- *Corresponding Author:
- Maiqiu Wang
School of Biological and Chemical Engineering, Zhejiang University of Science and Technology, Hangzhou, Zhejiang Province 310023, China
E-mail: maiqiu_wang@zust.edu.cn
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “39-44” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Insomnia stands out as a prevalent sleep disorder, with recent research highlighting the significance of psychological factors in its initiation and progression. Nonetheless, the precise biological mechanisms underlying insomnia remain elusive. Although genome-wide association studies have pinpointed numerous genetic loci linked to insomnia, elucidating the underlying biological rationale necessitates further investigation. We utilized the summary data-based Mendelian randomization approach to integrate genomewide association studies with expression quantitative trait loci studies and methylation quantitative trait loci studies. Additionally, we employed the heterogeneity in dependent instruments test to enhance our comprehension of the study findings and to identify and mitigate potential sources of bias or misinterpretation, thereby enhancing the credibility and accuracy of our study. We conducted expression quantitative trait loci analysis using the summary data-based Mendelian randomization method, uncovering 119 loci linked to gene expression. Concurrently, methylation quantitative trait loci analysis identified 491 loci associated with DNA methylation. Encouragingly, nine single nucleotide polymorphisms were found to overlap in both analyses. Subsequently, we conducted additional summary data-based Mendelian randomization analysis on these expression quantitative trait loci and methylation quantitative trait loci data, ultimately revealing 175 mediating models. These models elucidate the regulatory mechanism by which genetic variation influences DNA methylation, thereby modulating gene expression and ultimately influencing insomnia. This discovery offers valuable insights for a more comprehensive understanding of the genetic underpinnings of insomnia.
Keywords
Insomnia, Mendelian randomization, genome-wide association studies, expression quantitative trait loci, methylation quantitative trait loci
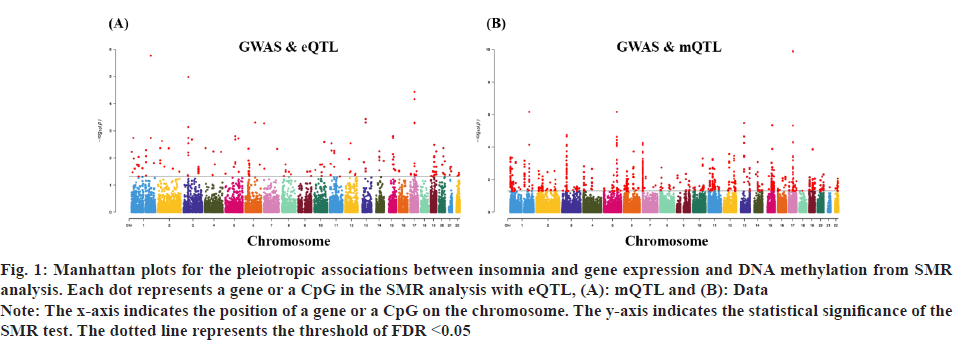
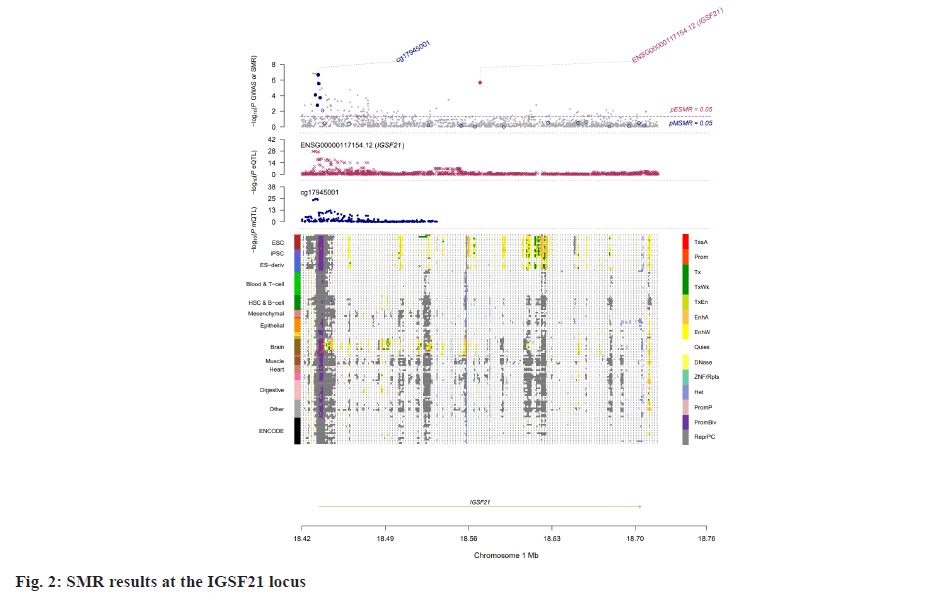
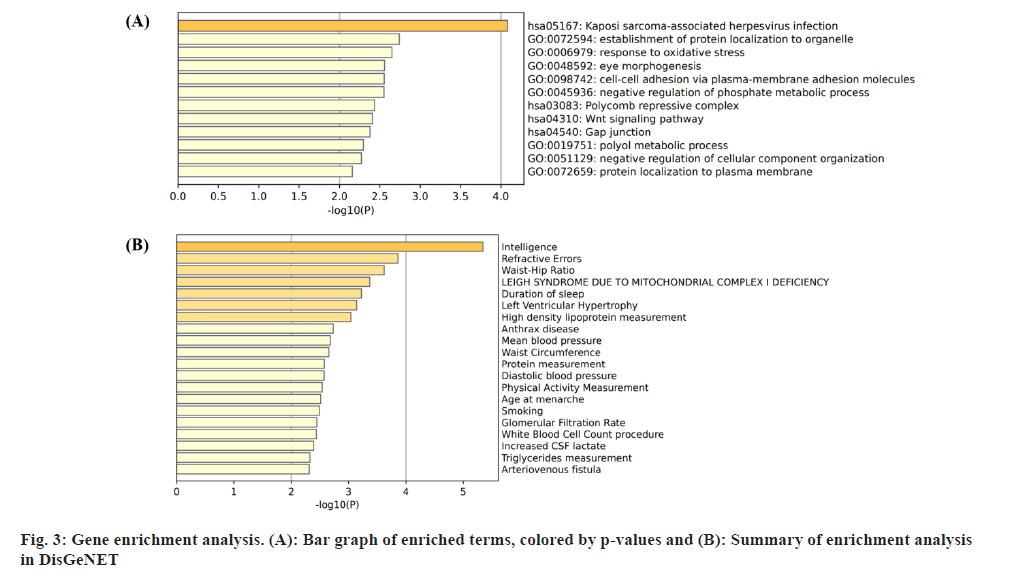
Insomnia stands as one of the prevailing, persistent sleep disorders, with its global prevalence currently estimated between 10 % and 20 % and projected to escalate amidst contemporary lifestyles[1]. Established risk factors for insomnia encompass advanced age, female gender, emotional disturbances, substance misuse, and environmental influences. Insomnia heightens susceptibility to anxiety, depression, cognitive impairment, cardiometabolic disorders[2], and contributes to diminished quality of life and productivity. Despite its epidemiological significance, the precise pathogenesis of insomnia remains largely elusive. Family-based genetic studies have elucidated that approximately 22 %-25 % of insomnia is heritable[3]. Large-scale Genome-Wide Association Studies (GWAS) have further identified 57 loci associated with insomnia[4]. However, the mechanisms through which these variants influence insomnia via specific genes or Deoxyribonucleic Acid (DNA) regulatory elements remain ambiguous. To address this puzzle, we introduced a novel approach, the Summary data-based Mendelian Randomization (SMR). This method integrates GWAS data with molecular trait data, including gene expression Quantitative Trait Loci (cis-eQTL) studies and DNA methylation (DNAm) cis-mQTL studies. Notably, SMR methods have previously been employed to uncover pleiotropic genes and DNAm loci in atrial fibrillation[5], yielding promising results in