- *Corresponding Author:
- Aiying Li
Department of Biochemistry and Molecular Biology, College of Basic Medicine, Hebei University of Chinese Medicine, Shijiazhuang, Hebei 050000, China
E-mail: aiyingli1963@163.com
| Date of Received | 04 Feb 2023 |
| Date of Revision | 09 May 2024 |
| Date of Acceptance | 08 August 2024 |
| Indian J Pharm Sci 2024;86(4):1418-1429 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
N-glycosylation, the predominant post-translational protein modification, significantly influences protein physicochemical properties and biological functions. Kifunensine, a N-glycosylation inhibitor, shows promise as a potential therapeutic agent for modulating the hyperactivation of transforming growth factor-beta 1 signalling pathways in fibrotic diseases. This study explored kifunensine's effects and mechanisms of action using an innovative in vitro cardiac fibrosis model involving primary rat cardiac fibroblasts treated with transforming growth factor-beta 1. Protein expression related to cardiac fibrosis was assessed via Western blotting and immunofluorescence assays. Kifunensine significantly protected these cells against transforming growth factor-beta 1 induced fibrosis, as indicated by decreased collagen I, collagen III, and alpha-smooth muscle actin expression. Interestingly, kifunensine did not alter transforming growth factor-beta receptor expression but reduced transforming growth factor-beta 1 ligand-receptor complex formation, inhibiting transforming growth factor-beta type I receptor and transforming growth factor-beta type II receptor activation and suppressing suppressor of mothers against decapentaplegic 2 and suppressor of mothers against decapentaplegic 3 phosphorylation. This underlines N-glycosylation's role in cardiac fibrosis and establishes the anti-fibrotic potential of moderate N-glycan alteration on transforming growth factor-beta receptors using kifunensine, suggesting a novel therapeutic strategy for myocardial fibrosis.
Keywords
Cardiac fibrosis, kifunensine, transforming growth factor-beta receptor, N-glycosylation
Cardiac fibrosis is a substantial pathological alteration in the heart, and its continuous development may ultimately lead to heart failure [1]. During cardiac fibrosis, a large amount of Extracellular Matrix (ECM) including collagen I and II is deposited, and Alpha-Smooth Muscle Actin (α-SMA) is also highly expressed [2,3]. Although the comprehensive molecular mechanisms underlying cardiac fibrosis remain incompletely understood, substantial evidence suggests that Transforming Growth Factor-Beta (TGF-β) signalling plays a pivotal role in fibrosis progression. TGF-β1 is a Hyper activation of TGF-β1 signalling pathways has been observed in various pathological conditions, including cardiovascular diseases and fibrotic disorders [6]. Therapeutic intervention to normalize disrupted TGF-β1 signalling is an emerging approach to restore cardiac function [7].
TGF-β1 exerts its effects on cells by binding with TGF-β type I Receptor (TβRI) and TGF-β type II Receptors (TβRII) [8-11]. The TβRIs and TβRIIs are single transmembrane-spanning proteins with an extracellular, cysteine-rich, ligand-binding domain, and an intracellular dual specificity kinase domain, often referred to as the serine-threonine kinase domain. Highly active TGF-β1 binds to TβRIIs and induces their assembly with TβRIs. TβRI, in turn, initiates intracellular signalling by phosphorylating the Receptor-regulated Suppressor of Mothers against Decapentaplegic (R-SMAD), such as SMAD2 and SMAD3, and these phosphorylated R-SMAD will bind with common mediator SMAD (co-SMAD) such as SMAD4, then the SMAD complexes import into the nucleus and regulate target genes expression and initiate the progression of myocardial fibrosis [12-17].
N-glycosylation represents the most abundant post-translational modification of proteins and influences their physicochemical properties and biological functions [18-20]. The roles of N-glycosylation in cardiac fibrosis are still unclear, but many reports have confirmed that N-glycosylation is necessary for TβRs. TβRI and TβRII are membrane proteins with high-level N-glycosylation, and N-glycosylation also regulates their function. Kim et al. [10] disrupted N-glycosylation of TβRII through site mutation and N-glycosylation inhibitor tunicamycin in the human lung adenocarcinoma cell line A549, and found that impaired TβRII was retained in the near-nuclear region and failed to transport to the cell surface [21]. Sun et al. [11] reported that α1,3-mannosyltransferase 3 (ALG3) overexpression promoted radioresistance and cancer stemness by inducing N-glycosylation of TGF-β receptor II in breast cancer cell lines, and tunicamycin can reverse this effect [22]. Park et al. [12] discovered that glucosamine hydrochloride potentially ameliorates renal fibrosis by inhibiting the N-linked glycosylation of TβRII [23]. The glucosamine hydrochloride inhibits this glycosylation, thereby suppressing the binding of TβRI to TβRII, leading to the attenuation of TGF-β/SMAD2/3 signalling, and ultimately ameliorating renal fibrosis in TGF-β1 treated renal cells. These results demonstrate that N-glycosylation is a necessary and functional modification of TβRI and TβRII, and N-glycosylation inhibitor is a potential therapeutic treatment for intervening hyperactivation of TGF-β1 signalling pathways in fibrosis.
Recent studies has reported that particularly modifying the structure of N-glycans yields more benefits than their complete elimination, as the later could lead to severe cellular damage or even death. In this study, we found that Kifunensine (KIF), a mannosidase I inhibitor that holds N-glycans at the high mannose stage, can effectively suppress TGF-β1 induced fibrosis in primary rat cardiac fibroblasts, which was seen from reduced expression of collagen I and II is deposited, and α-SMA. Mechanistically, KIF displays fundamental differences in comparison to tunicamycin, the most commonly utilized complete glycosylation inhibitor. In contrast to tunicamycin, which entirely inhibits N-glycosylation and consequently prevents the synthesis and transport of TβRs, KIF does not impact the expression of TβRs. Instead, we observed that KIF reduced the formation of TGF-β1 ligand-receptor complexes, subsequently inhibiting the activation of TβRI and TβRII, and suppressing the phosphorylation of downstream signalling molecules such as SMAD2 and SMAD3. These findings illustrate the role of N-glycosylation in cardiac fibrosis and exhibit the anti-fibrosis effectiveness of moderately alternation of N-glycans of TβRs using the N-glycosylation inhibitor KIF, potentially contributing to a novel therapeutic approach for myocardial fibrosis.
Materials and Methods
Isolation and culture of primary rat Cardiac Fibroblasts (CFs):
Primary CFs isolated from neonatal rat ventricular tissues were used in this study. Neonatal Sprague-Dawley (SD) rats of Specific Pathogen Free (SPF) grade, aged (1-3) d were purchased from Beijing Charles River Laboratory (Beijing, China). The animal study protocol was authorized by the Institutional Animal Use Committee of Hebei University of Chinese Medicine (No: DWLL2021003). The protocol of isolation of neonatal CFs was undertaken according to the paper [15]. The hearts’ apical tissue of neonatal rats were removed and shredded into almost an paste-like substance in D-hanks solution, then digested in digestion solution (0.5 mg/ml collagenase II, Solarbio Science and Technology Co., Ltd., Beijing, China) in a 37° water bath for 10 min. Repeat the above digestion steps 5-6 times until the tissue fragment digestion is compete. FBS was added to stop the digestion process, and then the mixture was centrifuged at 1000 rpm for 5 min at 4°. The supernatant was discarded, and high-glucose Dulbecco’s Modified Eagle medium (DMEM) containing 10 % Fetal Bovine Serum (FBS) and 1 % Penicillin-Streptomycin Liquid (Gibco, Invitrogen Inc., Carlsbad, California, United States of America (USA)) was added. The mixture was gently shacked until the pellet was reconstituted. Subsequently, the cell suspension was transferred to a 10 cm dish and cultured in a humidified atmosphere containing 5 % Carbon dioxide (CO2) at 37° for 2 h, which allowed CFs attach to the culture plates. The 2nd or 3rd generations of CFs were used in our experiments.
