- *Corresponding Author:
- H. Ding
Department of Clinical Laboratory, Tianjin Baodi Hospital, Baodi Clinical College of Tianjin Medical University, Baodi, Tianjin 301800, China
E-mail: 258860152@qq.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “79-85” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To elucidate the molecular mechanism underlying the high metastasis rate of brain for non-small cell lung cancer patients and to establish a comprehensive differential gene profile for non-small cell lung cancer brain metastasis. The differentially expressed genes for non-small cell lung cancer brain metastasis patients were systematically investigated. The enriched biological processes and signaling pathways were explored. The protein-protein interaction network was constructed. Compared with primary non-small cell lung cancer, the brain metastasis patients demonstrated 352 differentially expressed genes with the majority as down-regulated ones (342) and 10 up-regulated ones. Meanwhile, C-C motif chemokine ligand 21, C-C motif chemokine ligand 19, deleted in malignant brain tumors 1 gene, tryptase alpha/beta 1, lactotransferrin, complement component 4 binding protein alpha, C-C motif chemokine ligand 14, C-X-C motif chemokine ligand 9, cluster of differentiation 79A as well as membrane spanning 4-domains A1 were significant and differentially expressed (all down-regulated). The genes of cluster including C-C motif chemokine ligand 21, C-C motif chemokine ligand 19 played a pivotal role in non-small cell lung cancer brain metastasis. The cytokines C-C motif chemokine ligand 19 and C-C motif chemokine ligand 21 as well as their closely associated receptors, C-C chemokine receptor 7 could be detected as high expression levels in non-small cell lung cancer cell line (A549). Inhibition of C-C motif chemokine ligand 19 and C-C motif chemokine ligand 21 as well as C-C chemokine receptor 7 using small interfering ribonucleic acid obviously repressed the cell invasion and metastasis but not for cell proliferation and apoptosis. Meanwhile, the procedure was independent of micro ribonucleic acid lethal-7a regulation. This study hypothesized an innovative signaling axis (C-C motif chemokine ligand 21/C-C motif chemokine ligand 19-C-C chemokine receptor 7) for non-small cell lung cancer brain metastasis and provided a beneficial strategy for better understanding the lung metastases in clinic.
Keywords
Lung cancer, non-small cell lung cancer, metastasis, protein-protein interaction analysis
Lung cancer is characterized as the most abundant cancer and the leading cause of cancer death for men. Meanwhile, it is also the third most common cancer (just after breast and colorectal cancers) and the second leading cause of cancer death (just after breast cancer) for women worldwide[1]. As estimated, the worldwide incidence and mortality of lung cancer is about 1.82 and 1.6 million per year, contributing for 12.9 % of total incidence from carcinoma[2]. The lung cancer is classified into Small Cell Lung Cancer (SCLC) as well as Non-Small Cell Lung Cancer (NSCLC), in which NSCLC is the major type accounting for more than 80 % of all lung cancer cases and encompasses squamous cell carcinoma and adenocarcinoma[3]. Currently, smoking cessation is still the dominant risk factor for lung cancer[4]. In addition, other potential factors including outdoor or indoor air pollution, occupational exposure as well as hereditary susceptibility also raise the risk of lung cancer[4].
Metastasis is a term defined as the spread of tumor cells from primary sites to surrounding structures and distant sites. This process requires a complicate procedure including original detachment from the primary tumor sites, subsequent invasion into vessels as well as extravasation into the appropriate secondary site plus the development of surrounding microenvironment establishment[5]. The majority of lung cancer-associated deaths result from metastasis[6]. It has been reported that the brain is the primary site for NSCLC metastasis, which is responsible for approximately 50 % of all brain metastases[7,8].
