- *Corresponding Author:
- Jianbo Zhu
Department of Surgery, Taizhou Maternity Hospital, No. 568 Dongfeng South Road, Taizhou, Jiangsu 225300, China
E-mail: zjbyf.good@163.com
| This article was originally published in a special issue,“Biomedical applications in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2020:82(2) Spl issue 3;121-126 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of this study was to investigate liquid biopsies of ovarian cancer to provide a more adequate basis for its treatment. Forty four patients with ovarian cancer and 47 healthy people were enrolled in this study from the Second Affiliated Hospital of Soochow Universit Changed to “ Taizhou People’s Hospital between February 2017 and December 2018 for analysis. The expression of miRNA-21-5p and B-cell translocation gene 2 mRNA was determined using reverse transcription polymerase chain reaction, the target genes of miRNA-21-5p were predicted by databases and the expression of B-cell translocation gene 2 protein was detected by western blot. ES-2 cells were used as model group and normal ovarian epithelial cells served as the control group. The results showed that miRNA-21-5p was highly expressed in the serum of patients with ovarian cancer. Through the databases prediction and the intersection of multiple databases, B-cell translocation gene 2 was found to be a downstream target gene regulated by miRNA-21-5p and its expression was negatively correlated with B-cell translocation gene 2 by transfection of miRNA-21-5p inhibitor. MiRNA-21-5p is highly expressed in ovarian cancer cells, and it can be used to treat ovarian cancer by up-regulating the expression of the target gene B-cell translocation gene 2. MiRNA-21-5p can be used as a biological marker, which may inhibit the proliferation of cancer cells by up-regulating B-cell translocation gene 2 gene to treat ovarian cancer.
Keywords
Ovarian cancer, Liquid biopsies, Circulating tumor cells, Circulating tumor DNA, Circulating miRNA
The ovarian tumor is one of the common malignant tumors of female reproductive system, and the incidence rate is second only to cervical cancer and endometrial cancer[1]. Epithelial cancer is the most common in ovarian tumors, and the death rate of ovarian epithelial cancer is the first among various gynaecological tumors, which seriously threatens women’s lives[2]. Most cancers are detected in advanced stages, so it is important to find effective markers to determine the occurrence of the disease. MicroRNAs (miRNAs) constitute a class of small non-coding RNA molecules that function as post-transcriptional gene regulators and have been increasingly recognized as oncogenes or tumor suppressors[3]. Studies have shown that a variety of miRNAs could be involved in the regulation of occurrence and development of the same disease and the same miRNA could occur in multiple diseases[4]. Therefore, miRNAs can be used as a biological marker of disease to predict its occurrence.
Studies have shown that a variety of miRNAs are abnormally expressed in ovarian cancer and participate in the regulation of ovarian cancer-related genes[5,6], in this study, the difference of miRNA-21-5p between ovarian cancer patients and healthy subjects was detected by RT-PCR, B-cell translocation gene 2 (BTG-2) was predicted as a target gene of miRNA- 21-5p using databases. The BTG-2 gene is a member of the TOB/BTG family. It has been found to have antiproliferative properties and to function as a transient early response protein, which plays an important regulatory role in differentiation and apoptosis. However, studies on BTG-2 as a target gene of miRNA- 21-5p in ovarian cancer have not been reported. The potential mechanism of ovarian cancer was explored by transfection of miRNA-21-5p inhibitor, which provided sufficient theoretical basis for miRNA-21-5p as a biomarker of ovarian cancer.
Materials and Methods
Patients and groups:
Blood samples were selected from 44 patients with ovarian cancer and 47 healthy people in the Taizhou People’s Hospital, and the samples were divided into the model group and the control group. At the time of admission, the basic information and various evaluation indicators were recorded and samples were taken for testing.
Cell culture:
ES-2 cells and normal ovarian epithelial cells were purchased from Shanghai Enzyme Research Biotechnology Co., Ltd. Cells were seeded in a 10 cm cell culture dish and the medium were made up of 10 % fetal bovine serum (FBS, Gibco) and 1 % P/S at 37° in a humidified atmosphere of 5 % CO2 and 95 % air. The expression of miRNA-21-5p in two groups was detected by RT-PCR[7,8].
