- *Corresponding Author:
- Yifan Huang
Department of Pathology, School of Basic Medicine, North Sichuan Medical College, Nanchong, 637000, China
E-mail: huangyifan2021225@163.com
| This article was originally published in a special issue,“Therapeutic Perspectives in Biomedical Research and Pharmaceutical Sciences and their Nursing Methods” |
| Indian J Pharm Sci 2021:83(4)Spl issue “207-213” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the effect of circular RNA_0025033 on the proliferation, migration and invasion of cervical cancer HeLa cells and its molecular mechanism. The cancer tissues and adjacent tissues of 39 patients with cervical cancer in our hospital from January 2017 to December 2020 were selected. The expression levels of circular RNA_0025033 and microRNA-1256 were detected by Quantitative reverse transcription polymerase chain reaction. Cell counting kit 8 was used to detect cell viability. Plate cloning test was conducted to detect the number of cell clones. Scratch test was carried out to detect cell scratch healing rate. Transwell chamber was used to detect cell invasion. Western blot was adopted to detect protein expression. Dual luciferase reporter assay was carried out to detect the targeting relationship between circular RNA_0025033 and microRNA-1256. The expression level of circular RNA_0025033 in cervical cancer tissues was increased and the expression level of microRNA-1256 was decreased (p<0.05). After inhibiting the expression of circular RNA_0025033 or overexpression of microRNA-1256, cell viability was decreased and the number of cell clone formation was decreased. The healing rate of cell scratches was decreased and the number of invasive cells was decrease. E-cadherin expression was increased and N-cadherin expression was decreased (p<0.05). Circular RNA_0025033 targeted regulation of the expression of microRNA-1256 was carried out; interference with microRNA-1256 expression reversed the inhibition of circular RNA_0025033 on the proliferation, migration and invasion of cervical cancer HeLa cells. Inhibition of circular RNA_0025033 expression may inhibit the proliferation, migration and invasion of cervical cancer HeLa cells through targeted regulation of microRNA-1256.
Keywords
Circular RNA Circ_0025033, microRNA 1256, cervical cancer, chemotherapy
The mortality rate of cervical cancer patients ranks in the forefront of female malignant tumors, which seriously threatens women's life and health. The emerging targeted drugs are expected to improve the treatment effect and living quality of cervical cancer patients[1,2]. Studies have shown that circular RNA (circRNA) plays an important role in tumor progression and can be used as a diagnostic or therapeutic target for various types of cancers[3]. It is reported that circ_0025033 is overexpressed in ovarian cancer tissues and cells and promotes the development of ovarian cancer by regulating microRNA (miR)-330- 5p/Kallikrein-related peptidase-4 (KLK4) axis[4]. Upregulating circ_0025033 promotes the proliferation and invasion of papillary thyroid carcinoma cells by sponging miR-1231 and miR-1304[5]. However, the effect and mechanism of circ_0025033 on the proliferation, migration and invasion of cervical cancer cells are still unclear. According to the prediction of biological software, circ_0025033 has a binding site with miR-1256. It is reported that the high expression of miR-1256 can inhibit the cell proliferation and cell cycle progression of papillary thyroid carcinoma[6]. The expression of miR-1256 is down-regulated in nonsmall cell lung cancer tissues and the overexpression of miR-1256 inhibits the proliferation and migration of non-small cell lung cancer cells[7]. Circ-DCAF6 promotes the growth and invasion of gastric cancer cells by inhibiting the expression levels of miR-1231 and miR-1256[8]. The effect of miR-1256 on proliferation, migration and invasion of cervical cancer cells and whether circ_0025033 regulates miR-1256 are still unclear. Therefore, this experiment aims to study the effect of circ_0025033 on the proliferation, migration and invasion of cervical cancer cells as well as whether the mechanism is related to miR-1256.
Materials and Methods
Data:
From January 2017 to December 2020, 39 patients with cervical cancer, aged from 32 to 75 y were selected. All patients were diagnosed as cervical cancer by pathology and had not received radiotherapy, chemotherapy and biological immunotherapy before operation; the tissues adjacent to cancer were taken as control. All patients were informed and agreed.