Cell treatment:
KIF (MCE, China) was dissolved in Dimethyl Sulfoxide (DMSO), resulting in a final DMSO concentration (Solarbio, China) of 1/1000. TGF-β1 (Proteintech, Rosemont, Illinois, USA) was used at a concentration of 10 ng/ml. The dose of DMSO was kept constant across different groups to eliminate confounding factors. After administration, the cells were cultured for an additional 48 h prior to use in subsequent experiments
Ribonucleic Acid (RNA) isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR):
Total RNA was extracted from cells or tissues using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. Real-time PCR was performed using the QuantStudio 1 real-time detection instrument (Thermo Fisher Scientific, Massachusetts, USA). Total RNA was extracted from CFs using the Omega Total RNA Kit II (Omega, R6934-01, Shanghai, China). Subsequently, complementary Deoxyribonucleic Acid (cDNA) was synthesized using the MonScript™ RTIII All-in-One Mix with a dsDNase kit (Monad, MR05101, Suzhou, China). The expression levels of TβRI and TβRII were assessed using the MonAmp ChemoHS qPCR Mix kit (Monad, MQ00401, Suzhou, China).The relative expression levels were calculated using the 2-ΔΔCT method, and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) was used as the internal reference gene.
The primers were designed according to the gene database of NCBI, as follows, GAPDH sense: GACAGCCGCATCTTCTTGTG; GAPDH antisense: TTCTCAGCCTTGACTGTGCC; TβRI sense: CATTTCAGAGGGCACCACCTT; TβRI antisense: CCAAACTTCTCCAAACCGACC; TβRII sense: CCCAAGTCGGTTAACAGCGA and TβRII antisense: GTGAAGCCGTGGTAGGTGAA.
Western blotting:
Western blotting assay was conducted to measure the expression of collagen I, collagen III, α-SMA, TβRI, TβRII, phospho-TβRI, phospho-TβRII, SMAD2, SMAD3, SMAD4, and phospho-SMAD2, SMAD3; GAPDH was used as an internal reference. CFs cells were lysed with Radio-Immunoprecipitation Assay (RIPA) lysis buffer (Beyotime, Shanghai, China) supplemented with Phenylmethyl Sulfonyl Fluoride (PMSF), protease inhibitor cocktail (Roche, Switzerland), and phosphatase inhibitors (Servicebio Technology Co., Ltd., Wuhan, China). The protein concentration was measured using the Bicinchoninic Acid (BCA) assay kit (Solarbio, Beijing, China). An equal amount of protein sample was separated by 10 % Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) gel electrophoresis and transferred to Polyvinylidene Difluoride (PVDF) membranes (Invitrogen, Carlsbad, California, USA). However, to observe the reduction in the molecular weight of the TβRs induced by KIF treatment, 8 % SDS-PAGE separating gels were used for electrophoresis. The membranes were blocked with 5 % skimmed milk at room temperature, and then incubated with the following primary antibodies; anti-collagen I (Abcam, ab270993, 1:1000), anti-collagen III (Affinity, AF0136, 1:1000), anti-α-SMA (Abcam, ab32575, 1:1000), anti-TβRI (Abcam, ab331013, 1:1000), anti-phospho-TβRI (Affinity, AF 8080, 1:1000), anti-TβRII (Abcam, ab186838, 1:1000), anti-phospho-TβRII (Affinity, AF 8191, 1:1000), anti-SMAD2, SMAD3, and SMAD4 (Servicebio, GB11172, 1:1000; Abcam, ab40854, Abcam, ab40759, 1:1000), and anti-phospho-SMAD2 and SMAD3 (Affinity, AF3450, 1:1000; Affinity, AF3362, 1:1000), GAPDH (Affinity, AF7021, 1:20 000) and Sodium/Potassium (Na+/K+)-ATPase (Cell Signaling, #3010?1:1000) antibody were used as internal control. The signals were detected using Enhanced Chemiluminescence (ECL) reagents (Invitrogen, Carlsbad, California, USA) and the OmegaLumC multifunction imaging system (Aplegen, American). Subsequently, the protein expression was evaluated using ImageJ software (GE Healthcare, Beijing, China).
Immunofluorescence assay:
After 48 h of administration, the DMEM of rat primary CFs was removed. Subsequently, the cells were fixed using 4 % paraformaldehyde for 15 min at room temperature. After washing three times with Phosphate Buffered Saline (PBS) (pH 7.4), the CFs cells were blocked in 3 % bovine serum albumin for 1 h at room temperature. Subsequently, the CFs cells were incubated overnight at 4° with the primary rabbit monoclonal antibody against TGF-?1 (ab31013, Abcam, Cambridge, United Kingdom, 1:50). Subsequently, the cells were incubated with the secondary antibody, goat anti-rabbit IgG H&L Alexa Fluor® 488 (ab150077, Abcam), for 1 h. Cell nuclei were stained with DAPI. The samples were examined using a fluorescence microscope system (Leica Microsystems, Wetzlar, Germany). The fluorescence results were quantified using ImageJ software (National Institutes of Health (NIH), Bethesda, Maryland, USA).
Extraction of membrane proteins:
The extraction of membrane proteins was performed according to the protocol provided by the Membrane Proteins Extraction Kit (Solarbio Science & Technology Co., Ltd., Beijing, China). Briefly, after washing twice with cold PBS, CFs were lysed by lysis buffer mixed with a protease inhibitor cocktail on ice for 30 min. The cell lysis supernatant was obtained after centrifugation at 12 000 ×g for 5 min at 4°. The supernatant was then transferred into another clean centrifuge tube and incubated at 37° for (5-10) min, followed by centrifugation at 37°, 1000 g for 5 min. At this point, the solution separated into two layers, with the lower layer containing the membrane proteins. The lower membrane protein fraction was fully dissolved using the purification solution, followed by mixing and an ice bath for 2 min. This was then subjected to a 37° water bath for 5-10 min, and centrifugation at 37°, 1000 g for 5 min. The solution separated into two layers once again, with the lower layer containing the membrane proteins being retained.
Co-Immunoprecipitation (Co-IP):
Co-IP was performed using a Capturem IP and Co-IP Kit (Takara, Kusatsu, Japan) following the manufacturer's instructions. Briefly, CFs were lysed in lysis/equilibration buffer mixed with a protease inhibitor cocktail on ice for 30 min. Next, the supernatant or mixed recombinant proteins were incubated overnight at 4° with the TβRI antibody or isotype IgG (Cell Signaling Technology, Boston, Massachusetts, USA). The cell lysate was centrifuged at 17 000 g for 10 min, and the supernatant was collected. The following day, samples pre-incubated with the antibody were centrifuged using an equilibrated spin column and washed with 100 μl of wash buffer. The eluted proteins were analysed by Western blot using the primary TβRI and TβRII antibody, as described previously.
Statistical analysis:
GraphPad Prism 8 software (GraphPad Software, California, USA) was used to create the figures. All experiments were conducted at least three times, and the results were presented as mean± Standard Error of Mean (SEM). Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software (version 25, IBM Corp., Armonk, New York, USA). Statistical significance was determined using one-way ANOVA followed by Tukey's post hoc test. A value of p<0.05 was considered statistically significant. The levels of significance were presented as *p<0.05, **p<0.01, and ***p<0.001.