Bioinformatics analyses:
The Starbase, Target Scan, Tarbase[9-11] databases were used to predict the downstream target genes regulated by miRNA-21-5p. The downstream target genes of miRNA-21-5p were predicted using these three databases and introduced into the Venn diagram generator respectively, and the target included in 3 groups were taken out. Targets associated with ovarian cancers were selected for follow-up studies.
Luciferase reporter assay:
Luciferase reporter assay was performed to identify the relationship between miRNA-21-5p and BTG-2. In brief, ES-2 cells at 80 % confluence were co-transfected with wild-type or mutant BTG-2 3′-UTR reporters together with miRNA-21-5p or negative control using Lipofectamine 2000. The plasmid (Promega) encoding luciferase was used to control for transfection efficiency. Cells were lysed 24 h after transfection and tested for luciferase activities using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions[12].
Inhibitor transfection:
MiRNA-21-5p inhibitor oligonucleotide (Ribobio, 20 μM) was transfected into cells using Lipofectmine 2000 (5 μl) in a 6-well plate. Cells were divided into 3 groups, the control group, the model group and the inhibitor group. Cells were prepared for the following experiments.
RT-PCR:
Extraction of total RNA was performed with TRIzol reagent (Life Technologies) following the manufacturer’s instruction, extracted RNA was quantified. The expression of miR-21-5p was normalized to the expression of small nuclear RNA U6 as an endogenous control. BTG-2 expression was examined via standard RT-PCR and normalized to β-actin expression as an endogenous control[13].
Western blot:
Cells were lysed in the radioimmune precipitation assay buffer with the protease inhibitor cocktail (Sigma), separated on sodium dodecyl sulfate polyacrylamide gels and transferred to a polyvinylidene fluoride membrane. The membrane was incubated with antiBTG-2 and antiβ-actin (Abcam, Cambridge, MA, USA) at 4° overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibody for 1 h. Bands were visualized with ECL[14].
Statistical analysis:
All data’s were analysed by SPSS version 13.0 (SPSS, IL, USA) and graphs were drawn by GraphPad Prim 5.0 software. The normal distribution of the data was detected by Box plot. The data conforming to the normal distribution was expressed as mean±standard deviation (SD) and the comparison between groups were analysed by variance and the t-test was used for comparison between two groups. If the measurement data is non-normally distributed, the results were expressed as median and non-parametric test; the count data is expressed by rate or composition ratio and a chi-square test was performed. p<0.05 was considered statistically significant.
Results and Discussion
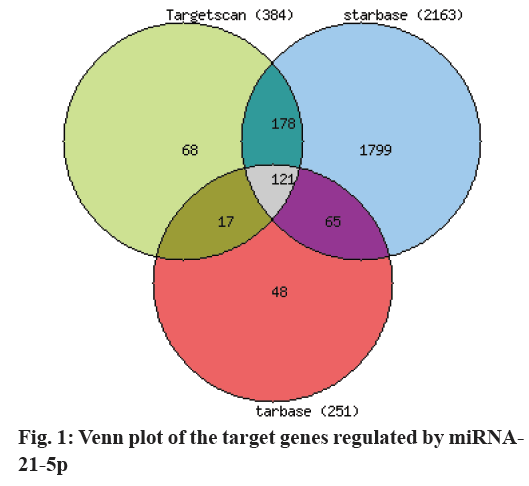
Through database search, the Starbase database retrieved 2163 downstream target genes regulated by miRNA-21-5p, 384 target genes were detected by Target Scan database, and 251 target genes regulated by miRNA-21-5p were searched by Tarbase database. The results of the databases were used to integrate the miRNA-21-5p-regulated target genes shared by the 3 databases. Among them, the miRNA-21-5p shared target genes were searched by the 3 databases (fig. 1). There are 121 target genes regulated by miRNA-21-5p and its sequence binding site is 2039-2045 (Table 1).
