultured in a cell incubator. The medium was changed every 2-3 d and visible clonal cells were formed about 2 w later. After washing with phosphate buffered saline (PBS) buffer, the cells were fixed with 4 % paraformaldehyde, stained with 0.1 % crystal violet solution and photographed.
Cell migration and invasion assays:
Matrigel was diluted with a serum free medium and added to the matrigel chamber in the transwell chamber. The cells of each group were inoculated in a small chamber at a density of 1.5×105 cells/well. A serum free medium was added in the upper chamber, while a complete medium (600 l) was added to the lower chamber. The cells were cultured in a constant temperature incubator. After 24 h, the chamber was taken out, washed with PBS solution, fixed with methanol for 30 min and then stained with crystal violet. Five fields were randomly selected for observation in each well and 3 replicates were used for each well.
Double luciferase assay:
The well-grown cells were inoculated into a 6-well plate at a density of 5×105 cells/well. Then, TLR-4 fragments were inserted into the luciferase vector to construct TLR-4 three prime untranslated region (3'UTR) wild-type and mutant plasmids, which were co-transfected with miR-198 mimics and miR-NC respectively and then cultured in a carbon dioxide incubator for 48 h. Luciferase activity in each group was assayed with a luciferase reporter assay kit. Three replicates were used for each well.
Western blotting:
The cells were lysed with radio-immunoprecipitation assay (RIPA) cell lysing kit, followed by centrifugation and the protein contents of the lysates were measured with bicinchoninic acid (BCA) kit. Equal amounts of protein samples (50 µg) were subjected to 10 % sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane which was subsequently blocked with 5 % skimmed milk powder for 2 h. Then, the membrane was incubated with TLR-4 primary antibody (diluted 1:1000) overnight at 4°, followed by incubation with horse radish peroxidase-labeled sheep anti-rabbit secondary antibody (diluted 1:500) at room temperature for 2 h. Thereafter, the 3,3'-diaminobenzidine (DAB) chromogenic solution was added for color development and protein strip images were obtained using a gel imaging system for gray level analysis.
Statistical analysis:
Statistical analysis was done with statistical package for the social sciences (SPSS) 22.0 statistical software. Data are expressed as mean±standard deviation (x̄±s). A comparison between two independent samples was carried out with a t-test. Values of p<0.05 were taken as indicative of the statistical significance of differences.
Results and Discussion
In order to determine the expression of miR-198 in NSCLC cell lines and clinical tissue samples, RT-PCR was used. The highest degree of downregulation was found in A549 and H1703 cells. Similarly, miR-198 expression in tumor tissues was significantly lower than that in para-cancerous tissues (p<0.05), indicating that miR-198 was downregulated in NSCLC. These results are shown in fig. 1.
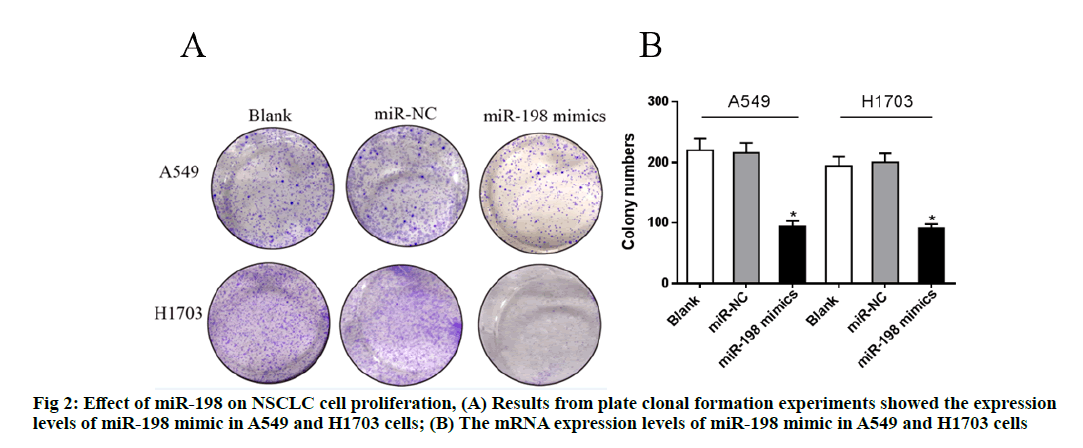
The relative expression level of miR-198 in each group was determined with RT-PCR. As shown in fig. 2, the expression levels of miR-198 mimic in A549 and H1703 cells were significantly lower than that of the miR-NC (negative control) group (p<0.05), while there were no significant differences in the expression levels of miR-198 between the miR-NC groups and the blank groups (p>0.05), indicating successful transfection.Plate clonal formation experiments showed that the number of clones of both cells decreased significantly after overexpression of miR-198 (p<0.05), suggesting that miR-198 inhibited the proliferation of the NSCLC cells.