Results and Discussion
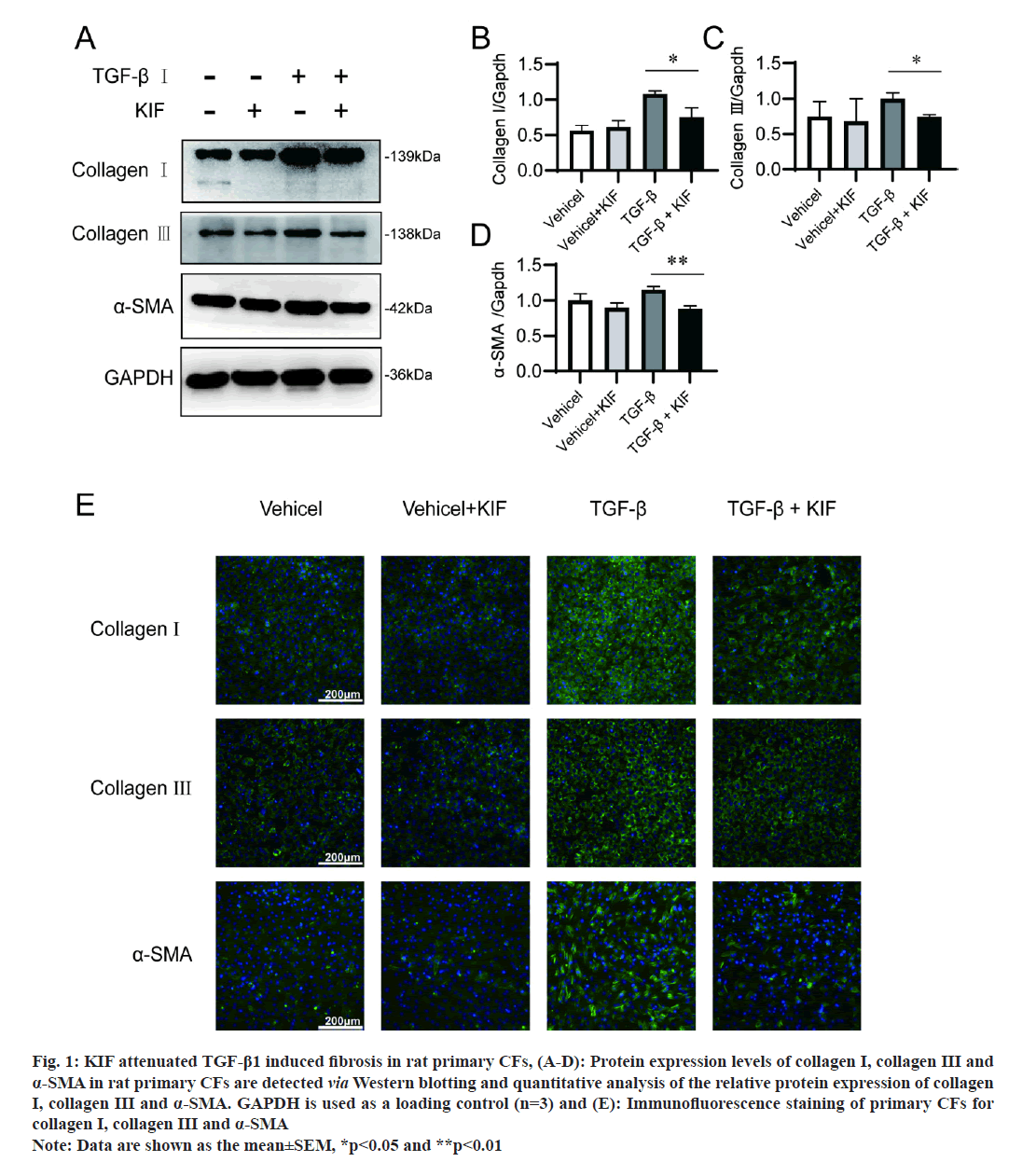
In this study, we simulated cardiac fibrosis by culturing rat primary CFs with TGF-β1. Fibrosis is characterized by an abundant increment of collagen, predominantly collagen types I and III, and α-SMA [16]. The results of Western blotting in our study revealed a significant upregulation of collagen I (p=0.028) and III (p=0.007), and α-SMA (p=0.002) in rat primary CFs after TGF-β stimulation (fig. 1A-fig. 1D), which is consistent with the report from existing literature that TGF-β1 is an important fibrogenic factor. And immunofluorescence assays confirmed those results (fig. 1E). We then assessed the impact of KIF on fibrosis. The outcomes from Western blotting and immunofluorescence assays indicated that KIF mitigated the upregulation of fibrosis-associated marker expressions induced by TGF-β1. In detail, without TGF-β1 treatment, fibrotic signalling was not activated, and collagen I, collagen III and α-SMA were expressed at the basal level. Meanwhile, additional KIF treatment did not affect the basal expression of collagen I, collagen III and α-SMA (fig. 1). However, with TGF-β1 treatment, the expression of collagen I, collagen III and α-SMA significantly increased, and the additional KIF treatment notably attenuates these increments. These results demonstrate that KIF attenuates TGF-β1 induced cardiac fibrosis.
Fig 1:KIF attenuated TGF-β1 induced fibrosis in rat primary CFs, (A-D): Protein expression levels of collagen I, collagen III and α-SMA in rat primary CFs are detected via Western blotting and quantitative analysis of the relative protein expression of collagen I, collagen III and α-SMA. GAPDH is used as a loading control (n=3) and (E): Immunofluorescence staining of primary CFs for collagen I, collagen III and α-SMA Note: Data are shown as the mean±SEM, *p<0.05 and **p<0.01
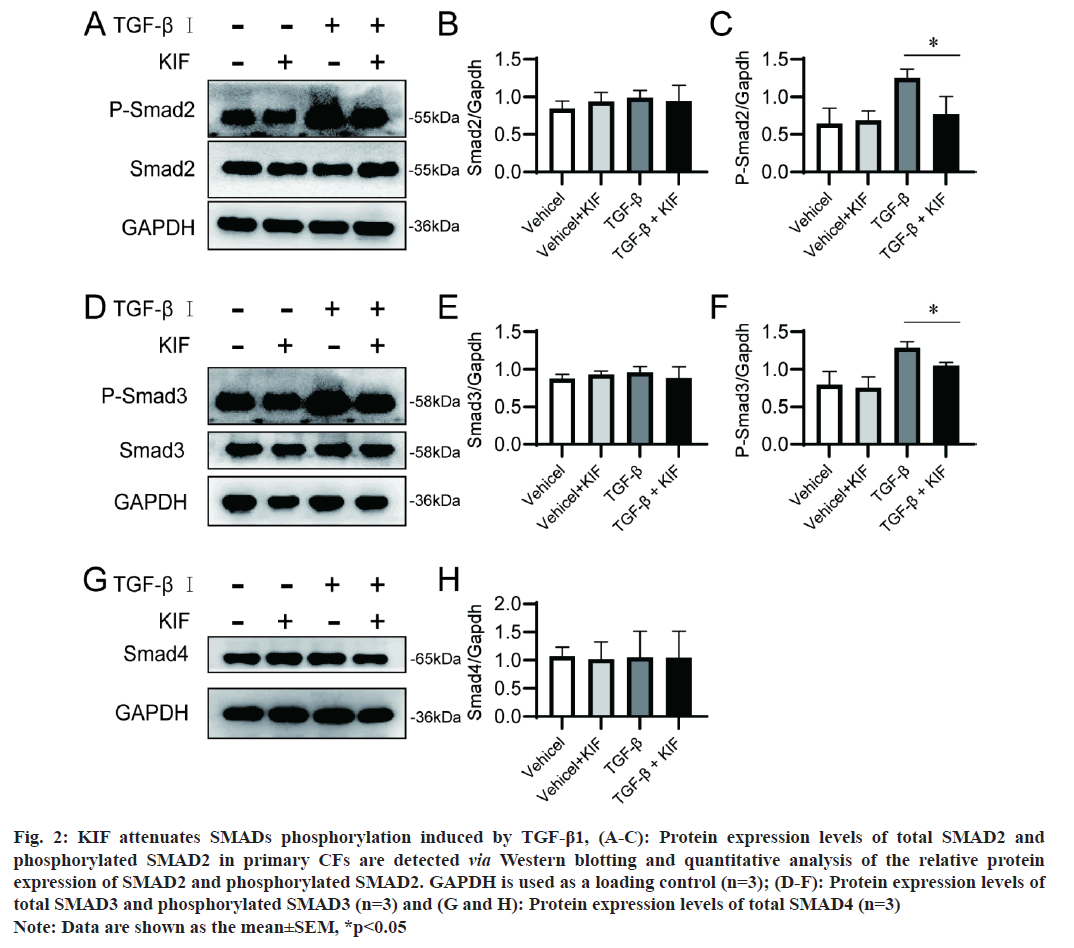
R-SMADS including SMAD2, SMAD3, and co-SMAD such as SMAD4, are pivotal downstream signaling mediators of TGF-β in cardiac fibrosis [17]. In this study, we investigated the impact of KIF on the expression and phosphorylation of SMAD proteins. The findings indicated that KIF did not alter the protein expression of SMAD2, SMAD3, and SMAD4, regardless of the presence or absence of TGF-β stimulation (fig. 2A-fig. 2C). However, KIF was observed to attenuate the phosphorylation levels of SMAD2 (p=0.003) and SMAD3 (p=0.001) induced by TGF-β (fig. 2D-fig. 2H). In the context of cardiac fibrosis, TGF-β is upregulated in response to myocardial injury and stress. TGF-β then binding to TβRs, the activated TβRs subsequently phosphorylates R-SMAD. Once phosphorylated, SMAD2 and SMAD3 form a complex with the common mediator SMAD4. This SMAD complex then translocates