| Position 2039-2045 of BTG2 3' UTR | 5'-GUAGUAGUAUGUUUGUAAGCUAU... |
| hsa-miR-21-5p | 3'-AGUUGUAGUCAGACUAUUCGAU... |
Table 1: The Binding Site between Mirna-21-5p and Btg-2
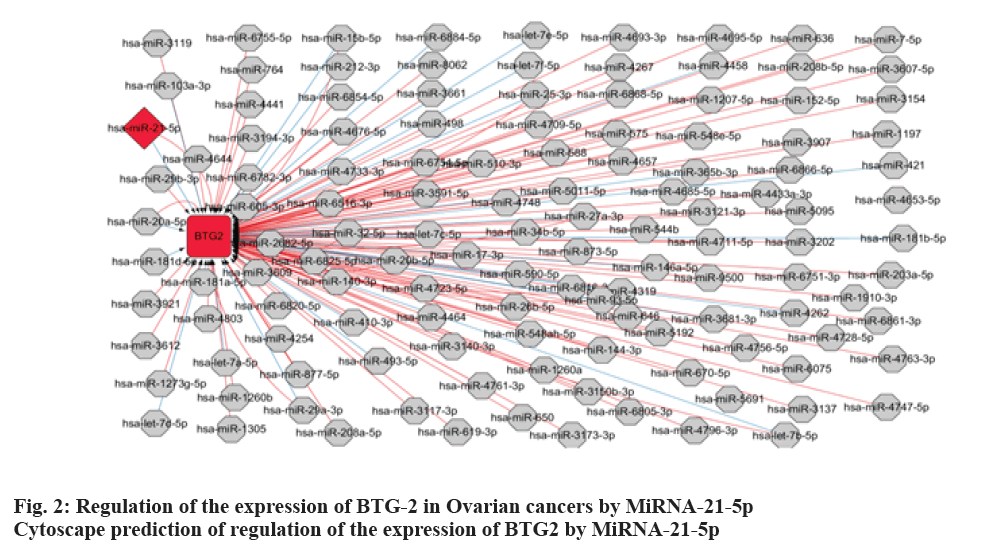
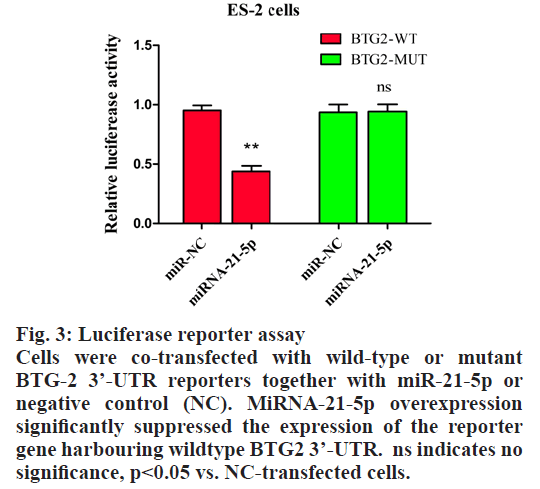
According to the database search results, there were 121 genes regulated by miRNA-21-5p, and BTG-2 was a gene closely related to the development of ovarian cancers, it was predicted that miRNAs regulated BTG-2 expression. The results showed that miRNA- 21-5p is one of the miRNAs to regulate BTG-2 (fig. 2). To validate that the miR-21-5p’s function is to directly regulate BTG-2 expression, a binding site was found between miRNA-21-5p and BTG-2 to identify its mechanism. The binding site between miRNA-21- 5p and BTG-2 were mutated to determine the binding ability of these 2 genes. The results of luciferase reporter assay showed that the miRNA-21-5p could bind to BTG2-WT and decrease the fluorescence intensity, but the mutation of BTG-2 cannot bind to miRNA-21-5p, and the fluorescence intensity is not affected by the amount of miRNA-21-5p expression (fig. 3). These results indicated that BTG-2 is a target gene directly regulated by miRNA-21-5p.
Figure 3: Luciferase reporter assay
Cells were co-transfected with wild-type or mutant
BTG-2 3’-UTR reporters together with miR-21-5p or
negative control (NC). MiRNA-21-5p overexpression
significantly suppressed the expression of the reporter
gene harbouring wildtype BTG2 3’-UTR. ns indicates no
significance, p<0.05 vs. NC-transfected cells.