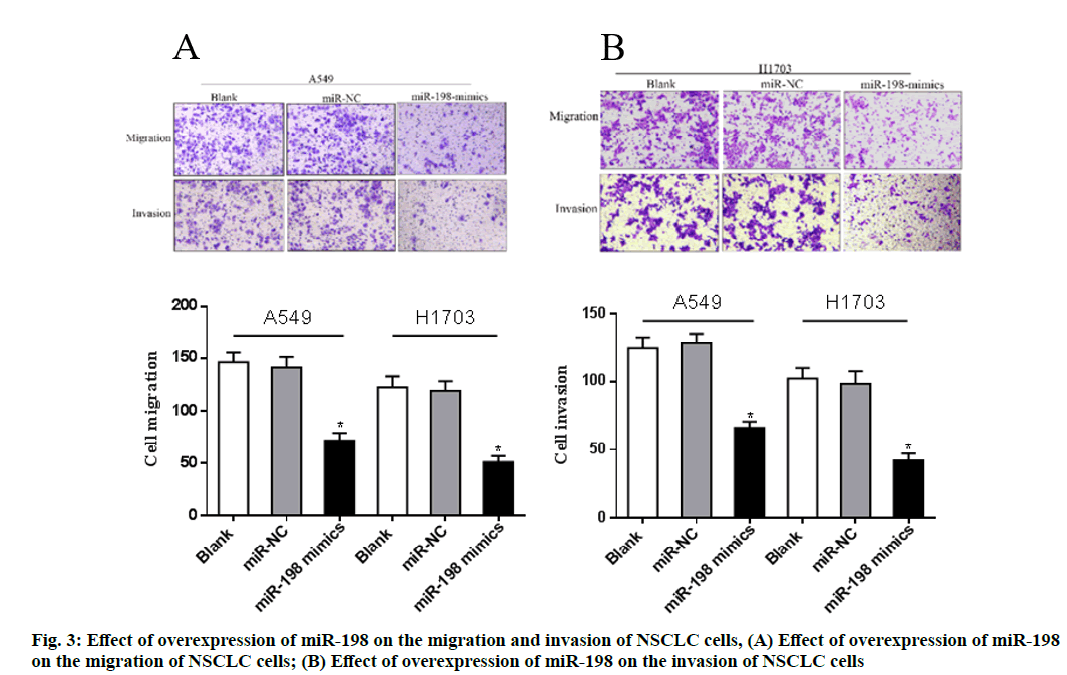
Transwell assay was further used to determine the migration and invasion of cells in each group. The results showed that the average number of cells migrating and invading in the miR-198 mimics groups was significantly lower than that in miR-NC groups (p<0.05). This further confirms that miR-198 inhibited the migration and invasion of NSCLC cells (A549 and H1703). These results are shown in fig. 3.