Cells and main reagents:
Cervical cancer HeLa cells were purchased from the American type culture collection (ATCC), the USA; Roswell Park Memorial Institute (RPMI) 1640 medium was purchased from Gibco, the USA; Trizol reagent and fluorescent quantitative kit were purchased from Takara Co., Ltd., Japan; LipofectamineTM 2000 transfection reagent was purchased from Invitrogen, the USA; cell counting kit 8 (CCK-8) was purchased from Tongren Institute, Japan; Transwell cell and matrix glue were purchased from BD Company, the USA; RIPA protein lysis buffer was purchased from Beyotime Biotechnology Research Institute; double luciferase reporter gene detection kit was purchased from Shanghai Beinuo Biotechnology Co., Ltd.
Cell processing and grouping:
The cervical cancer HeLa cells were cultured in RPMI- 1640 medium and were collected in the logarithmic phase. The collected cells were transfected with negative control siRNA (siNC), si-circ_0025033, miRNC and miR-1256 respectively into HeLa cells, which were recorded as si-NC group, si-circ_0025033 group, miR-NC group and miR-1256 group; si-circ_0025033 was co-transfected with anti-miR-NC and antimiR- 1256, respectively into HeLa cells and recorded as the si-circ_0025033+anti-miR-NC group and the sicirc_ 0025033+anti-miR-1256 group.
Detection of the expression levels of circ_0025033 and miR-1256 by Quantitative reverse transcription polymerase chain reaction (RT-qPCR):
The surgically removed cervical cancer tissues and adjacent tissues were stored in a sterilized cryopreservation tube in a liquid nitrogen tank. 50 mg tissue was weighed and taken. The tissue was cut and ground, then 1 ml Trizol reagent was added for lysis; the cultured cells (1×106 cells) were directly lysed by adding 1 ml Trizol reagent. Total RNA was extracted and reversely transcribed into complementary DNA (cDNA) and then PCR was carried out according to the instructions of the kit. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal reference. The upstream primer sequence of circ_0025033 was 5'-GTGAGCCAGCTTGAGAACAC-3' and its downstream primer sequence was 5 ' - T TGAGAAT C A G T G G C C G A C G - 3 ' ; the upstream primer sequence of GAPDH was 5'-CCACATCGCTCAGACACCAT-3 and its downstream primer sequence was 5'-CCAGGCGCCCAATACG-3'; the upstream primer sequence of miR-1256 was 5'-GGCGCGATTTTAGTTTATC-3' and its downstream primer sequence was 5 ' - T T TAAT TACCAACCGAATA C G - 3 ' ; the upstream primer sequence of U6 was 5'-CTCGCTTCGGCAGCACA-3' and its downstream primer sequence was 5'-AACGCTTCACGAATTTGCGT-3'; the relative expression was calculated by 2-△△Ct.
Detection of cell proliferation by CCK-8:
The cells of each group were cultured for 48 h and 10 μl CCK-8 reagent was added to each well. The cells were cultured in a constant temperature incubator with 5 % CO2 at 37° for 4 h and the absorbance value (OD) of each well was measured with a microplate reader at 450 nm.
Detection of number of cell clone formation by plate cloning test:
After digestion of each group of cells, 2×103 cells of each group were re-inoculated into a 6-well plate and cultured in a constant temperature incubator with 5 % CO2 at 37°. After about 14 d, the culture was stopped. After washing the cells with Phosphate-buffered saline (PBS), paraformaldehyde was added to fix the cells and then Giemsa was added for staining for 30 min. After staining, the cells were washed, dried and counted under a microscope.
Detection of cell scratch healing rate by scratch test:
After digestion, the cells of each group were inoculated in a 6-hole culture plate for culture. After 24 h, the cells were scratched with a 200 μl Tip gun. At the 0th h and the 24th h after scratching, photos were taken for observation. The scratch area and the scratch healing rate were calculated by Image-Pro Plus6.0 software; scratch healing rate=(scratch area at the 0th h-scratch area at the 24th h)/scratch area at the 0th h×100 %.