identifying potential risk genes and pathways for neuropsychiatric and substance use disorders[6]. Utilizing SMR methods, we discerned functional genes or DNAm loci pertinent to insomnia, amalgamating extensive insomnia GWAS summary statistics with cis-eQTL/cis-mQTL data in the brain. Furthermore, through the amalgamation of consistent pleiotropic associations across DNAm and insomnia, gene expression and insomnia, and DNAm and gene expression, we delineated numerous mediating models for genes associated with insomnia susceptibility. Specifically, our findings underscore that the influence of genetic variation on insomnia is mediated through DNAm regulation of gene transcription. These findings not only furnish pivotal insights for forthcoming investigations but also afford a deeper understanding of the functional mechanisms underlying DNA mutations implicated in insomnia, thus furnishing novel perspectives for the development of therapeutic interventions. Summary statistics for GWAS on insomnia were acquired from the United Kingdom (UK) Biobank, and data were accessible via the sleep disorders knowledge portal data download[4]. The study comprised individuals of European ancestry from the UK Biobank (n=453, 379), who were queried regarding their frequency of experiencing insomnia, specifically, "Do you have trouble falling asleep at night or waking up in the middle of the night?". Notably, 29 % of respondents reported frequent (usually) episodes of insomnia. Female participants, older individuals, shift workers, and those reporting shorter sleep durations exhibited a higher prevalence of insomnia symptoms. The brain eQTL data utilized in this study were sourced from the meticulously curated BrainMeta v2 cis-eQTL summary dataset, accessible via the SMR website. These eQTL data were derived from the brain cortex tissue of 2443 unrelated individuals of European ancestry, each with available genotype and Ribonucleic Acid sequencing (RNA-seq) data[7]. A total of 1 962 048 cis-eQTL associations were identified, with a significance threshold of p<5×10-8, encompassing 16 704 genes. Furthermore, the brain mQTL data were obtained from the meticulously prepared BrainMeta mQTL dataset, also accessible on the SMR website. These mQTL data were derived from a comprehensive meta-analysis[8] encompassing three independent studies; Religious Orders Study and Memory and Aging Project (ROSMAP) data (brain cortical region, n=468)[9], Hannon et al.[10] data (fetal brain, n=166), and Jaffe et al.[11] data (frontal cortex region, n=526). Notably, all samples included in these studies were of European descent, and DNAm levels were quantified using the Illumina Infinium HumanMethylation450K array. The SMR analysis was employed to capitalize on the estimated effects of instrumental Single Nucleotide Polymorphisms (SNPs) on both outcome and exposure, aiming to detect pleiotropic associations. A detailed description of this methodology can be found elsewhere[12]. In the context of testing for genes/Cancer Predisposition Genes (CPGs), exposure pertained to gene expression/DNAm, while outcome referred to insomnia. Only genes/CpGs with at least one eQTL/mQTL surpassing the threshold of p<5×10-8 were included in the analysis. Specifically, cis-eQTL/mQTL within a 2 Mb distance from each gene/CpG were considered for analysis. However, it’s crucial to note that the SMR analysis cannot entirely exclude the possibility of two variants in Linkage Disequilibrium (LD) independently influencing two traits; for instance, one may affect gene expression while the other impacts insomnia. Therefore, we additionally employed the Healthcare Effectiveness Data and Information Set (HEDIS) method[12]. A gene/CpG was considered a potential causal candidate when it passed both the SMR test (False