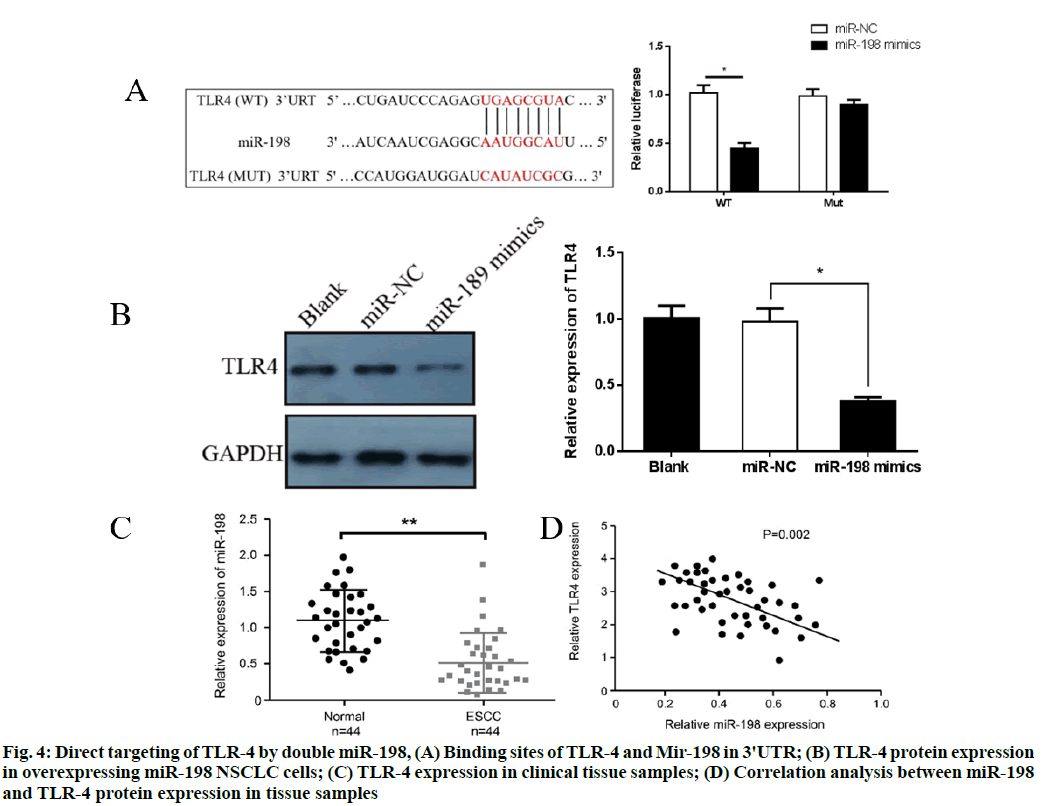
The target genes of miR-198 were predicted with the bioinformation prediction websites TargetScan and miRanda and the binding sites of 3'UTR of TLR-4 and miR-198 were found. However, they had no significant effect on the luciferase activity of mutant plasmid (p>0.05), indicating a direct targeted regulatory relationship between miR-198 and TLR-4, as shown in fig. 4A. Western blot results showed that overexpression of miR-198 significantly inhibited protein expressions of TLR-4 in NSCLC cells (p<0.05) (fig. 4B). Furthermore, TLR-4 expression in clinical tissue samples was analyzed. Western blot results showed that TLR-4 expression in tumor tissues was significantly higher than that in adjacent normal tissues (p<0.05) (fig. 4C). Pearson's correlation analysis showed that miR-198 was negatively correlated with protein expression of TLR-4 in tissue samples (p<0.01) (fig. 4D).