Detection of cell invasion by Transwell chamber:
50 μl of matrix glue diluted at 1:6 was added into the upper chamber of transwell chamber and 100 μl of cell suspension with a concentration of 4×105 cells/ml was inoculated after coagulation. After 24 h of culture, the cells that did not pass through the membrane were wiped off. The remaining cells were stained with 0.1 % crystal violet, rinsed clean, observed and counted under an inverted microscope.
Detection of protein expression by Western blot:
After culture for 24 h, the cells of each group were collected. The lysate was added to extract the total cell protein. Then, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel electrophoresis was carried out, with the protein loading amount of each group being 30 μg. After electrophoresis, membrane transfer was carried out. The membrane was sealed with 5 % skim milk at room temperature for 1 h, incubated at 4° overnight with primary antibody and then incubated at room temperature for 2 h with secondary antibody. After that, it was exposed for development and fixation in a dark room. The gray value of protein band was analyzed. The ratio of target band to GAPDH band was taken as the protein expression level.
Dual luciferase reporter assay:
Wild-type and mutant dual luciferase reporter plasmids circ_0025033 were constructed and cotransfected into HeLa cells with miR-NC and miR-1256 respectively. Luciferase activity was detected according to the instructions. pcDNA, pcDNA-circ_0025033, si-NC and si-circ_0025033 were transfected into HeLa cells and the expression level of miR-1256 was detected by RT-qPCR.
Statistical analysis:
Statistical analysis was performed using statistical package for the social sciences (SPSS) 20.0 software.
The measurement data that conformed to the normal distribution were expressed as the meanstandard deviation (x±s). The t-test was performed for comparison between two groups and one-way analysis of variance (ANOVA) was used for comparison between multiple groups. p<0.05 was considered as the difference with statistical significance.
Results and Discussion
Compared with the adjacent tissues, the expression level of circ_0025033 was increased in the cervical cancer tissues and the expression level of miR-1256 was decreased (p<0.05) (Table 1).
| Group | circ_0025033 | miR-1256 |
|---|---|---|
| Adjacent tissues | 1.00±0.07 | 1.00±0.08 |
| Cervical cancer tissues | 4.51±0.34* | 0.39±0.04* |
| t | 63.146 | 42.591 |
| p | 0.000 | 0.000 |
Note: Compared with the adjacent tissues, *p<0.05
Table 1: Expressions of Circ_0025033 and Mir-1256 in Cervical Cancer Tissue (X̄±S, N=39)
Compared with the si-NC group, the expression level of circ_0025033 in the si-circ_0025033 group was decreased. In addition, the cell activity was decreased and the number of cell clone formation was decreased (p<0.05) (Table 2).
| Group | circ_0025033 | OD value (450 nm) | Number of cell clone formation |
|---|---|---|---|
| si-NC | 1.00±0.00 | 0.93±0.07 | 88.92±7.54 |
| si-circ_0025033 | 0.43±0.04* | 0.42±0.04* | 34.11±3.56* |
| t | 42.750 | 18.977 | 19.720 |
| p | 0.000 | 0.000 | 0.000 |
Note: Compared with the si-NC group, *p<0.05
Table 2: The Effect of Inhibiting the Circ_0025033 Expression on the Proliferation of Cervical Cancer Hela Cells
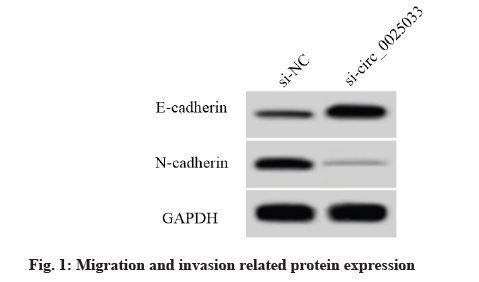
Compared with the si-NC group, the scratch healing rate and the number of invasive cells in the sicirc_ 0025033 group were decreased. The expression level of E-cadherin protein was increased and the expression level of N-cadherin protein was decreased (p<0.05) (fig. 1 and Table 3).