The 5 y survival rate of lung cancer patients with brain metastasis has been greatly hampered with a median survival rate less than 6 mo[9]. Moreover, accumulating evidences have supported an increased tendency for lung cancer brain metastasis[10]. In spite of plethora of studies, the knowledge of the genetic basis for lung cancer brain metastasis especially brain metastasis is still dismal. This is mainly due to the existence of Blood-Brain Barrier (BBB), which impacts the accessibility of chemical therapeutic agents to the brain parenchyma[11]. With decades of efforts, several risk factors have slowly surfaced. For instance, properties of lung cancer patient age, the presence of clinical bulky mediastinal lymph nodes (˃2 cm), Karnofsky performance status as well as the absence of extracranial metastases are closely correlated with brain metastasis[12-15]. However, the molecular mechanisms underlying the lung cancer brain metastasis is still controversial. To understand the molecular mechanisms in this study in-depth, a comprehensive analysis of differentially expressed genes was performed between conventional lung cancer (NSCLC) and NSCLC brain metastasis patients. The potential genes were further investigated by enrichment analysis as well as external functional experiments.
Materials and Methods
Data source:
The differentially gene expression profile was established from Gene Expression Omnibus (GEO) database, numbering GSE161116, which included 28 tissues from primary and brain metastases of NSCLC (13 from conventional NSCLC and 15 from NSCLC brain metastasis patients respectively). The whole genome expression profiles of the above samples were detected by Affymetrix Human Genome U133 plus 2.0 array chip platform for differential gene analysis.
Differential gene analysis:
The limma package in R language was developed to analyze the differentially expressed messenger Ribonucleic Acid (mRNAs) between the two groups, taking the absolute value of the log-transformed differential expression multiple Logarithm of Fold Change (Log2FC)>1 and p-value<0.05 as a standard for analysis.
Functional enrichment analysis:
For the obtained differentially expressed genes, we used the “clusterProfiler” function package in R language for enrichment analysis of Gene Ontology (GO) (including biological process, molecular function and cellular component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. When p-value<0.05, we considered the corresponding entries to be significantly enriched.
Protein-Protein Interaction (PPI) network establishment:
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database is the one for analyzing and predicting protein functional associations and protein interactions. In this study, we utilized STRING (https://STRING-db.org/, version 11.0) to analyze protein functional associations and protein interactions[16]. The Cytoscape (version 3.7.2) was used to visualize PPI network and the cytoHubba plugin in Cytoscape was used to screen the primary candidate genes in PPI network based on the algorithm of Maximum Neighborhood Component (MNC)[17].
Cell culture:
The NSCLC cell line (A549) was purchased from Tongpai Biotechnology Co., Ltd (Shanghai, China) and cultured in F-12K medium (HyClone, United States of America (USA)) supplemented with 10 % fetal bovine serum (Gibco, USA). The mimics of antago Lethal-7a (Let-7a) and its corresponding negative controls (antago microRNA-Negative Control (miR-NC)) were designed and synthesized by Life Technologies Corporation. At the same time, the C-C Motif Chemokine Ligand 21 (CCL21), C-C Motif Chemokine Ligand 19 (CCL19) and C-C Chemokine Receptor 7 (CCR7) small interfering RNA (siRNAs) as well as their corresponding Negative Control (NC siRNA) were constructed by Genewiz Corporation Co. (Tianjin, China).
Luciferase reporter assay:
The 3’ non-coding region of CCR7 was synthesized and inserted into Xho I and Not Ⅰ sites of the psiCHECKTM-2 luciferase vector. The wild-type or mutant plasmid and NC or let-7a was co-transfected. The luciferase analysis was performed by dual luciferase reporter analysis system (Promega, USA).
Cellular function measurements:
The cell proliferation was evaluated using Cell Counting Kit-8 (CCK-8) kit (Fisher, China). At the same time, the cell apoptosis was examined based on flow cytometry analysis after Annexin V Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) double staining. The scratch assay was performed to examine A549 cell migration. At the same time, the cell invasion assays were measured using Transwell chamber assay (Costar, USA). All the experiments were conducted in triplicate.