Discovery Rate (FDR) <0.05) and the HEIDI test (p>0.05). Subsequently, significant genes and CpGs were utilized to explore potential mediation mechanisms, wherein the effect of a SNP on insomnia was mediated by gene expression through DNAm. In the context of testing for epigenetic mediation mechanisms, exposure was defined as DNAm, while outcome was defined as gene expression. Gene-based enrichment analyses were conducted on the genes that passed both the SMR and HEIDI tests. CpGs identified through the mQTL data were mapped to UCSC_RefGene_Name using the annotation file provided by the manufacturer to assign them to genes. Subsequently, we performed enrichment analyses on the obtained genes using Metascape, an effective online tool for uncovering biological functions and pathway enrichment information for gene sets[13]. This analysis offers valuable insights into the underlying biological processes and pathways associated with these genes, thus providing crucial clues for investigating the pathophysiological mechanisms of insomnia. The eQTL and mQTL data derived from brain tissue were initially employed to identify potential risk genes and CpGs associated with insomnia through SMR analysis. Specifically, only cis-eQTL/mQTL was considered in the analysis to minimize the potential for pleiotropy. The outcomes of the SMR analysis, conducted at both the gene expression and DNAm levels, are illustrated in fig. 1A and fig. 1B. The SMR analysis revealed 171 genes significant at an FDR threshold of <0.05, among which 119 genes passed the null hypothesis (indicating a single causal variant influencing both gene expression and insomnia) of the HEIDI test (p>0.05). For detailed information regarding these results. In the DNAm analysis, the SMR analysis identified 963 CpGs significant at an FDR threshold of <0.05, with 491 CpGs (corresponding to the 17 closest genes) passing the null hypothesis (indicating a single causal variant influencing both DNAm and insomnia) of the HEIDI test (p>0.05). DNAm is a prevalent epigenetic modification known to regulate gene expression. In our study, we aimed to investigate whether DNAm could influence insomnia by modulating gene expression. To explore this potential mediation mechanism, we integrated the 119 identified genes and 491 CpGs into the SMR and HEIDI analysis, with DNAm considered as the exposure and gene expression as the outcome. Notably, the expression levels of 15 genes showed significant associations with at least one CpG at an FDR threshold of <0.05 in the SMR test. Additionally, 175 CpG-gene pairs passed the null hypothesis (indicating a single causal variant influencing both gene expression and DNAm) of the HEIDI test (p>0.05). For example, the cg17945001-Immunoglobin Superfamily Member 21 (IGSF21)-insomnia axis represents a plausible mediation model. Here, cg17945001 refers to a CpG located in the 5’ end of the IGSF21 gene fig. 2. It has been demonstrated that IGSF21 selectively regulates inhibitory presynaptic differentiation and plays a crucial role in synaptic inhibition in the brain[14]. The SMR and HEIDI analysis revealed 136 unique genes, which were subsequently subjected to gene-based enrichment analysis. Utilizing Metascape, we conducted pathway and process enrichment analysis and found that these genes are significantly associated with pathways such as Kaposi sarcoma-associated herpes virus infection and establishment of protein localization to organelle, among others (fig. 3A). Additionally, our analysis in DisGeNET demonstrated strong associations between the identified genes and traits such as intelligence and duration of sleep (fig. 3B)[15]. These findings underscore the potential involvement of these genes in diverse biological processes and provide valuable insights into the pathophysiological