| Group | Scratch healing rate (%) | Invasive cells | E-cadherin protein | N-cadherin protein |
|---|---|---|---|---|
| si-NC | 61.68±5.33 | 125.89±10.80 | 0.29±0.02 | 0.59±0.04 |
| si-circ_0025033 | 24.19±2.18* | 56.56±5.37* | 0.69±0.05* | 0.17±0.02* |
| t | 19.531 | 17.244 | 22.283 | 28.174 |
| p | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with the si-NC group, *p<0.05
Table 3: The Effect of Inhibiting the Circ_0025033 Expression on the Migration and Invasion of Cervical Cancer Hela Cells (X̄±S, N=9)
Circular RNA interactome software predicted that the sequence of circ_0025033 contained a nucleotide sequence complementary to miR-1256 (fig. 2). The results of the dual luciferase reporter assay showed that the luciferase activity of cells co-transfected with wild type (WT)-circ_0025033 and miR-1256 was decreased, while the luciferase activity of cells co-transfected with MUT-circ_0025033 and miR-1256 was not significantly changed (Table 4). Compared with the pcDNA group, the expression level of miR-1256 in the pcDNA-circ_0025033 group was decreased. Compared with the si-NC group, the expression level of miR-1256 in the si-circ_0025033 group was increased (p<0.05) (Table 5).
| Group | WT-circ_0025033 | MUT-circ_0025033 |
|---|---|---|
| miR-NC | 1.04±0.07 | 1.01±0.06 |
| miR-1256 | 0.52±0.04* | 1.05±0.04 |
| t | 19.349 | 1.664 |
| p | 0.000 | 0.116 |
Note: Compared with the miR-NC group, *p<0.05
Table 4: Dual Luciferase Reporter Assay (X̄±S, N=9)
| Group | miR-1256 |
|---|---|
| pcDNA | 1.00±0.00 |
| pcDNA-circ_0025033 | 0.33±0.03* |
| si-NC | 0.99±0.06 |
| si-circ_0025033 | 3.11±0.26# |
| F | 730.631 |
| p | 0.000 |
Note: Compared with the miR-NC group, *p<0.05
Table 5: Circ_0025033 Regulates the Expression of Mir-1256 (X̄±S, N=9)
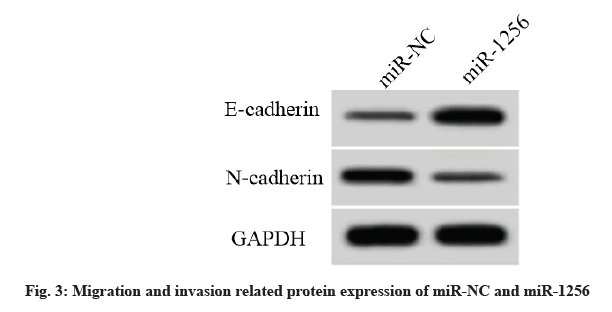
Compared with the miR-NC group, the miR-1256 expression level in the miR-1256 group was increased and the cell activity and the number of cell clone formation were decreased. The cell scratch healing rate and the number of invasive cells were decreased. The E-cadherin protein expression level was increased and the N-cadherin protein expression level was decreased (p<0.05) (fig. 3, Table 6).