Results and Discussion
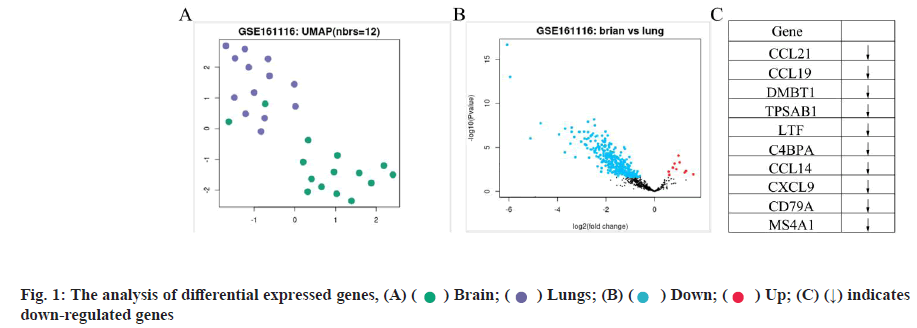
Differentially expressed genes for NSCLC brain metastasis was shown in fig. 1. In this study, we first compared the overall expression pattern between primary and brain metastases of NSCLC by Uniform Manifold Approximation and Projection (UMAP) algorithm analysis. The two groups of specimens demonstrated totally different expression models (fig. 1A). In the volcano map of differentially expressed genes, the horizontal axis represents the multiple of differential expression (Log2FC), the vertical axis represents -log10 (False Discovery Rate (FDR)), while the blue dot indicates down-regulated genes and the red dot indicates up-regulated genes respectively. Compared with convention NSCLC, NSCLC patients with brain metastases showed 352 differentially expressed genes, with the majority as down-regulated ones (342) and 10 up-regulated ones (fig. 1B). The top 10 primary differentially expressed genes for NSCLC brain metastasis were compared with conventional NSCLC. Among these, CCL21, CCL19, Deleted in Malignant Brain Tumors 1 (DMBT1), Tryptase Alpha/Beta 1 (TPSAB1), Lactotransferrin (LTF), Complement Component 4 Binding Protein Alpha (C4BPA), C-C Motif Chemokine Ligand 14 (CCL14), C-X-C Motif Chemokine Ligand 9 (CXCL9), Cluster of Differentiation 79A (CD79A) as well as Membrane Spanning 4-Domains A1 (MS4A1) were significant and differentially expressed (all down-regulated, indicated as arrows) as shown in fig. 1C.
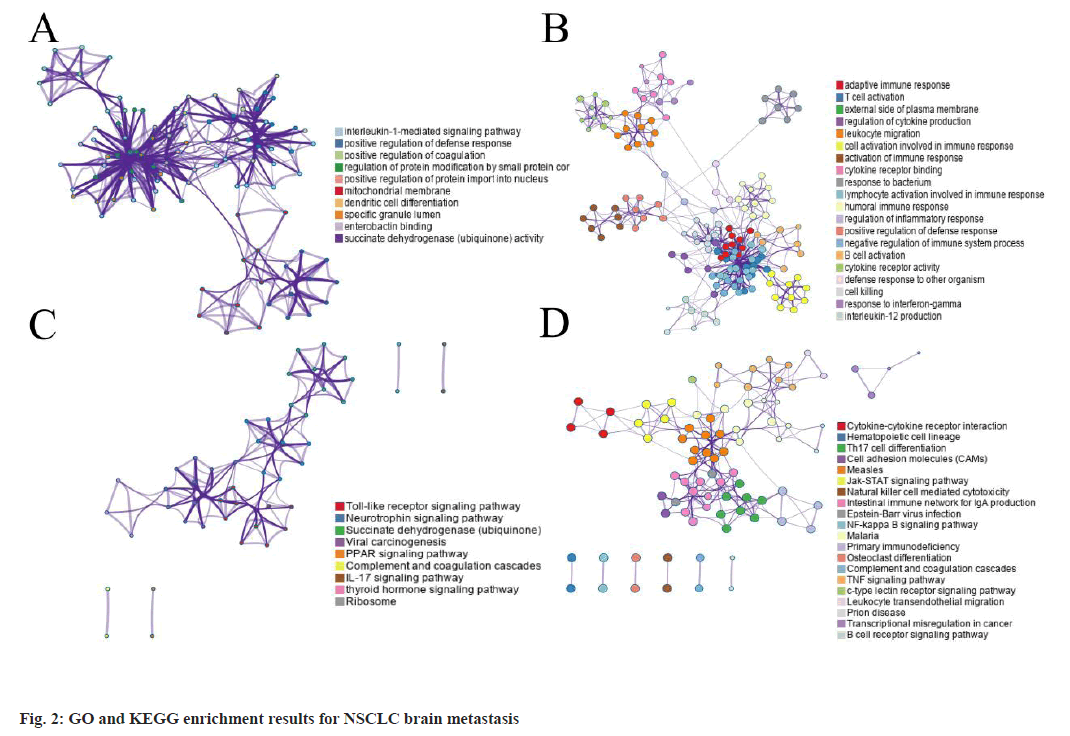
GO and KEGG enrichment analysis results were shown in fig. 2. The top up-regulated GO term enrichment results with the largest number of genes were shown here. In the figure, the horizontal axis represents the number of enriched genes and the vertical axis represents the name of each GO term respectively. Based on the GO and KEGG enrichment analysis of the 352 differentially expressed genes, it could be implicated that the