- *Corresponding Author:
- N. Chauhan
Department of Pharmacology, L. M. College of Pharmacy, Navrangpura, Ahmedabad-380 009
E-mail: neelam@niperahm.edu.in
| Date of Submission | 28 May 2013 |
| Date of Revision | 19 December 2013 |
| Date of Acceptance | 26 December 2013 |
| Indian J Pharm Sci 2014; 76(1): 72-77 |
Abstract
The present study was aimed to investigate the role of plasma glucose concentration as a phenotypic marker and to study the frequency distribution of CYP2C9 genetic variants in Gujarat state diabetic population. One hundred and nine unrelated diabetes mellitus patients treated with sulfonylureas were genotyped for CYP2C9*2 and CYP2C9*3 alleles. Their pre- and posttreatment postprandial blood glucose levels were recorded and mean glucose drop per milligram of drug values were calculated and further used as an index for phenotypic correlation. The frequencies of CYP2C9*1, CYP2C9*2 and CYP2C9*3 alleles in the Gujarat state diabetic population were 0.84, 0.07 and 0.09, respectively. The distribution of CYP2C9*1/*1, CYP2C9*1/*2, CYP2C9*1/*3, CYP2C9*2/*2, CYP2C9*2/*3 and CYP2C9*3/*3 genotypes were 0.73, 0.08, 0.13, 0.0, 0.06 and 0.0, respectively. Patients with CYP2C9*1/*2 genotype did not show any significant difference in the mean glucose drop per milligram of drug values when compared with wild-type patients in glipizide-treatment group. Patients with CYP2C9*1/*3 genotype showed greater mean glucose drop per milligram of drug values than patients with CYP2C9*1/*1 wild-type genotype for both glipizide and glimepiride while patients with CYP2C9*2/*3 genotype showed greater drop than patients with CYP2C9*1/*1 genotype only in the glipizide-treatment group. The presence of CYP2C9*3 allele significantly affected plasma glucose drop per milligram of drug values in patients taking glipizide and glimepiride, while effects of CYP2C9*2 allele were insignificant. Further studies are needed to confirm the effects of CYP2C9*2 allele on plasma glucose drop per milligram of drug values. However, plasma glucose concentration is a complex physiological marker that cannot be used to establish perfect genotype-phenotype correlation. Hence studies exploring robust phenotypic markers must be initiated.
Keywords
CYP2C9, sulfonylurea, plasma glucose, phenotypic marker, diabetes
Diabetes mellitus (DM) is a metabolic disease, characterised by hyperglycaemia resulting from defects in insulin secretion, insulin action or both. The chronic hyperglycaemia of diabetes is associated with long-term damage, dysfunction and failure of various organs, especially the eyes, kidneys, nerves, heart and blood vessels [1]. Type 2 DM is the most common form of diabetes constituting 90% of the diabetic population. The global prevalence of diabetes is estimated to increase, from 4.0% in 1995 to 5.4% by the year 2025 [2]. A national survey of diabetes conducted in six major cities in India in the year 2000 showed that the prevalence of diabetes in urban adults was 12.1%. Prevalence of impaired glucose tolerance (IGT) was also high (14.0%) [3]. Type 2 DM is a multifactorial disease having genetic and nongenetic components, which interact to precipitate the diabetic phenotype [4]. Factors contributing to type 2 DM such as obesity, sedentary lifestyle, smoking and certain drugs are well known, but less is known about the genetic predisposition to disease susceptibility, treatment failures or adverse drug reactions (ADRs) [5].
Sulfonylureas (glyburide, glimepiride, glipizide and gliclazide) are the most frequently prescribed oral hypoglycaemic agents used in the treatment of type 2 DM. Severe sulfonylurea-associated hypoglycaemic episodes are fatal in 1.4-10% of cases and necessitate long and costly hospital visits [6]. Moreover, the risk of hypoglycaemia may lead physicians to adjust blood glucose concentrations to above those identified as optimal for the prevention of microvascular and macrovascular complications [7]. Thus, the identification and concern of individual’s risk for sulfonylureaassociated hypoglycaemia may be of great importance for the optimisation of treatment.
Cytochrome P450 2C9 (CYP2C9) enzyme, is the most abundant of the CYP2C enzyme family and comprises approximately one-third of the total hepatic P450 content [8,9]. It is involved in the metabolism of more than 100 drugs, including coumarin, anticoagulants, sulfonylureas and some nonsteroidal antiinflammatory drugs, but is largely responsible for the metabolism of oral hypoglycaemic agents such as tolbutamide, glibenclamide, glimepiride, glipizide and nateglinide [6,10-14]. Many CYP2C9 variants have been associated with reduced enzyme activity, with CYP2C9*2 and CYP2C9*3, having the most clinical relevance (http://www.cypalleles.ki.se) [15,16]. However, the effect of functional CYP2C9 polymorphisms on the risk of ADRs with oral hypoglycaemic therapy in patients has not yet been widely studied.