| Group | miR-1256 | OD value (450 nm) | Number of cell clone formation | Scratch healing rate (%) | Invasive cells | E-cadherin protein | N-cadherin protein |
|---|---|---|---|---|---|---|---|
| miR-NC | 1.00±0.00 | 0.98±0.08 | 87.72±8.45 | 63.54±5.28 | 122.59±13.01 | 0.27±0.03 | 0.55±0.04 |
| miR-1256 | 2.66±0.23* | 0.63±0.05* | 44.68±4.11* | 34.61±3.39* | 65.49±5.15* | 0.62±0.05* | 0.24±0.03* |
| t | 21.652 | 11.130 | 13.741 | 13.832 | 12.243 | 18.007 | 18.600 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with the miR-NC group, *p<0.05
Table 6: Effect Of Mir-1256 Overexpression On The Proliferation, Migration And Invasion Of Cervical Cancer Hela Cells (X?±S, N=9)
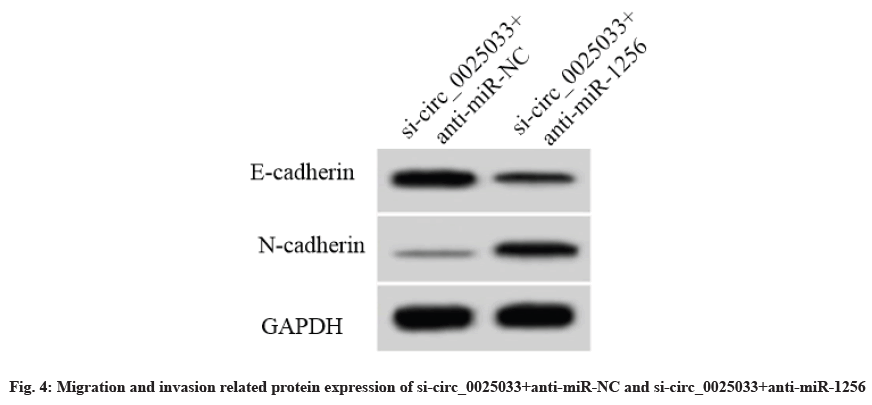
Compared with the si-circ_0025033+anti-miR-NC group, the miR-1256 expression level of the sicirc_ 0025033+anti-miR-1256 group was decreased and the cell activity was increased. The formation number of cell clones, the cell scratch healing rate and the number of invaded cells were increased. The E-cadherin protein expression level and the N-cadherin protein expression level were decreased (p<0.05) (fig. 4, Table 7).
| Group | miR-1256 | OD value (450 nm) | Number of cell clone formation | Scratch healing rate (%) | Invasive cells | E-cadherin protein | N-cadherin protein |
|---|---|---|---|---|---|---|---|
| si-circ_0025033+anti-miR-NC | 1.00±0.00 | 0.41±0.04 | 33.94±3.93 | 22.86±2.53 | 55.38±4.94 | 0.68±0.06 | 0.16±0.02 |
| si-circ_0025033+anti-miR-1256 | 0.36±0.04* | 0.80±0.07* | 78.55±6.58* | 52.05±4.81* | 99.93±8.41* | 0.38±0.04* | 0.47±0.04* |
| t | 48.000 | 14.512 | 17.462 | 16.113 | 13.703 | 12.481 | 20.795 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: compared with the si-circ_0025033+anti-miR-NC group, *p<0.05
Table 7: Interference with Mir-1256 Expression Reversed the Effect of Inhibiting Circ_0025033 on the Proliferation, Migration and Invasion of Cervical Cancer Hela Cells (X̄±S, N=9)
Molecular targeted therapy has achieved good results in the treatment of gynecological malignant tumors and the increase of targeted drugs in clinical practice has a positive promoting effect on the treatment of cervical cancer[9,10]. Therefore, it is urgent to explore more new targets to develop new targeted drugs. CircRNA plays an important role in a variety of cancers and can become a promising biomarker for the diagnosis of various cancers[11]. It has been reported that knockdown of circ_0025033 inhibits the colony formation, migration/ invasion and glycolytic metabolism in ovarian cancer cells[12]. The down-regulation of CIRC_0025033 inhibits tumor growth of papillary thyroid carcinoma in vitro and in vivo[13]. The results of this experiment showed that the expression level of circ_0025033 in cervical cancer tissues was increased. It indicated that circ_0025033 might play a pro-cancer role in cervical cancer. After the expression of circ_0025033 was inhibited, the activity of cervical cancer HeLa cells was decreased and the number of cell clonal formation was reduced; the cell scratch healing rate and the number of invasive cells were decreased; the expression level of E-cadherin protein was increased and the expression level of N-cadherin protein was decreased; it indicated that inhibition of the expression of circ_0025033 could inhibit the proliferation, migration and invasion of cervical cancer cells.