differentially expressed genes were significantly enriched in GO terms such as interleukin-1 mediated signaling pathway, positive regulation of defense response etc., (up-regulation) (fig. 2A) and adaptive immune response, T-cell activation etc., (down-regulation) and the top down-regulated GO term enrichment results (fig. 2B). The enrichment results of the up-regulated KEGG pathways with the largest number of genes were shown in fig. 2C. The horizontal axis in the figure indicates the number of genes enriched and the vertical axis indicates the name of each KEGG pathway respectively. At the same time, the toll-like receptor signaling pathways as well as neurotrophin signaling pathways were significantly up-regulated (fig. 2C) while cytokine-cytokine receptor interaction as well as other signaling pathways were down-regulated based on the KEGG analysis. The enrichment results of the down-regulated KEGG pathways with the largest number of genes was shown in fig. 2D.
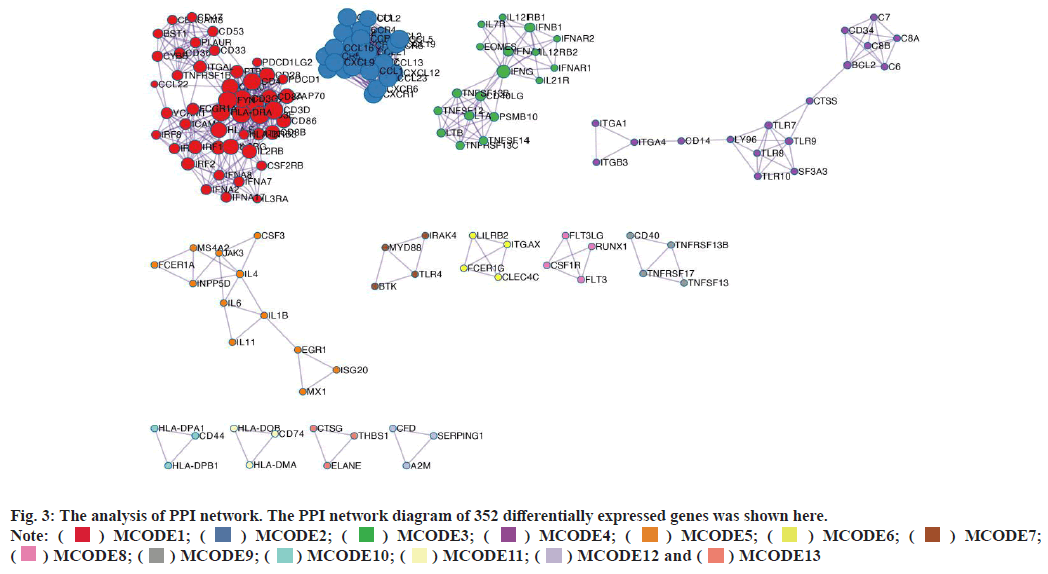
PPI network construction was shown in fig. 3. A STRING database was developed to construct a PPI network for the 352 differentially expressed genes and the gene interactions with confidence score≥0.4 were selected for visualization with Cytoscape software. The Molecular Complex Detection (MCODE) plugins were performed to identify significant clustering modules. As shown in fig. 3, the differentially expressed genes were grouped into 13 clusters. The clusters were further screened with MCODE score>2 as the threshold and 3 clusters were screened out (1-3). It is worth noting that cluster 2 included various C-X-C Motif Chemokine Ligand (CXCL) as well as Chemokine C-C Motif Ligand (CCL) protein family members. Since CCL21, CCL19, CCL14 as well as CXCL9 from cluster 2 were also the prominent differentially expressed genes (fig. 1C). We believed that the genes of cluster 2 may be the key targets for the progress of NSCLC brain metastasis.