to the nucleus, where it interacts with other transcription factors and co-activators to regulate the expression of target genes, including those encoding ECM components and profibrotic factors. This represents the typical mechanism of the TGF-β/SMAD signalling pathway. Our findings about the impact of KIF on SMAD, suggest that KIF continues to exert an anti-fibrotic effect through the classical TGF-β/SMAD signalling pathway. And we speculate that KIF modifies the activity of TβRs, consequently altering the phosphorylation of SMAD proteins.
Fig 2:KIF attenuates SMADs phosphorylation induced by TGF-β1, (A-C): Protein expression levels of total SMAD2 and phosphorylated SMAD2 in primary CFs are detected via Western blotting and quantitative analysis of the relative protein expression of SMAD2 and phosphorylated SMAD2. GAPDH is used as a loading control (n=3); (D-F): Protein expression levels of total SMAD3 and phosphorylated SMAD3 (n=3) and (G and H): Protein expression levels of total SMAD4 (n=3) Note: Data are shown as the mean±SEM, *p<0.05
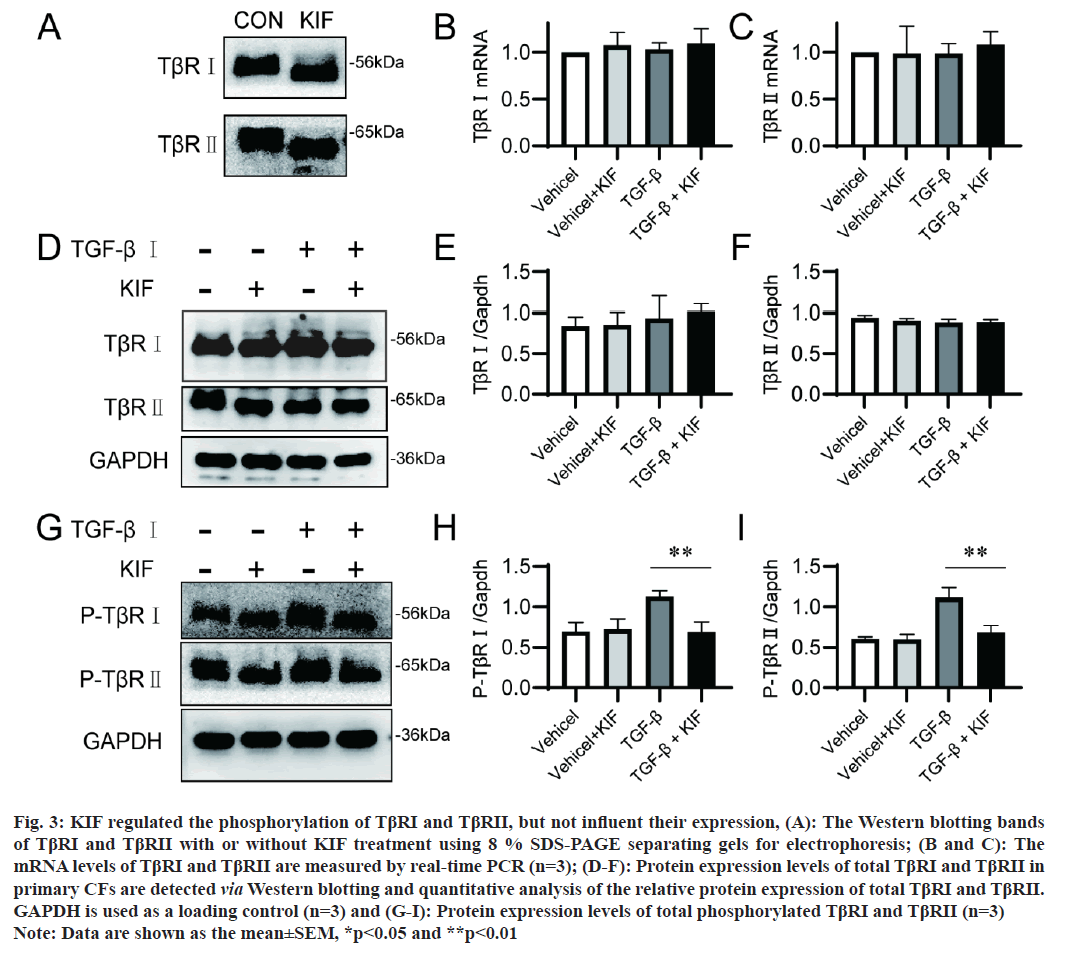
The aforementioned results refocus our attention on the impact of KIF on TβRs. The activation of TGF-β1 signaling pathway requires the cooperation of TβRI and TβRII, neither TβRI nor TβRII alone possess TGF-β1 signaling pathway activity [18]. In our study, we investigated the effect of KIF on both TβRs. The change of protein molecular weight is a direct manifestation of the effects of N-glycosylation inhibitors, the molecular weight of both TβRI and TβRII were reduced following KIF application (fig. 3A). This is consistent with the fact that both TβRI and TβRII belong to the N-glycosylation membrane proteins. KIF prevents the formation of complex and heavy N-glycans on them, leading to a decrease in protein molecular weight. Subsequently, we examined the effect of KIF on the transcription of TβRI and TβRII. Real-time PCR results revealed that the mRNA levels of TβRI and TβRII were not altered by KIF (fig. 3B and fig. 3C), which is consistent with previous reports that N-glycosylation inhibitors commonly do not change the mRNA levels of target proteins. Unexpectedly, Western blotting results showed that the expression of TβRI and TβRII was also unchanged by KIF (fig. 3D-fig. 3F). The efficacy of the receptor is not dependent on the expression level of the receptor, but rather on its activation level. The phosphorylation levels of TβRI and TβRII were sequentially examined. Results showed that TGF-β application significantly enhanced the phosphorylation level of TβRI and TβRII, and KIF notably attenuated this enhancement (fig. 3G-fig. 3I). Therefore, KIF did not affect the expression of TβRI and TβRII, but it did attenuate their activation induced by TGF-β.