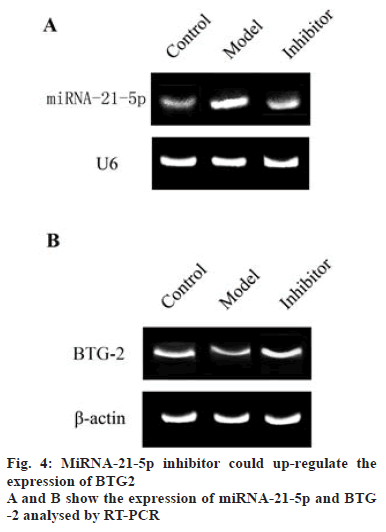
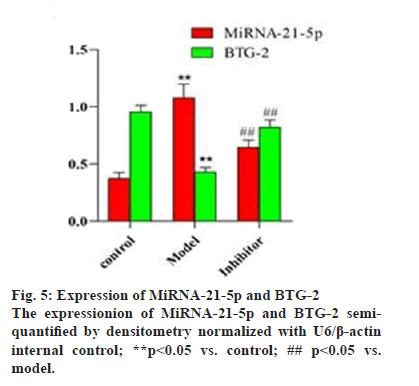
To identify the influence of miRNA-21-5p on the expression of BTG2, the miRNA-21-5p inhibitor was transfected into the ES-2 cells, the expression of miRNA-21-5p and BTG2 was determined (figs. 4A and 4B), The results showed that miRNA-21-5p was overexpressed in the model group compared to the control group (p<0.05). The inhibitor successfully inhibited the expression of miRNA-21-5p. The detected results of BTG-2 showed that the expression of BTG2 in the model group was lower than that in the control group (p<0.05, fig. 5), MiRNA-21-5p inhibitor increased BTG2 expression while miRNA-21-5p expression was low. Therefore, low expression of miRNA-21-5p could activate the expression of the target gene BTG-2, which indicated that miRNA-21-5p, is negatively correlated with BTG2.
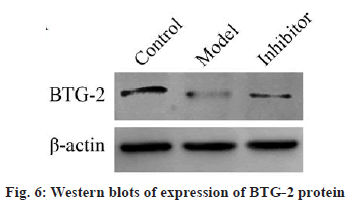
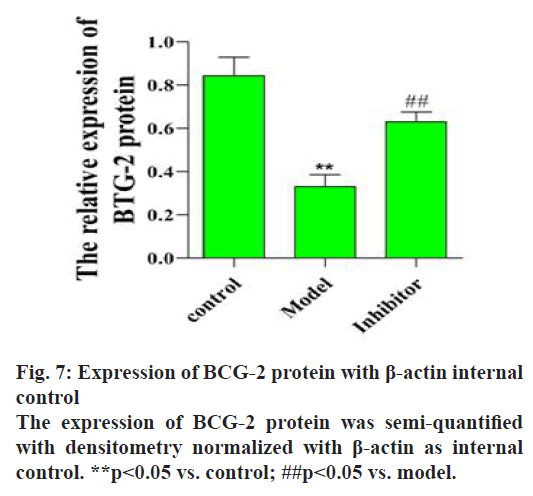
To investigate the effect of miRNA-21-5p on BTG-2 protein expression, the BTG-2 protein was detected using western blot (fig. 6). The results showed that BTG-2 protein expression was low in the model group compared to the control group (p<0.05, fig. 7). The miRNA-21-5p inhibitor could increase the BTG-2 protein expression while miRNA-21-5p expression was low. These results indicated that BTG-2 was under expressed in ES-2 cells and low expression of miRNA-21-5p could activate the expression of BTG-2, indicating that miRNA-21-5p and BTG2 are negatively correlated.
The development of cancer is a multi-gene, multi-step complex biological process[15]. Due to the diversity of morphological and biological characteristics of ovarian tissue, ovarian cancer is considered as a heterogeneous malignant tumor disease rather than a single disease. Currently, ovarian cancer is a common malignant tumor in female reproductive system. About 75 % of patients are diagnosed after the cancer is advanced and metastasized throughout the peritoneum, pleura or even liver[16] and the specific mechanisms and initiating factors are still not fully understood. Therefore, it is of great significance to study the early biological markers of ovarian cancer.
MiRNAs are endogenous small RNA that can treat diseases to inhibit its translation or promote its degradation by binding to the target gene[17]. The expression of some miRNAs is closely related to the occurrence and development of diseases. Among them, tumor-related diseases are one of the hottest areas of research, involving tumor proliferation[18], migration[19], apoptosis[20] and drug tolerance. In general, one miRNA molecule may have multiple target genes and one gene may also be regulated by multiple miRNAs molecules simultaneously.