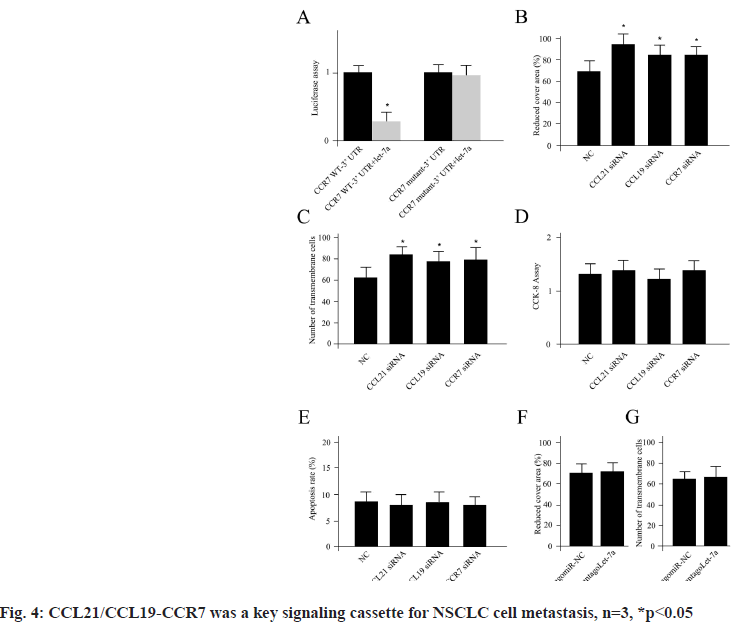
CCL21/CCL19-CCR7 was a key signaling cassette for NSCLC cell metastasis which was shown in fig. 4. We have demonstrated that the genes of cluster 2 including CCL21, CCL19 played a pivotal role in NSCLC brain metastasis. Next, we sought to verify the connections using A549 cell line. The cytokines CCL19 and CCL21 as well as their closely associated receptors CCR7 were analyzed using the A549 cells. Previously, it had been reported that CCR7 was a target for miRNA let-7a[18]. First, we confirmed the direct binding connection in the NSCLC cells using luciferase measurement (fig. 4A). Inhibition of CCL19 and CCL21 as well as CCR7 using siRNAs obviously enhanced the cell invasion and metastasis (fig. 4B and fig. 4C). However, no significant differences for cell proliferation and apoptosis could be detected for CCL19 and CCL21 as well as CCR7 siRNAs transfections compared with NC siRNA treatment (fig. 4D and fig. 4E). While, the NSCLC cells transfected with mimics of antagoLet-7a did not display any significant difference for cell invasion and metastasis compared with its corresponding negative controls (antagomiR-NC) (fig. 4F and fig. 4G), which indicated the NSCLC metastasis regulation was independent of let-7a, n=3, *p<0.05.