Fig 3: KIF regulated the phosphorylation of TβRI and TβRII, but not influent their expression, (A): The Western blotting bands of TβRI and TβRII with or without KIF treatment using 8 % SDS-PAGE separating gels for electrophoresis; (B and C): The mRNA levels of TβRI and TβRII are measured by real-time PCR (n=3); (D-F): Protein expression levels of total TβRI and TβRII in primary CFs are detected via Western blotting and quantitative analysis of the relative protein expression of total TβRI and TβRII. GAPDH is used as a loading control (n=3) and (G-I): Protein expression levels of total phosphorylated TβRI and TβRII (n=3) Note: Data are shown as the mean±SEM, *p<0.05 and **p<0.01
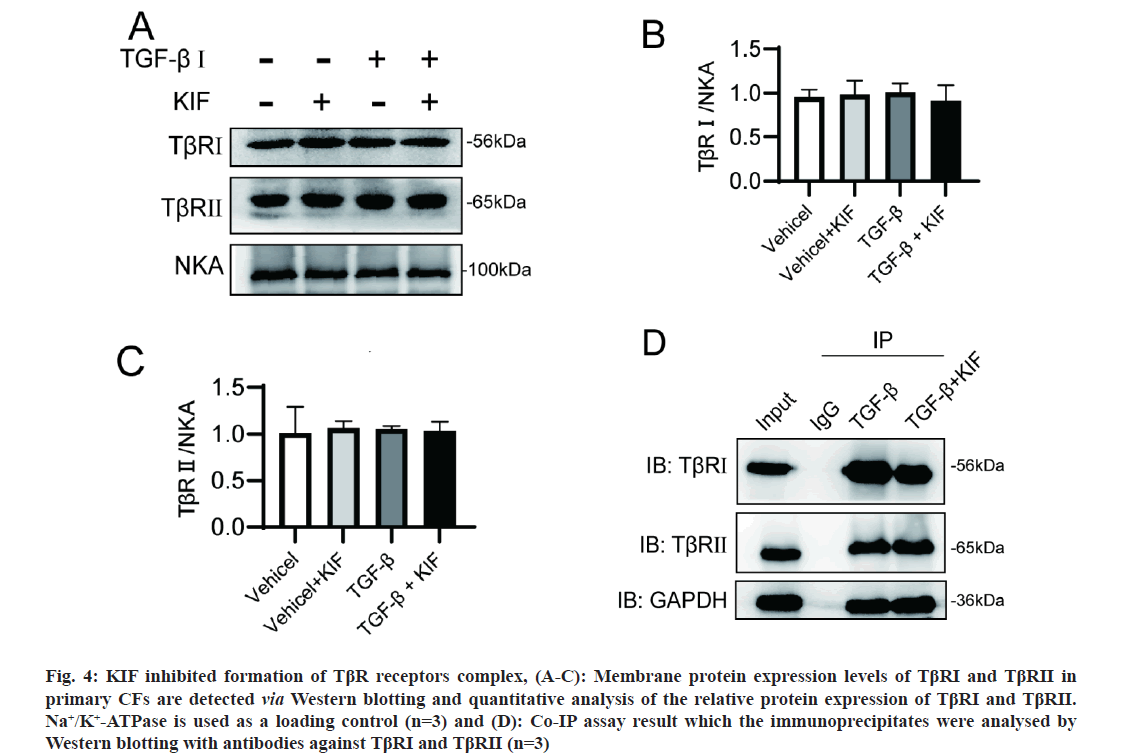
The initiation of the TGF-β/SMAD pathway is the formation of TGF-β ligand-receptor heterotetramer complexes. Consequently, the cell membrane expression and interaction of TβRs are crucial. Considering the crucial roles of N-glycosylation in the synthesis and trafficking of membrane proteins, we initially hypothesized that KIF might disturb the membrane expression of TβRs. This hypothesis was examined through membrane protein Western blotting. However, the results indicated that the cell surface localization of TβRI and TβRII is unaffected by KIF (fig. 4A- fig. 4C). This finding aligns with a previous report, where TβRII proteins were primarily localized on the cell surface in both untreated and KIF-treated HeLa cells, however, in tunicamycin-treated cells, these proteins predominantly accumulated in the perinuclear region [10]. Subsequently, we postulated that the formation of the TβR heterotetramer is of pivotal importance. Prior to activation, TβRI and TβRII are present on the cell surface as homodimers. When TGF-β1 is present, TGF-β1, TβRI, and TβRII aggregate into ligand-receptor complexes, and the interaction between TβRI and TβRII is necessary for the phosphorylation of R-SMAD. To investigate the interaction between TβRI and TβRII, we performed a Co-IP assay. Our Co-IP results demonstrated a strong interaction between TβRI and TβRII under TGF-β1 application (fig. 4D). However, this interaction was reduced with KIF treatment. This suggests that the interaction between TβRI and TβRII, or in other words, the formation of ligand-receptor complexes, was impaired by KIF treatment. This impairment could potentially explain why KIF attenuates TGF-β1 induced fibrosis.
Fig 4: KIF inhibited formation of TβR receptors complex, (A-C): Membrane protein expression levels of TβRI and TβRII in primary CFs are detected via Western blotting and quantitative analysis of the relative protein expression of TβRI and TβRII. Na+/K+-ATPase is used as a loading control (n=3) and (D): Co-IP assay result which the immunoprecipitates were analysed by Western blotting with antibodies against TβRI and TβRII (n=3) Note: Data are shown as the mean±SEM, *p<0.05
Cardiac fibrosis is a prevalent manifestation of chronic heart disease, and TGF-β1 signalling known to play a crucial role in fibrosis. As reported in the literature, TGF-β1 levels are elevated in many heart diseases, such as diabetic cardiomyopathy and myocardial infarction [19,20]. TGF-β1 stimulates myofibroblasts transdifferentiation, enhances ECM protein synthesis, increases the expression of integrin’s, and exerts potent matrix-preserving actions on CFs [21]. Consequently, TGF-β1 is regarded as the most well-characterized fibrogenic growth factor [22]. Given the pivotal role of TGF-β1 in cardiac fibrosis, inhibiting the TGF-β1 signalling pathway is considered a promising strategy to combat cardiac fibrosis. Several antifibrotic therapeutics targeting TGF-β1 signalling pathways have been investigated. Gene knockout of TβRI and TβRII in fibroblasts has been shown to attenuated cardiac fibrosis in an animal model of left ventricular pressure overload. Additionally, SM16, an orally active TβRI inhibitor, and pirfenidone, an oral drug that can decrease TGF-β expression, have been found to attenuate the development of cardiac fibrosis, reduce collagen crosslinking, and improve diastolic function in pressure overload-induced fibrosis [23]. Despite the challenges posed by their toxicity and limited clinical evidence, these anti-fibrotic drugs targeting TGF-β1 signalling pathway have attracted significant attention and investment due to their substantial therapeutic potential. We noticed that TβRI and TβRII undergo high level N-glycosylation modification. Consequently, we hypothesized that N-glycosylation inhibitors may affect TβRs and suppress the fibrotic process induced by TGF-β1. Our results demonstrate that the N-glycosylation inhibitor KIF significantly attenuated the upregulation of collagen I, collagen III and α-SMA induced by TGF-β1 in primary rat CFs. These findings suggest that the regulation of N-glycosylation of TβRs may be an effective anti-fibrotic strategy, positioning KIF as a potential therapeutic agent for cardiac fibrosis.