As one of the most well-known oncogenic miRNAs, miRNA-21 is highly expressed in many tumor tissues[21]. It has been reported that miRNA-21 is highly expressed in ovarian cancer, indicating that miRNA-21 plays an role important in the development of ovarian cancer[22]. MiRNAs can regulate the expression levels of many protein-coding genes more accurately and have become one of the research hotspots in the medical field. Intensive research on miRNAs may bring substantial changes in clinical cancer treatment. Recent studies have shown that a variety of miRNAs play an important role in the development of ovarian cancer and clinical detection of the expression of certain or some miRNAs can be used as an independent biological marker[23].
BTG-2 is a member of the BTG/TOB antiproliferative gene family. Studies have shown that the BTG-2 gene may be involved in tumor cell proliferation, metastasis and regulation of cell cycle progression[24]. Therefore, the BTG-2 gene may be a target gene for the treatment The results of this study suggested that BTG-2 may be an ideal target gene for the treatment of ovarian cancer. Rapid proliferation of cancer cells is the ultimate step in the development of solid tumors and is the most common cause of death in cancer patients. Controlling the unrestricted proliferation of tumor cells can improve the survival rate of cancer patients, and BTG-2 plays an important role in the development of tumors.
In this study, the target genes regulated by miRNA-21- 5p were predicted by databases. About 121 genes were found to be regulated by miRNA-21-5p and BTG-2 is one of the genes, which closely related to ovarian cancer. The miRNAs, which regulated the expression of BTG-2 were predicted by Cytoscape and the results were consistent with the databases prediction. The targeting relationship was determined by luciferase reporter assay, the results showed that miRNA-21-5p could regulate the expression of BTG-2 directly. To investigate the mechanism of miRNA-21-5p on ovarian cancer, ES-2 cells were used as the model group and the normal ovarian epithelial cells were served as the control group. The miRNA-21-5p inhibitor was transfected into cells and it was found that low-expression of miRNA- 21-5p could increase the BTG-2 at the same time. These results indicated that miRNA-21-5p can regulate the expression of BTG-2 and the expression of these 2 genes is negatively correlated. MiRNA-21-5p inhibitor can activate the expression of BTG-2 and reduce the progression of ovarian cancer by activating BTG-2.
Acknowledgements
We appreciate the support of the Taizhou People’s Hospital.
Conflict of Interest
All authors report no conflicts of interest in this work.
References
- Kuhl CK, Heuck A, Kreft BP, Luckhaus S, Reiser M, Schild HH. Combined intrauterine and ovarian pregnancy mimicking ovarian malignant tumor: imaging findings. Am J Roentgenol 1995;165:369-70.
- Sopo M, Anttila M, Hämäläinen K, Kivela A, Yla-Herttuala S, Kosma VM, et al. Expression profiles of VEGF-A, VEGF-D and VEGFR1 are higher in distant metastases than in matched primary high grade epithelial ovarian cancer. BMC cancer 2019;19:584.
- Lin Y, Liang X, Yao Y, Xiao H, Shi Y, Yang J. Osthole attenuates APP-induced Alzheimer's disease through up-regulating miRNA-101a-3p. Life Sci 2019;225:117-31.
- Alevizos I. MicroRNAs as biomarkers in rheumatic diseases. Nature reviews. Rheumatology 2010;6:391-8.
- Yan H, Li H, Li P, Li X, Lin J, Zhu L, et al. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J Exp Clin Cancer Res 2018;37:237.
- Yu N, Yong S, Kim HK, Choi YL, Jung Y, Kim D, et al. Identification of tumor suppressor microRNAs by integrative microRNA and mRNA sequencing of matched tumor-normal pairs in lung adenocarcinoma. Mol Oncol 2019; 13:1356-68.
- Chen MZ, Zhong L, Wei DM. Effects of Cellular Density on the Induction of Suspension Globe of Ovarian Cancer Stem Cells. Sichuan Da Xue Xue Bao Yi Xue Ban 2017;48:758-62.