For a long period, researchers have been trying to find breakthroughs in the prevention, early screening, diagnosis and treatment of lung cancer. However, the morbidity and mortality of lung cancer keep in a high level due to the tumor metastasis. Actually, metastasis has been considered as a tremendous factor for all types of tumors. For example, in breast cancer, the overall 5 y survival drops significantly from 96 % to 21 % with distant metastasis. Meanwhile, patients of colorectal cancer with lung or liver metastasis have a 5 y survival of less than 10 % compared with 91 % with conventional colorectal cancer[19]. No matter from the aspect of structure or the function, the metastatic tumor is completely distinct from the primary tumor, which could be approved by our outcomes (fig. 1A). The primary tumors could metastasize to distinct organs, which is so called “organotropism” or “organ-specific metastasis”[20]. Two fundamental hypotheses have been initiated to explain the complicate procedure. The first hypothesis, “the hemodynamic or mechanical hypothesis”, which prefers the spreading of tumor cells into the lymphatic system or circulating system was a critical factor for the metastatic patterns[21]. Clearly, this is not suitable for NSCLC brain metastasis patients. The brain is the primary site for lung cancer metastasis, which is up to 50 %. However, the liver, which receives an equal amount of blood, displays much less frequency of lung cancer metastasis. The second hypothesis, referring to “seed-and-soil”, suggests that the primary lung cancer cells are functional as a “seed”, which only blossom in the permissive secondary tissues (“soil”)[22]. Nowadays, the second hypothesis is more acceptable for researchers. Meanwhile, it is beneficial to explore the organ-specific “seed” and “soil” for distinct types of cancer.
In addition to the “seed” and “soil”, the tumor microenvironment may be functional as a “fertilizer”, which consists of Extracellular Matrix (ECM), various inflammatory cytokines as well as other constructional remodeling enzymes. The tumor development and metastasis are closely related to the structure and function of the tumor microenvironment, of which the relationships between cytokine and tumor metastasis have attracted much attention. CCL represents the Chemokine (C-C) Motif Ligand, which is a group of chemotactic cytokines known as CCL1-CCL28[23]. Their shared characteristics are the N-terminal CC domain and digits in their symbols depend on the order of discovery[24]. The CCL family members were subdivided into two clusters: Anti and pro-cancer based on their function for carcinogenesis according to the “Human Protein Atlas” (https://www.proteinatlas. org/). The anti-cancer property of CCL mainly depends on the recruitment of anti-cancer Tumor- Infiltrating Lymphocytes (TIL), which destroy cancer cells[25]. Inversely, the pro-cancer functions of CCL are correlated with the recruitment of cells, which are characterized as factors to stimulate tumor growth, activate cell proliferation, increase tumor cell migration as well as inhibit the functions of tumor repressors[26,27]. Yet, some members may demonstrate both pro and anticancer activities. However, the correlation between CCL and tumor metastasis is not fully understood yet.