Figure 4: Direct targeting of TLR-4 by double miR-198, (A) Binding sites of TLR-4 and Mir-198 in 3'UTR; (B) TLR-4 protein expression in overexpressing miR-198 NSCLC cells; (C) TLR-4 expression in clinical tissue samples; (D) Correlation analysis between miR-198 and TLR-4 protein expression in tissue samples
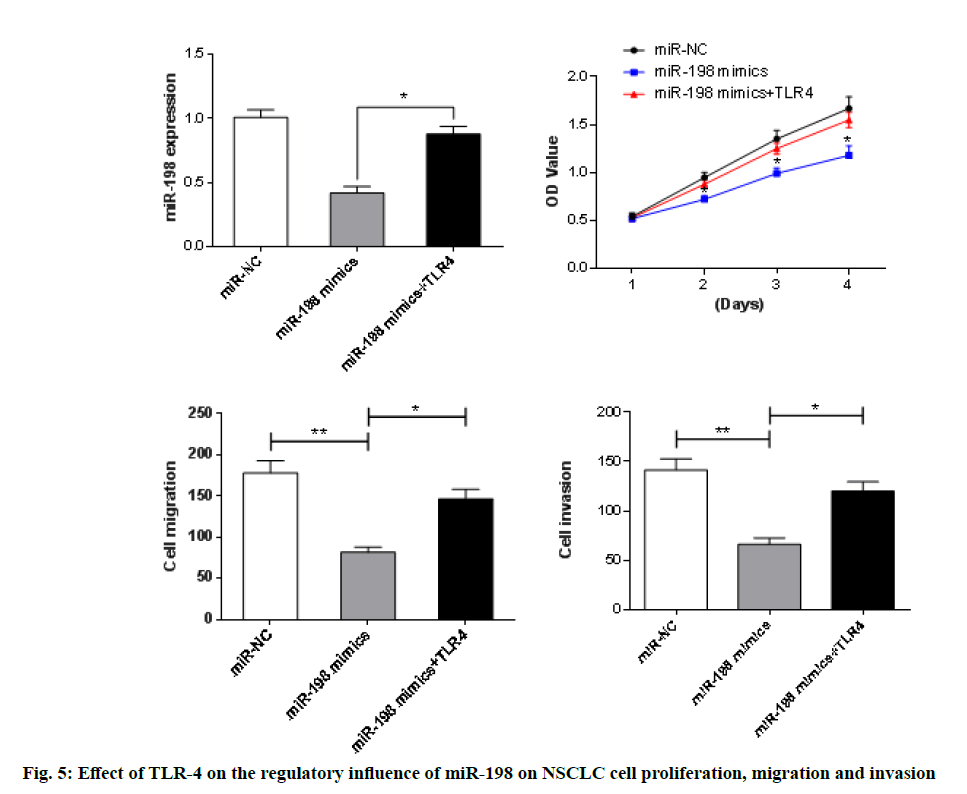
The TLR-4 was further overexpressed in A549 cells in the miR-198 mimics group. As presented in fig. 5, results from CCK-8 and Transwell assays showed that overexpression of TLR-4 resulted in significant enhancement of the inhibition of proliferation, migration and invasion of A549 cells, when compared with the miR-198 mimic groups (p<0.05). This indicates that overexpression of TLR-4 reversed the regulatory effect of miR-198 to some extent.
Lung cancer occurs mainly due to environmental factors such as chemical, physical and viral carcinogens which cause DNA mutations or sequential activation of oncogenes and/or inactivation of tumor suppressor genes. They may also change the expressions of gene products, thereby causing malignant transformations in cells [9]. The miRNAs are a class of highly conserved single-stranded non-coding small molecular RNAs that inhibit the translation of target gene mRNA from the post-transcriptional level by complementing and pairing with the 3'-UTR of specific downstream target gene mRNA [10]. In recent years, more and more studies have confirmed that miRNA is differentially expressed in various malignant tumors and it affects tumor growth, invasion and metastasis, angiogenesis and sensitivity to radiotherapy. A variety of miRNAs are involved in the regulation of the occurrence and progression of NSCLCs. These miRNAs have important potential values in the early diagnosis, treatment and prognosis of NSCLC.