mechanisms underlying insomnia. To our knowledge, this study represents the first attempt to integrate GWAS, eQTL, and mQTL datasets in the investigation of insomnia. The SMR and HEIDI methodologies were employed to assess pleiotropic associations between gene expression level/DNAm and insomnia. Additionally, our endeavor aimed to elucidate which CpGs might influence insomnia through the regulation of gene expression. In total, we have identified 136 genes whose expression is linked to insomnia. Notably, some of these genes have been previously associated with insomnia or related sleep disorders. For instance, MYO1H was among the 57 loci previously identified by genome-wide association analysis as being linked to insomnia symptoms[4]. Additionally, a study concluded that Peroxisome proliferator-activated receptor-Gamma Coactivator (PGC) increases the risk of insomnia, which aligns with the directional effect of PGC expression observed in our findings (0.0025). Other genes associated with sleep disorders include SNCA-AS1, which exhibited differential expression across various brain regions in a study on rapid eye movement sleep behavior disorder and was further supported by co-localization analysis[16]. Moreover, Mediator Complex Subunit 20 (MED20) was found to be associated with sleep health scores in a comprehensive genetic analysis[17], while polymorphisms in Mitogen-Activated Protein 2 Kinase 5 (MAP2K5) and SKOR1 were associated with periodic leg movements during sleep[18]. Furthermore, alterations in protein abundance of Containing C2 (CSDC2) and Cyclin M2 (CNNM2) in the brain were linked to sleep apnea[19], and C7orf50 was associated with sleepiness[20]. Enrichment analysis of the 136 genes also revealed a robust association with sleep duration. In our analysis, we integrated models of insomnia susceptibility genes by considering consistent pleiotropic associations between DNAm and insomnia, between gene expression and insomnia, and between DNAm and gene expression. For instance, while the association of the IGSF21 locus with insomnia at the epigenetic level was previously unclear, our study shed light on this aspect. We identified a risk CpG (cg17945001) in the IGSF21 promoter region and established the cg17945001-IGSF21-Insomnia axis as a mediator model. The effect estimate of cg17945001-IGSF21 was -0.68, aligning with the typical inverse correlation between DNAm in promoter regions and gene expression. Although direct experimental evidence linking IGSF21 to insomnia is lacking, our findings, coupled with the known role of IGSF21 in synaptic function[14], suggest its plausibility as a susceptibility gene for insomnia. Thus, IGSF21 could potentially serve as a promising drug target for the prevention and treatment of insomnia. Our study has several limitations that should be acknowledged. Firstly, we exclusively analyzed eQTL and mQTL data derived from brain tissue. While the Central Nervous System (CNS) is undoubtedly pivotal in insomnia, pathways involving other tissues may also contribute. Secondly, the exclusion of trans-eQTL/mQTL from our analysis means that we might have overlooked some potential risk genes/CpGs. Lastly, our study solely incorporated eQTL and mQTL data, and future investigations could consider incorporating additional types of molecular traits, such as splicing QTL (sQTL) and protein QTL (pQTL), for a more comprehensive understanding. Our SMR analysis identified numerous genes and DNAm loci exhibiting pleiotropic or potentially causal associations with insomnia. Additionally, we elucidated plausible mechanisms by which genetic variation impacts insomnia through the genetic regulation of DNAm transcription. The genes and CpGs uncovered in our study represent promising candidates for future functional investigations.