In this study, we explored the potential key genes for the progress of NSCLC brain metastasis. Interestingly, most of them were associated with CCL family more or less (CCL21, CCL19, CCL14 and CXCL9). The CCL21 is the chemokine ligand for CCR7, which is mainly expressed by both dendritic cells and T cells for immune responses. Zhang and his colleagues reported that CCR7 expressed tumor cells could metastasize to lymphatic system by the similar way as immune cells[28]. Targeting CCL21/CCR7 axis to inhibit lymphatic metastasis, they remain potent and anti-tumor immune response has increasingly become a spot light of tumor immunotherapy. Later, as reported by Kim et al. they showed that the 3'-Untranslated Regions (3' UTR) of CCR7 is a direct target of microRNA (miRNA) let- 7a[18]. The let-7a downregulates CCR7 expression and directly influences the migration and invasion of breast cancer cells, which confirms targeting of CCL21-CCR7 signaling is a valid approach for breast cancer therapy. However, in this study, we did not detect the expression difference for let-7a compared with primary NSCLC and NSCLC with brain metastasis patients. This raised two possibilities: Either the brain metastasis for NSCLC is let-7a independent or the progress depends on manipulation of CCL21 instead of CCR7. It is worth noting that both of the prominent target genes implicated from this study (CCL21 and CCL19) are the ligands for CCR7, which indicated the central function of axis of CCL21/CCL19-CCR7 for progress of NSCLC brain metastasis. In this study, we emphasized the crucial activities of CCL21/CCL19-CCR7 for NSCLC cell metastasis. However, we could not completely rule out the possible functions of CCL21/CCL19-CCR7 for tumor cell proliferation and apoptosis since the experiments were conducted in NSCLC tumor cell line. To answer this question, the comparison between NSCLC tumor cell and non-tumor cell line might be required. Recently, it has been suggested that CCR7- CCL19/CCL21 manipulate the homing of T cells and dendritic cells to areas of the lymph nodes where T cell priming and the initiation of the adaptive immune response occur[29]. Another research also claimed misregulation of this signaling axis could contribute to pathobiology of chronic inflammation, tumorigenesis, metastasis as well as autoimmune diseases[30].
Based on the facts that NSCLC patients with brain metastasis encounter distinct clinical prognosis and treatment options compared to others with primary tumor of origin, almost more than 500 people die per day from metastatic tumors worldwide. This highlights the major limitations of the modern-day treatment options once the disease is widespread. To address this issue, in this integrate work, we deeply investigated the differentially expressed genes for the development of brain metastasis in NSCLC patients, which hypothesized various candidate genes for future study. Due to the high rate of brain metastasis as well as the existence of BBB, this provided an alternative way to better understand the mechanism underlying with brain metastasis as well as initiate a beneficial reference for clinical administration.
Author’s contributions:
Bo Xiao and Jing Li contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interest.
References
- Wang X, Adjei AA. Lung cancer and metastasis: New opportunities and challenges. Cancer Metastasis Rev 2015;34(2):169-71.
[Crossref] [Google scholar] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359-86.
[Crossref] [Google scholar] [PubMed]
- Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol 2018;13(1):395-412.
[Crossref] [Google scholar] [PubMed]
- Dempke WC, Suto T, Reck M. Targeted therapies for non-small cell lung cancer. Lung Cancer 2010;67(3):257-74.
[Crossref] [Google scholar] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7-34.
[Crossref] [Google scholar] [PubMed]
- Dragoj M, Milosevic Z, Bankovic J, Tanic N, Pesic M, Stankovic T. Targeting CXCR4 and FAK reverses doxorubicin resistance and suppresses invasion in non-small cell lung carcinoma. Cell Oncol 2017;40(1):47-62.
[Crossref] [Google scholar] [PubMed]
- Gavrilovic IT, Posner JB. Brain metastases: Epidemiology and pathophysiology. J Neurooncol 2005;75(1):5-14.
[Crossref] [Google scholar] [PubMed]
- Minchom A, Yu KC, Bhosle J, O'Brien M. The diagnosis and treatment of brain metastases in EGFR mutant lung cancer. CNS Oncol 2014;3(3):209-17.
[Crossref] [Google scholar] [PubMed]
- Maher EA, Mietz J, Arteaga CL, dePinho RA, Mohla S. Brain metastasis: Opportunities in basic and translational research. Cancer Res 2009;69(15):6015-20.
[Crossref] [Google scholar] [PubMed]
- Fox BD, Cheung VJ, Patel AJ, Suki D, Rao G. Epidemiology of metastatic brain tumors. Neurosurg Clin N Am 2011;22(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol 2015;7(1):a020412.
[Crossref] [Google scholar] [PubMed]
- Ceresoli GL, Reni M, Chiesa G, Carretta A, Schipani S, Passoni P, et al. Brain metastases in locally advanced nonsmall cell lung carcinoma after multimodality treatment: Risk factors analysis. Cancer 2002;95(3):605-12.