Studies have shown that miR-198 exerts a specific tumor inhibitory effect on a variety of cancers. The expression of miR-198 is downregulated in cervical cancer, while overexpression of miR-198 significantly reduces the proliferation, migration and invasion ability of cervical cancer cells through a mechanism involving targeted expression of mitogen-activated protein kinase 1 (MAPK1) [11]. Li et al. have shown that miR-198 inhibited the proliferation of breast cancer cells and promoted their apoptosis by targeting the expression of CUB domain-containing protein 1 (CDCP1) [12]. Wang et al. found that miR-198 expression in colorectal tumor tissues was significantly lower than that in adjacent normal tissues, and its expression was significantly correlated with the patients survival [13]. Other studies have reported that miR-198 reduced the migration and invasion of osteosarcoma cells by targeting Rho associated coiled-coil containing protein kinase 1 (ROCK1) expression [14].
It is known that TLR-4, a type I transmembrane receptor on the cell surface, belongs to the toll-like receptor family which is widely expressed on various immune cells [15]. Lipopolysaccharide and the anti-tumor drug paclitaxel are specific ligands for TLR-4 and TLR-4 also recognizes endogenous ligands such as heat shock protein (HSP), fibrinol.ytic protein and hyaluronic acid oligosaccharides [16,17]. Some recent studies have confirmed that the high expression of TLR-4 is closely related to tumor metastasis and drug resistance. Liu et al. found that the expression level of TLR-4 in postoperative specimens of NSCLC patients was significantly higher than that in patients with benign lesions and its expression level was closely related to lymph node metastasis and tumor differentiation [18]. Yeh et al. overexpressed TLR-4 in lung cancer cell lines and observed enhanced proliferation and reduced apoptosis of the cells and further studies found that TLR-4 enhanced vascular endothelial growth factor (VEGF) expression [19].
In this study, it was found that the expression of miR-198 was significantly upregulated in tumor tissues and cell lines of NSCLC patients, indicating that miR-198 is involved in the pathogenesis of NSCLC and may be a tumor suppressor gene. The degree of downregulation was highest in A549 and H1703 cells. Therefore, these two cell lines were selected for use in subsequent studies. Overexpression of miR-198 in A549 and H1703 cells were successfully stabilized via liposome transfection. The proliferation, migration and invasion of A549 and H1703 cells were significantly reduced after overexpression of miR-198, when compared with the control group, indicating that miR-198 inhibited the proliferation, migration and invasion of NSCLC cell lines. Results from the application of bioinformatics software revealed that miR-198 had complementary binding sites for TLR-4 3'URT. Double luciferase experiments confirmed that miR-198 inhibited luciferase activity of wild-type plasmids, while overexpression of miR-198 reduced the protein expression of TLR-4 in A549 cells. Analysis of clinical samples revealed a negative correlation between miR-198 and TLR-4 protein expression, thereby further confirming the targeted inhibition of TLR-4.
In summary, miR-198 is downregulated in NSCLC, while overexpression of miR-198 significantly inhibits the proliferation, migration and invasion of NSCLC cells through a mechanism most likely involving targeted inhibition of TLR-4 expression. These findings suggest that miR-198 has the potential for use as a marker for the diagnosis and treatment of NSCLC.
Acknowledgments:
This work was supported by The Project for Science and Technology in Yangzhou City (YZ2017071) and Jiangsu Provincial Commission of Health and Family Planning Research Project (Z201314).
Author’s contribution:
All work was done by the author named in this article and the authors accept all liability resulting from claims which relate to this article and its contents. The study was conceived and designed by Shichun Lu and Hongcan Shi; Weiguo Jin, Yusheng Shu, Xiaolin Wang, Shichun Lu, Hongcan Shi collected and analysed the data, Weiguo Jin and Yusheng Shu wrote the text and all authors have read and approved the text prior to publication. Weiguo Jin and Yusheng Shu contributed equally to this work and should be considered as co-first authors. Shichun Lu and Hongcan Shi are the cocorresponding authors.
Conflicts of interest:
There are no conflicts of interest in this study.
References
- Liao J, Shen J, Leng Q, Qin M, Zhan M, Jiang F. MicroRNA‐based biomarkers for diagnosis of non-small cell lung cancer (NSCLC). Thorac Cancer 2020;11(3):762-8.