- Ukaji T, Lin Y, Banno K, Okada S, Umezawa K. Inhibition of IGF-1-Mediated Cellular Migration and Invasion by Migracin A in Ovarian Clear Cell Carcinoma Cells. PloS one 2015;10:e0137663.
- Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v20: decoding miRNA-ceRNA, miRNA-lncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7.
- Shi Y, Yang F, Wei S. Identification of Key Genes Affecting Results of Hyperthermia in Osteosarcoma Based on Integrative ChIP-Seq/TargetScan Analysis. Med Sci Mon Int Med J Exp Clin Res 2017;23:2042-8.
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239-45.
- Qu Y, Zhang YP, Wu J, Jie LG, Deng JX, Zhao DB, et al. Down regulated microRNA-135a ameliorates rheumatoid arthritis by inactivation of the phosphatidylinositol 3-kinase/AKT signalling pathway via phosphatidylinositol 3-kinase regulatory subunit 2. J Cell Physiol 2019; 234:17663-76.
- Wu SZ, Xu HC, Wu XL, Liu P, Shi YC, Pang P, et al. Dihydrosanguinarine suppresses pancreatic cancer cells via regulation of mut-p53/WT-p53 and the Ras/Raf/Mek/Erk pathway. Phytomedicine 2019;59:152895.
- Ding L, Yuan X, Yan J, Huang Y, Xu M, Yang Z, et al. Nrf2 exerts mixed inflammation and glucose metabolism regulatory effects on murine RAW2647 macrophages. Int Immune Pharmacol 2019;71:198-204.
- Neben CL, Zimmer AD, Stedden W, van den Akker J, O'Connor R, Chan RC, et al. Multi-Gene Panel Testing of 23,179 Individuals for Hereditary Cancer Risk Identifies Pathogenic Variant Carriers Missed by Current Genetic Testing Guidelines. J Mol Diagn 2019;21:646-57.
- Weston C, Hales D. Ovarian Cancer Risk in Laying Hens Is Reduced by Dietary Polyunsaturated Fatty Acids: A Case for Redox Homeostasis and Reprogrammed Mitochondrial Metabolism in Ovarian Tumors. Curr Dev Nutr 2019; 3 nzz030. OR04-05-19.
- Chen Z, Zhuang W, Wang Z, Xiao W, Don W, Li X, et al. MicroRNA-450b-3p inhibits cell growth by targeting phosphoglycerate kinase 1 in hepatocellular carcinoma. J Cell Biochem 2019;120:18805-15.
- Gao J, Feng X, Wang F, Wang J, Wang H, Li H, et al. microRNA-448 inhibits the progression of non-small-cell lung cancer through regulating IRS2. J Cell Biochem 2019;120:13453-63.
- Zhang X, Zhao X, Li Y, Zhou Y, Zhang Z. Long noncoding RNA SOX21-AS1 promotes cervical cancer progression by competitively sponging miR-7/VDAC1. J Cell Physiol 2019;234:17494-504.
- Lu M, Huang H, Yang J, Li J, Zhao G, Li W, et al. miR-338-3p regulates the proliferation, apoptosis and migration of SW480 cells by targeting MACC1. Exp Ther Med 2019;17:2807-14.
- Carabia J, Carpio C, Abrisqueta P, Jiménez I, Purroy N, Calpe E, et al. Microenvironment regulates the expression of miR-21 and tumor suppressor genes PTEN, PIAS3 and PDCD4 through ZAP-70 in chronic lymphocytic leukemia. Sci Rep 2017;7:12262.
- Mahmoud EH, Fawzy A. Serum MicroRNA-21 Negatively Relates to Expression of Programmed Cell Death-4 in Patients with Epithelial Ovarian Cancer. Asian Pac J Cancer Prev 2018;19:33-8.
- Xu YZ, Xi QH, Ge WL. Identification of serum microRNA-21 as a biomarker for early detection and prognosis in human epithelial ovarian cancer. Asian Pac J Cancer Prev 2013;14:1057-60.
- Yang DL, Gan ML, Tan Y, Ge GH, Li Q, Jiang YZ, et al. MiR-222-3р Regulates the Proliferation and Differentiation of C2C12 Myoblasts by Targeting BTG2. Mol Biol 2019;53:44-52.