Fig. 1: Manhattan plots for the pleiotropic associations between insomnia and gene expression and DNA methylation from SMR
analysis. Each dot represents a gene or a CpG in the SMR analysis with eQTL, (A): mQTL and (B): Data.
Note: The x-axis indicates the position of a gene or a CpG on the chromosome. The y-axis indicates the statistical significance of the
SMR test. The dotted line represents the threshold of FDR <0.05.
Conflict of interests:
The authors declared no conflict of interests.
References
- Morin CM, Benca R. Chronic insomnia. Lancet 2012;379(9821):1129-41.
- Hoevenaar-Blom MP, Spijkerman AM, Kromhout D, van Den Berg JF, Verschuren WM. Sleep duration and sleep quality in relation to 12-year cardiovascular disease incidence: The MORGEN study. Sleep 2011;34(11):1487-92.
[Crossref] [Google Scholar] [PubMed]
- Lind MJ, Gehrman PR. Genetic pathways to insomnia. Brain Sci 2016;6(4):64.
[Crossref] [Google Scholar] [PubMed]
- Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet 2019;51(3):387-93.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Li B, Ma Y, Huang Y, Ouyang F, Liu Q. Mendelian randomization integrating GWAS, eQTL, and mQTL data identified genes pleiotropically associated with atrial fibrillation. Front Cardiovasc Med 2021;8:745757.
[Crossref] [Google Scholar] [PubMed]
- Gedik H, Nguyen TH, Peterson RE, Chatzinakos C, Vladimirov VI, Riley BP, et al. Identifying potential risk genes and pathways for neuropsychiatric and substance use disorders using intermediate molecular mediator information. Front Genet 2023;14:1191264.
[Crossref] [Google Scholar] [PubMed]
- Qi T, Wu Y, Fang H, Zhang F, Liu S, Zeng J, et al. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat Genet 2022;54(9):1355-63.
[Crossref] [Google Scholar] [PubMed]
- Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun 2018;9(1):2282.
[Crossref] [Google Scholar] [PubMed]
- Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, et al. An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat Neurosci 2017;20(10):1418-26.
[Crossref] [Google Scholar] [PubMed]
- Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 2016;19(1):48-54.
[Crossref] [Google Scholar] [PubMed]
- Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016;19(1):40-7.
[Crossref] [Google Scholar] [PubMed]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016;48(5):481-7.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1523.
[Crossref] [Google Scholar] [PubMed]
- Tanabe Y, Naito Y, Vasuta C, Lee AK, Soumounou Y, Linhoff MW, et al. IgSF21 promotes differentiation of inhibitory synapses via binding to neurexin2α. Nat Commun 2017;8(1):408.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucl Acids Res 2017;45(D1):D833-9.
[Crossref] [Google Scholar] [PubMed]
- Krohn L, Heilbron K, Blauwendraat C, Reynolds RH, Yu E, Senkevich K, et al. Genome-wide association study of REM sleep behavior disorder identifies polygenic risk and brain expression effects. Nat Commun 2022;13(1):7496.
[Crossref] [Google Scholar] [PubMed]
- Yao Y, Jia Y, Wen Y, Cheng B, Cheng S, Liu L, et al. Genome-wide association study and genetic correlation scan provide insights into its genetic architecture of sleep health score in the UK biobank cohort. Nat Sci Sleep 2022:1-2.
[Crossref] [Google Scholar] [PubMed]
- Moore IV H, Winkelmann J, Lin L, Finn L, Peppard P, Mignot E. Periodic leg movements during sleep are associated with polymorphisms in BTBD9, TOX3/BC034767, MEIS1, MAP2K5/SKOR1, and PTPRD. Sleep 2014;37(9):1535-42.
[Crossref] [Google Scholar] [PubMed]
- Gui J, Meng L, Huang D, Wang L, Yang X, Ding R, et al. Identification of novel proteins for sleep apnea by integrating genome-wide association data and human brain proteomes. Sleep Med 2024;114:92-9.
[Crossref] [Google Scholar] [PubMed]
- Barfield R, Wang H, Liu Y, Brody JA, Swenson B, Li R, et al. Epigenome-wide association analysis of daytime sleepiness in the Multi-ethnic study of atherosclerosis reveals African-American-specific associations. Sleep 2019;42(8):zsz101.
[Crossref] [Google Scholar] [PubMed]