- Gober MK, Flight RM, Lambert J, Moseley H, Stromberg A, Black EP. Deregulation of a network of mRNA and miRNA genes reveals that CK2 and MEK inhibitors may synergize to induce apoptosis KRAS-active NSCLC. Cancer Inform 2019;18:1176935119843507.
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol 2017;15(1):1-4.
- Zaporozhchenko IA, Rykova EY, Laktionov PP. The Fundamentals of miRNA Biology: Structure, Biogenesis, and Regulatory Functions. Russ J Bioorgan Chem 2020;46(1):1-3.
- Lopez-Rincon A, Martinez-Archundia M, Martinez-Ruiz GU, Schoenhuth A, Tonda A. Automatic discovery of 100-miRNA signature for cancer classification using ensemble feature selection. BMC Bioinformat 2019;20(1):1-7.
- Cui Z, Zheng X, Kong D. Expression of Concern to: Decreased miR-198 expression and its prognostic significance in human gastric cancer. World J Surg Oncol 2019;17(1):1.
- Liu W, Zhao J, Jin M, Zhou M. circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol Ther Nucleic Acids 2019;14:609-16.
- Duan X, Jiang B, Yang J, Zhou L, Tian B, Mao X. FOXP 3 inhibits MYC expression via regulating miR‐198 and influences cell viability, proliferation and cell apoptosis in HepG2. Cancer Med 2018;7(12):6182-92.
- Novello S, Barlesi F, Califano R, Cufer T, Ekman S, Levra MG, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annal Oncol 2016;27:1-27.
- Heller G, Altenberger C, Steiner I, Topakian T, Ziegler B, Tomasich E, et al. DNA methylation of microRNA-coding genes in non-small-cell lung cancer patients. J Pathol 2018;245(4):387-98.
- Xu F, Ni M, Li J, Cheng J, Zhao H, Zhao J, et al. Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun 2020;526(1):14-20.
- Niu L, Zhou Y, Zhang W, Ren Y. Long noncoding RNA LINC00473 functions as a competing endogenous RNA to regulate MAPK1 expression by sponging miR-198 in breast cancer. Pathol Res Pract 2019;215(8):152470.
- Li LX, Lam IH, Liang FF, Yi SP, Ye LF, Wang JT, et al. MiR-198 affects the proliferation and apoptosis of colorectal cancer through regulation of ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci 2019;23(4):1487-93.
- Zhang S, Zhao Y, Wang L. MicroRNA-198 inhibited tumorous behaviors of human osteosarcoma through directly targeting ROCK1. Biochem Biophys Res Commun 2016;472(3):557-65.
- Li L, Pan J, Cai X, Gong E, Xu C, Zheng H, et al. Human umbilical cord mesenchymal stem cells suppress lung cancer via TLR-4/NF-κB signalling pathway. Biotechnol Biotechnol Equipment 2020;34(1):24-9.
- Sun Y, Zheng J, Xu Y, Zhang X. Paraquat-induced inflammatory response of microglia through HSP60/TLR-4 signaling. Human Exp Toxicol 2018;37(11):1161-8.
- Fei D, Meng X, Yu W, Yang S, Song N, Cao Y, et al. Fibronectin (FN) cooperated with TLR2/TLR-4 receptor to promote innate immune responses of macrophages via binding to integrin β1. Virulence 2018;9(1):1588-600.
- An J, Li Z, Dong Y, Ren J, Guo K. Methicillin-Resistant Staphylococcus Aureus infection exacerbates NSCLC cell metastasis by up-regulating TLR-4/MyD88 pathway. Cell Mol Biol 2016;62(8):1-7.
- Zhang L, Tao X, Fu Q, Ge C, Li R, Li Z, et al. Curcumin inhibits cell proliferation and migration in NSCLC through a synergistic effect on the TLR-4/MyD88 and EGFR pathways. Oncol Rep 2019;42(5):1843-55.