Glycosylation is arguably the most diverse post-translational modification of proteins [24]. N-glycosylation, the most common type of glycosylation of membrane and secreted proteins, has been shown to play an important role in protein synthesis, transport, and degradation [24,25]. The roles of N-glycosylation in cardiac fibrosis remain unclear, but numerous reports have confirmed that N-glycosylation is necessary for TβRs. TβRs has been reported to possess N-linked oligomannoside [11]. The complex type modification of TβRs is required for its successful transportation to the cell surface. The elimination of N-glycosylation or mutation in N-glycosylation sites retains TβRII in the perinuclear region, thereby inhibiting the TGF-β1/SMAD signalling pathways [11]. Similar to TβRII, the elimination of N-glycosylation can reduce the amounts of TβRI on the cell surface [26]. Moreover, the core fucosylation of TβRII and TβRI is crucial for optimal TGF-β receptor interactions and R-SMAD phosphorylation [27]. Abnormal glycosylation of TGF-β receptors generally impedes its transportation to the cell surface and it’s binding to the TGF-β ligand, thereby affecting TGF-β downstream signaling. It should be emphasized that N-glycosylation maybe have cell- and tissue-specificity [28]. The regulation of the glycoproteome has reached an unprecedented level of complexity. While no gene encodes N-glycan structures, approximately 2 % of the mammalian genome is estimated to encode genes involved in the synthesis of these N-glycan structures [29]. Numerous cellular parameters determine N-glycan structural diversity, such as regulation at the transcriptional (transcription factors, epigenetic regulation), post-transcriptional (microRNA (miRNA)), translational (speed of synthesis), and post-translational (conformation, modifications, interaction with other proteins) levels [29]. As highly N-glycosylated proteins, TβRs possess large glycans. When N-glycosylation is complete, TβRII exhibits multiple molecular weights, presenting multiple bands of high molecular weight over approximately 72 kDa in Human Kidney proximal tubular epithelial Cell line (HKC-8) cells, however, if all N-glycans are eliminated from TβRII, only a single band at roughly 64 kDa is observed [12]. However, the mechanisms through which interventions can modulate these glycans to induce structural differences, thereby resulting in functional alterations in TβRs, remain an area waiting for further exploration. Regardless, N-glycan is a necessary and functional component of TβRI and TβRII, and regulation of N-glycosylation is a promising strategy to manipulate the TGF-β1 signaling pathway and pathological changes associated with it.
N-glycosylation inhibitors have been explored as potential therapeutic agents for some diseases. Tunicamycin, a GlcNAc phosphotransferase inhibitor, can prevent the attachment of N-glycans to nascent polypeptides, thereby completely eliminating the formation of N-glycosylation. Sun et al. [11] reported that tunicamycin can mitigate the radio resistance and cancer stem ness in breast cancer cells induced by Alpha 1,3-Mannosyltransferase 3 (ALG3) overexpression by inhibiting N-glycosylation of TβRII [30]. Similarly, Helei et al. demonstrated that tunicamycin inhibited the TGF-β mediated the epithelial-mesenchymal transition in hepatocellular carcinoma cells through the inhibition of CD44s and the ERK1/2 signaling pathway. However, the complete elimination of N-glycosylation often leads to severe functional abnormality. In contrast, modification of N-glycans’ structure also alters the properties of glycoproteins with more subtle adverse effects. Core fucosylation, catalysed by Fucosyltransferases (FUTs), is a crucial component of N-glycans, some studies reports that inhibiting FUT8, FUT3, and FUT6 can bring benefits to serial disease. Wen et al. reported that blocking core fucosylation using FUT8 siRNA does not affect the protein expression levels of TβRI and TβRII. However, it significantly decreased SMAD2/3 phosphorylation and alleviated calcium and phosphorus deposition in rat vascular smooth muscle cells [31]. In a rat model of peritoneal fibrosis, Longkai et al. observed that blocking core fucosylation via FUT8 knockdown ameliorates glucose dialysate-induced peritoneal fibrosis. They found that inhibiting FUT8 deactivates the TGF-β/SMAD signalling pathway, without affecting the expression levels of TβRI and TβRII [32]. Similarly, in a rat model of unilateral ureteral obstruction, Shen reported that blocking core fucosylation also attenuates renal interstitial fibrosis by suppressing TGF-β/SMAD signalling, without impacting the expression levels of TβRI and TβRII [33]. Zhao also reported that inhibiting FUT8 via 1-deoxynojirimycin suppressed core fucosylation of TβRII and inhibits diabetic cardiomyopathy-associated fibrosis in db/db mice [34]. In metastatic colorectal cancer, Hirakawa et al. [35] reported that the inhibiton of FUT3 and FUT6 diminishes TGF-β induced EMT in metastatic colorectal cancer cells, and this effect is primarily reliant on TβRI, rather than TβRII [35]. Despite the role of core fucosylation, the branched N-glycan structure also plays a significant role in TGF-β1 signalling, depletion of 1,6 N-Acetylglucosaminyltransferase V (Mgat5) would inhibits the formation of β1,6-GcNAc branched N-glycans and reduces the sensitivity of TGF-β1 to TβRs [36]. In summary, modification of composition of N-glycan also has the potential to alter the function of TβRs, and has been considered as a potential therapeutic target for some diseases.
In the present study, we provide evidence that KIF substantially attenuated TGF-β induced cardiac fibrosis. KIF is a mannosidase I inhibitor that can maintain N-glycans at the high mannose stage, thereby preventing the formation of complex-type and hybrid-type N-glycan structures [37]. And some studies have identified KIF as a promising pharmacological agent or a useful tool for N-glycosylation studies. Ran et al. [38] report that KIF is an powerful immunomodulatory, and could potentially be a candidate drug for the treatment of chronic thromboembolic pulmonary hypertension. Cacheux et al. [39] reported that KIF could also serve as a potential treatment for recessive forms of Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT) caused by tradin mutation, KIF corrects the CPVT phenotype and restores cardiac function through the overexpression of calsequestrin 2. You et al. [40] have demonstrated that pre-treatment with KIF can inhibit proliferation of hepatocellular carcinoma cells, mediated by the alteration the receptor tyrosine kinases properties. Furthermore, KIF has been shown to disrupt the trafficking of glucose transporter GLUT1 during the glucose uptake process, leading to a reduction in the GLUT1 membrane expression and glucose uptake inhibition [41]. KIF is also effective against TβRs. Young- Kim et al. observed that KIF reduced the molecular weight of TβRII in lung cancer cell line A549. They reported that KIF did not affect TβRII’s trafficking, it only reduces sensitivity of TβRII in FLAG-TβRII transfected A549 cells [10]. Interestingly, they also reported that KIF not works on TβRII’s sensitivity in transfected cells. In our study, we observed KIF decreasing the molecular weight of both TβRI and TβRII in primary CFs, indicating KIF’s effectiveness against TβR’s. Similarly, our Western blotting and immunofluorescence results also demonstrated that KIF does not change the expression and trafficking of TβRI and TβRII in rat primary CFs. Rather, it inhibits the phosphorylation of downstream molecules, such as SMAD2 and SMAD3, which are stimulated by TGF-β1.