[Crossref] [Google scholar] [PubMed]
- Gaspar L, Scott C, Rotman M, Asbell S, Phillips T, Wasserman T, et al. Recursive partitioning analysis (RPA) of prognostic factors in three radiation therapy oncology group (RTOG) brain metastases trials. Int J Radiat Oncol Biol Phys 1997;37(4):745-51.
[Crossref] [Google scholar] [PubMed]
- Hanibuchi M, Kim SJ, Fidler IJ, Nishioka Y. The molecular biology of lung cancer brain metastasis: An overview of current comprehensions and future perspectives. J Med Invest 2014;61(3.4):241-53.
[Crossref] [Google scholar] [PubMed]
- Schwartz AG, Cote ML. Epidemiology of lung cancer. Adv Exp Med Biol 2016;893:21-41.
[Crossref] [Google scholar] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607-13.
[Crossref] [Google scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google scholar] [PubMed]
- Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, et al. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of CC chemokine receptor type 7. Breast Cancer Res 2012;14(1):1-2.
[Crossref] [Google scholar] [PubMed]
- Jones GS, Baldwin DR. Recent advances in the management of lung cancer. Clin Med 2018;18(2):s41-6.
[Crossref] [Google scholar] [PubMed]
- Tayyeb B, Parvin M. Pathogenesis of breast cancer metastasis to brain: A comprehensive approach to the signaling network. Mol Neurobiol 2016;53(1):446-54.
[Crossref] [Google scholar] [PubMed]
- Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, et al. Metastatic patterns in adenocarcinoma. Cancer 2006;106(7):1624-33.
[Crossref] [Google scholar] [PubMed]
- Obenauf AC, Massagué J. Surviving at a distance: Organ-specific metastasis. Trends Cancer 2015;1(1):76-91.
[Crossref] [Google scholar] [PubMed]
- Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity 2000;12(2):121-7.
[Crossref] [Google scholar] [PubMed]
- Hughes CE, Nibbs RJ. A guide to chemokines and their receptors. FEBS J 2018;285(16):2944-71.
[Crossref] [Google scholar] [PubMed]
- Lança T, Costa MF, Gonçalves-Sousa N, Rei M, Grosso AR, Penido C, et al. Protective role of the inflammatory CCR2/CCL2 chemokine pathway through recruitment of type 1 cytotoxic γδ T lymphocytes to tumor beds. J Immunol 2013;190(12):6673-80.
[Crossref] [Google scholar] [PubMed]
- Mou T, Xie F, Zhong P, Hua H, Lai L, Yang Q, et al. MiR-345-5p functions as a tumor suppressor in pancreatic cancer by directly targeting CCL8. Biomed Pharmacother 2019;111:891-900.
[Crossref] [Google scholar] [PubMed]
- Dagouassat M, Suffee N, Hlawaty H, Haddad O, Charni F, Laguillier C, et al. Monocyte chemoattractant protein‐1 (MCP‐1)/CCL2 secreted by hepatic myofibroblasts promotes migration and invasion of human hepatoma cells. Int J Cancer 2010;126(5):1095-108.
[Crossref] [Google scholar] [PubMed]
- Zhang L, Wang F, Yao X, Ma S, Zhang L, Qin Z. Progress in targeting therapy of cancer metastasis by CCL21/CCR7 axis. Sheng Wu Gong Cheng Xue Bao 2020;36(12):2741-54.
[Crossref] [Google scholar] [PubMed]
- Hauser MA, Legler DF. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J Leukoc Biol 2016;99(6):869-82.
[Crossref] [Google scholar] [PubMed]
- Purvanov V, Matti C, Samson GP, Kindinger I, Legler DF. Fluorescently tagged CCL19 and CCL21 to monitor CCR7 and ACKR4 functions. Int J Mol Sci 2018;19(12):3876.
[Crossref] [Google scholar] [PubMed]
 indicates
down-regulated genes
indicates
down-regulated genes