Biological software predicted the presence of a binding site for circ_0025033 and miR-1256. It has been reported that miR-1256 is low expressed in colorectal cancer and it may be an independent prognostic parameter for predicting poor prognosis in colorectal cancer patients[14]. The results of this experiment showed that the expression level of miR-1256 in cervical cancer tissues was decreased. Overexpression of miR-1256 could reduce cell activity, the number of colony cell formation and the number of invasive cells and reduce the rate of cell scratch healing; these results indicated that the overexpression of miR-1256 could inhibit the proliferation, migration and invasion of cervical cancer cells. In addition, it has been reported that circ_0026134 regulates the proliferation and invasion of non-small cell lung cancer cells by sponging miR-1256 and miR-1287[15]. CircHECTD1 promotes glutamine decomposition by targeting miR-1256 and activating β-catenin/c-Myc signaling to promote the progress of gastric cancer[16]. The results of this experiment showed that circ_0025033 realized targeted regulation of the expression of miR-1256; interference with the expression of miR-1256 reversed the effects of inhibiting circ_0025033 on the proliferation, migration and invasion of cervical cancer HeLa cells. In summary, inhibiting the expression of circ_0025033 may inhibit the proliferation, migration and invasion of cervical cancer HeLa cells by targeted regulation of miR-1256.
Conflicts of interest:
The authors declared no conflicts of interest.
References
- Li HY, Cai J, Zhu HL. Research progress of clinical treatment of cervical cancer. J Ningxia Med Univ 2020;42(4):101-5.
- Vora C, Gupta S. Targeted therapy in cervical cancer. ESMO open 2018;3:e000462.
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176(4):869-81.
- Cheng H, Wang N, Tian J, Li Y, Ren L, Shi Z. Circular RNA Circ_0025033 promotes the evolvement of ovarian cancer through the regulation of miR-330-5p/KLK4 axis. Cancer Manag Res 2020;12(1):2753.
- Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun 2019;510(2):334-8.
- Wu C, Ma L, Wei H, Nie F, Ning J, Jiang T. MiR-1256 inhibits cell proliferation and cell cycle progression in papillary thyroid cancer by targeting 5-hydroxy tryptamine receptor 3A. Human cell 2020;33(3):630-40.
- Liu W, Wan X, Mu Z, Li F, Wang L, Zhao J, et al. MiR-1256 suppresses proliferation and migration of non-small cell lung cancer via regulating TCTN1. Oncol Lett 2018;16(2):1708-14.
- Wu L, Liu D, Yang Y. Enhanced expression of circular RNA circ-DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR-1231 and miR-1256 in gastric cancer. Exp Mol Pathol 2019;11(1):104273.
- Leilin, She JJ, Guo DT. Clinical research progress of molecular targeted therapy for gynecologic malignant tumor. Electronic J Clin Med Literature 2019;6(24):204.
- Zou YM. Research progress of targeted drugs for advanced cervical cancer. Electronic J Clin Med Literature 2020;7(25):197-8.
- Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol 2020;235(2):718-24.
- Hou W, Zhang Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract 2021;217(1):153275.
- Ye M, Hou H, Shen M, Dong S, Zhang T. Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma by sponging miR-1179 and regulating HMGB1 expression. Mol Ther Nucleic Acids 2020;19(1):741-50.
- Liu ZY, Yang L, Chang HY. Clinicopathologic and prognostic relevance of miR-1256 in colorectal cancer: a preliminary clinical study. Eur Rev Med Pharmacol Sci 2018;22:7704-9.
- Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother 2019;112:108743.
- Cai J, Chen Z, Wang J, Wang J, Chen X, Liang L, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis 2019;10(8):1-5.