Canonical SMAD-dependent cascades are pivotal mediators of TGF-β contributing significantly to fibroblast activation in fibrotic hearts. Specifically, SMAD3 signalling plays critical role in activation of reparative fibroblasts, thereby inducing ECM protein synthesis, integrin transcription, and α-SMA expression [42]. SMADs often serve as indicators of TGF-β1 signalling activation. In this study, we examined the impact of KIF on SMAD proteins’ expression and phosphorylation. Our findings revealed that KIF did not affect the expression or molecular weight of SMAD2, SMAD3, and SMAD4, irrespective of TGF-β1 stimulation. This result in keeping with the previous studies, the N-glycosylation modifications always do not directly change the expression of SMADs. However, we found that KIF suppressed the phosphorylation level of SMAD2 and SMAD3 induced by TGF-β1 stimulation. This finding prompts us to refocus on the activation of TβRs, which are responsible for phosphorylation of SMADs. As we did not observe any alteration in membrane expression level of TβRI or TβRII, we hypothesized that KIF only influences the function, not the expression or trafficking, of TβRs, a notion similar to the core fucosylation. A critical and early step in the TβRs activation is the formation of tetrameric receptor complex consisting of two TβRII units and two TβRI units. TβRI is activated once the complex is complete, subsequently initiating intracellular signalling by phosphorylating downstream effectors, such as SMADs [43,44]. We postulated that the reduction in complex-type and hybrid-type N-glycans due to KIF could interfere with the formation of the receptor complex, thereby decreasing the efficiency of TGF-β1 signalling [45,46]. Our Co-IP results support this hypothesis, as the TβRII bonded to TβRI was reduced after KIF treatment. It suggests that the interaction between TβRI and TβRII, or in other words, the formation of receptor complexes, was impaired following KIF treatment. This is may explain why KIF impairs TGF-β1 signalling.
In conclusion, we have demonstrated that KIF is an effective strategy to attenuate the cardiac fibrosis induced by TGF-β1 stimulation. KIF significantly suppressed the expression of collagen I, collagen III and α-SMA by inhibiting TGF-β/SMAD signalling in rat primary CFs. Furthermore, we established that KIF impairs the formation of the TβRs complex without affecting the expression and trafficking of TGF-β receptors. These findings enhance the understanding of N-glycosylation in cardiac fibrosis, and provide novel insights suggesting that moderate alternation of N-glycans of TβRs is sufficient to modify TGF-β1 signalling. This could potentially contribute to the development of new therapies for cardiac fibrosis.
Author’s contributions:
Juanjuan Zhang, Linna Zhao and Yangong Liu have contributed equally to this work.
Acknowledgements:
This work was supported by Hebei Natural Science Foundation (No: H2021423019) to Juanjuan Zhang and Hebei Natural Science Foundation (No: H2021423069) to Gaoshan Yang.
Conflict of interests:
The authors declared no conflict of interests.
References
- Tani H, Sadahiro T, Yamada Y, Isomi M, Yamakawa H, Fujita R, et al. Direct reprogramming improves cardiac function and reverses fibrosis in chronic myocardial infarction. Circulation 2023;147(3):223-38.
[Crossref] [Google Scholar] [PubMed]
- Hsieh PL, Chu PM, Cheng HC, Huang YT, Chou WC, Tsai KL, et al. Dapagliflozin mitigates doxorubicin-caused myocardium damage by regulating AKT-mediated oxidative stress, cardiac remodeling, and inflammation. Int J Mol Sci 2022;23(17):10146.
[Crossref] [Google Scholar] [PubMed]
- Song HF, He S, Li SH, Wu J, Yin W, Shao Z, et al. Knock?out of microRNA 145 impairs cardiac fibroblast function and wound healing post?myocardial infarction. J Cell Mol Med 2020(16):9409-19.
[Crossref] [Google Scholar] [PubMed]
- Jia S, Meng A. TGFβ family signaling and development. Development 2021;148(5):188490.
[Crossref] [Google Scholar] [PubMed]
- Goumans MJ, Dijke TP. TGF-β signaling in control of cardiovascular function. Cold Spring Harb Perspect Biol 2018;10(2):022210.
[Crossref] [Google Scholar] [PubMed]
- Hinck AP. Structural studies of the TGF-βs and their receptors-insights into evolution of the TGF-β superfamily. FEBS Lett 2012;586(14):1860-70.
[Crossref] [Google Scholar] [PubMed]
- Khalil H, Kanisicak O, Prasad V, Correll RN, Fu X, Schips T, et al. Fibroblast-specific TGF-β-SMAD2/3 signaling underlies cardiac fibrosis. J Clin Invest 2017;127(10):3770-83.
[Crossref] [Google Scholar] [PubMed]
- Hata A, Chen YG. TGF-β signaling from receptors to Smads. Cold Spring Harbor Perspect Biol 2016;8(9):a022061.
[Crossref] [Google Scholar] [PubMed]
- Stambuk T, Klasic M, Zoldos V, Lauc G. N-glycans as functional effectors of genetic and epigenetic disease risk. Mol Aspects Med 2021;79:100891.
[Crossref] [Google Scholar] [PubMed]
- Kim YW, Park J, Lee HJ, Lee SY, Kim SJ. TGF-β sensitivity is determined by N-linked glycosylation of the type II TGF-β receptor. Biochem J 2012;445(3):403-11.
[Crossref] [Google Scholar] [PubMed]
- Sun X, He Z, Guo L, Wang C, Lin C, Ye L, et al. ALG3 contributes to stemness and radioresistance through regulating glycosylation of TGF-β receptor II in breast cancer. J Exp Clin Cancer Res 2021;40(1):149.
[Crossref] [Google Scholar] [PubMed]
- Park J, Lee SY, Ooshima A, Yang KM, Kang JM, Kim YW, et al. Glucosamine hydrochloride exerts a protective effect against unilateral ureteral obstruction-induced renal fibrosis by attenuating TGF-β signaling. J Mol Med 2013;91:1273-84.
[Crossref] [Google Scholar] [PubMed]
- Shirakawa A, Manabe Y, Fukase K. Recent advances in the chemical biology of N-glycans. Molecules 2021;26(4):1040.
[Crossref] [Google Scholar] [PubMed]
- Mikami M, Takuya O, Yoshino Y, Nakamura S, Ito K, Kojima H, et al. Acorus calamus extract and its component α-asarone attenuate murine hippocampal neuronal cell death induced by l-glutamate and tunicamycin. Biosci Biotechnol Biochem 2021;85(3):493-501.
[Crossref] [Google Scholar] [PubMed]
- Lu J, Wang QY, Zhou Y, Lu XC, Liu YH, Wu Y, et al. Astragaloside? against cardiac fibrosis by inhibiting TRPM7 channel. Phytomedicine 2017;30:10-7.
[Crossref] [Google Scholar] [PubMed]
- Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 2000;103(2):295-309.
[Crossref] [Google Scholar] [PubMed]
- Hanna A, Humeres C, Frangogiannis NG. The role of Smad signaling cascades in cardiac fibrosis. Cell Signal 2021;77:109826.
[Crossref] [Google Scholar] [PubMed]
- Zhou J, Zhong DW, Wang QW, Miao XY, Xu XD. Paclitaxel ameliorates fibrosis in hepatic stellate cells via inhibition of TGF-β/Smad activity. World J Gastroenterol 2010;16(26):3330.
[Crossref] [Google Scholar] [PubMed]
- Chen Z, Zheng L, Chen G. 2-Arachidonoylglycerol attenuates myocardial fibrosis in diabetic mice via the TGF-β1/Smad pathway. Cardiovasc Drugs Ther 2023;37(4):647-54.
[Crossref] [Google Scholar] [PubMed]
- Hanna A, Frangogiannis NG. The role of the TGF-β superfamily in myocardial infarction. Front Cardiovasc Med 2019;6:140.
[Crossref] [Google Scholar] [PubMed]
- Saadat S, Noureddini M, Mahjoubin-Tehran M, Nazemi S, Shojaie L, Aschner M, et al. Pivotal role of TGF-β/Smad signaling in cardiac fibrosis: Non-coding RNAs as effectual players. Front Cardiovasc Med 2021;7:588347.
[Crossref] [Google Scholar] [PubMed]
- Frangogiannis NG. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspects Med 2019;65:70-99.
[Crossref] [Google Scholar] [PubMed]
- Engebretsen KV, Skårdal K, Bjørnstad S, Marstein HS, Skrbic B, Sjaastad I, et al. Attenuated development of cardiac fibrosis in left ventricular pressure overload by SM16, an orally active inhibitor of ALK5. J Mol Cell Cardiol 2014;76:148-57.
[Crossref] [Google Scholar] [PubMed]
- Esmail S, Manolson MF. Advances in understanding N-glycosylation structure, function, and regulation in health and disease. Eur J Cell Biol 2021;100(7-8):151186.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Ten Dijke P, Wuhrer M, Zhang T. Role of glycosylation in TGF-β signaling and epithelial-to-mesenchymal transition in cancer. Protein Cell 2021;12(2):89-106.
[Crossref] [Google Scholar] [PubMed]
- Koli KM, Arteaga CL. Processing of the transforming growth factor β type I and II receptors: Biosynthesis and ligand-induced regulation. J Biol Chem 1997;272(10):6423-7.
[Crosstref] [Google Scholar] [PubMed]
- Venkatachalam MA, Weinberg JM. New wrinkles in old receptors: Core fucosylation is yet another target to inhibit TGF-β signaling. Kidney Int 2013;84(1):11-4.
- Li X, Wang H, Zhu Y, Cao W, Song M, Wang Y, et al. Heritability enrichment of immunoglobulin G N-glycosylation in specific tissues. Front Immunol 2021;12:741705.
[Crosstref] [Google Scholar] [PubMed]
- Toustou C, Walet?Balieu ML, Kiefer?Meyer MC, Houdou M, Lerouge P, Foulquier F, et al. Towards understanding the extensive diversity of protein N?glycan structures in eukaryotes. Biol Rev 2022;97(2):732-48.
[Crosstref] [Google Scholar] [PubMed]
- Hou H, Ge C, Sun H, Li H, Li J, Tian H. Tunicamycin inhibits cell proliferation and migration in hepatocellular carcinoma through suppression of CD 44s and the ERK 1/2 pathway. Cancer Sci 2018;109(4):1088-100.
[Crossref] [Google Scholar] [PubMed]
- Wen X, Liu A, Yu C, Wang L, Zhou M, Wang N, et al. Inhibiting post-translational core fucosylation prevents vascular calcification in the model of uremia. Int J Biochem Cell Biol 2016;79:69-79.
[Crossref] [Google Scholar] [PubMed]
- Li L, Shen N, Wang N, Wang W, Tang Q, Du X, et al. Inhibiting core fucosylation attenuates glucose-induced peritoneal fibrosis in rats. Kidney Int 2018;93(6):1384-96.
[Crossref] [Google Scholar] [PubMed]
- Shen N, Lin H, Wu T, Wang D, Wang W, Xie H, et al. Inhibition of TGF-β1 receptor posttranslational core fucosylation attenuates rat renal interstitial fibrosis. Kidney Int 2013;84(1):64-77.
[Crossref] [Google Scholar] [PubMed]
- Zhao Q, Jia TZ, Cao QC, Tian F, Ying WT. A crude 1-DNJ extract from home made Bombyx batryticatus inhibits diabetic cardiomyopathy-associated fibrosis in db/db mice and reduces protein N-glycosylation levels. Int J Mol Sci 2018;19(6):1699.
[Crossref] [Google Scholar] [PubMed]
- Hirakawa M, Takimoto R, Tamura F, Yoshida M, Ono M, Murase K, et al. Fucosylated TGF-β receptors transduces a signal for epithelial-mesenchymal transition in colorectal cancer cells. Br J Cancer 2014;110(1):156-63.
- Partridge EA, Le Roy C, di Guglielmo GM, Pawling J, Cheung P, Granovsky M, et al. Regulation of cytokine receptors by golgi N-glycan processing and endocytosis. Science 2004;306(5693):120-4.
[Crossref] [Google Scholar] [PubMed]
- Zhou Q, Xie Y, Lam M, Lebrilla CB. N-Glycomic analysis of the cell shows specific effects of glycosyl transferase inhibitors. Cells 2021;10(9):2318.
[Crossref] [Google Scholar] [PubMed]
- Miao R, Dong X, Gong J, Wang Y, Guo X, Li Y, et al. Possible immune regulation mechanisms for the progression of chronic thromboembolic pulmonary hypertension. Thromb Res 2021;198:122-31.
[Crossref] [Google Scholar] [PubMed]
- Cacheux M, Fauconnier J, Thireau J, Osseni A, Brocard J, Roux-Buisson N, et al. Interplay between triadin and calsequestrin in the pathogenesis of CPVT in the mouse. Mol Ther 2020;28(1):171-9.
[Crossref] [Google Scholar] [PubMed]
- You RI, Wu WS, Cheng CC, Wu JR, Pan SM, Chen CW, et al. Involvement of N-glycan in multiple receptor tyrosine kinases targeted by Ling-Zhi-8 for suppressing HCC413 tumor progression. Cancers 2018;11(1):9.
[Crossref] [Google Scholar] [PubMed]
- Lodge EK, Bell JD, Roloff EM, Hamilton KE, Louters LL, Looyenga BD. Pharmacologic inhibition of N-linked glycan trimming with kifunensine disrupts GLUT1 trafficking and glucose uptake. Biochimie 2020;174:18-29.
[Crossref] [Google Scholar] [PubMed]
- Frangogiannis NG. Cardiac fibrosis. Cardiovasc Res 2021;117(6):1450-88.
- Goumans MJ, Ten Dijke P. TGF-β signaling in control of cardiovascular function. Cold Spring Harbor Perspect Biol 2018;10(2):022210.
[Crossref] [Google Scholar] [PubMed]
- Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGF-beta receptors and Smads. Cell Res 2009;19:21-35.
- Eickelberg O, Centrella M, Reiss M, Kashgarian M, Wells RG. Betaglycan inhibits TGF-β signaling by preventing type I-type II receptor complex formation: Glycosaminoglycan modifications alter betaglycan function. J Biol Chem 2002;277(1):823-9.
[Crossref] [Google Scholar] [PubMed]
- Nakano N, Tsuchiya Y, Kako K, Umezaki K, Sano K, Ikeno S, et al. TMED10 protein interferes with Transforming Growth Factor (TGF)-β signaling by disrupting TGF-β receptor complex formation. J Biol Chem 2017;292(10):4099-112.
[Crossref] [Google Scholar] [PubMed]