The CYP2C9*2 and CYP2C9*3 alleles have single base substitutions resulting in amino acid changes at residue 144 (Arg to Cys) and 359 (Ile to Leu), respectively [17]. In vitro and in vivo studies show that CYP2C9*3 is associated with a lower intrinsic clearance of substrate drugs than CYP2C9*2 [18-20]. The role CYP2C9*2 allele has in altering drug clearance is less clear with only some CYP2C9 substrates (e.g. warfarin and phenytoin) being affected in vivo. Studies have reported marked inter-ethnic variation in the distribution of CYP2C9 polymorphic alleles. Among Caucasians, over 30% of the population have one or two of these alleles, with the overall allele frequency of CYP2C9*2 and CYP2C9*3 being approximately 10% and 8% [12,21-24]. The distribution of CYP2C9 alleles varies with ethnicity, but the overall frequency of variant alleles CYP2C9*2 and CYP2C9*3 appears close to ~30% in the general population [25-29]. Pooled CYP2C9*2 and CYP2C9*3 allele frequency data is available for the south Indian population and is 4% and 8%, respectively [29]. The allelic frequencies of CYP2C9 gene variants in Gujarat’s healthy population have been documented by Sistonen et al. to be 4.4% for CYP2C9*2 and 9.6% for the CYP2C9*3 allele [30].
Studies on 29 healthy volunteers demonstrate that there is greater exposure to glyburide and glimepiride in healthy subjects heterozygous for CYP2C9*3 allele than in homozygous wild-type individuals. CYP2C9*2 allele does not significantly alter the pharmacokinetics of glyburide or glimepiride. Further, no significant differences are found in blood glucose variables of glyburide or glimepiride between the subjects with different hetero- and homozygous CYP2C9 genotypes [14]. Holstein et al. reported that individuals genetically determined for low CYP2C9 activity are at an increased risk of sulfonylurea-associated severe hypoglycaemia and that CYP2C9 genotyping could be a better predictive tool to determine to the susceptibility to adverse effects following oral hypoglycaemic treatment [7].
India contains an admixture of races. In most parts of India, the biological structure of the population, language, culture and religion bear the imprint of an intermixture of the Aryan, Dravidian, Kolarian and the Mongoloid races [31]. Although the populations of some states in south India share a common ethnic origin having descended from the Dravidians, it is difficult to distinctly trace back the origin of other Indian populations. An admixture of populations by inter-race marriage is prominent and leads to widespread genetic complexity. Thus defining a population primarily by geographic and common environmental boundaries is more logical and would provide rational population identity in terms of ‘specific geographical zones’ [26]. For this reason and considering the proximity of the research institution to the target population, selection of samples in the present study was restricted to the state of Gujarat, India.
In order to determine the influence of these CYP2C9 polymorphic mutant alleles in type 2 DM patients, the first step was to find the frequency of their prevalence in the diabetic population of Gujarat and then subsequently establish the genotype–phenotype correlation. Thus the present study was designed as a noninterventional, prospective, cohort study in Gujarat state diabetic population on sulfonylurea treatment for CYP2C9 genetic variants in order to investigate the role of plasma glucose concentration as a phenotypic marker, which is the first clinical sign for diabetes.
Materials and Methods
The study was conducted on 109 unrelated diabetic patients (43 males and 66 females, Table 1) on sulfonylurea or sulfonylurea and metformin combination treatment, residing in Gujarat especially Ahmedabad. All patients provided a written informed consent and the study was conducted as per the guidelines of the Institutional Ethics Committee, L. M. College of Pharmacy, Ahmedabad and B. J. Medical College and Civil Hospital Ethics Committee, Ahmedabad. The study was an exploratory, observational (noninterventional), open, prospective, cohort, multi-centric study to genotype type 2 DM patients on sulfonylurea treatment for CYP2C9 gene variants with the goal of establishing the phenotypic correlation.
| Demographic parameter | Females (n=66) | Males (n=43) | ||
|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | |
| Age (years) | 28–80 | 52.5 ± 10.2 | 39–86 | 57.6 ± 11.4 |
| Weight (kg) | 40–95 | 64.9 ± 11.7 | 35–98 | 70.1 ± 13.3 |
| Height (cm) | 143–169 | 159.0 ± 6.4 | 156–180 | 167.6 ± 5.6 |
| Body mass index (kg/m2) | 15.2–38.9 | 25.8 ± 4.8 | 13.7–32.9 | 24.8 ± 4.0 |
Table 1: Basic demographic details of diabetic Patients
Patients attending the diabetic outpatient department (OPD) of Civil Hospital, Vijayratna diabetes foundation, Dia-Care Clinic, and Diabetes Clinic, Ahmedabad, who met the inclusion and exclusion criteria, were screened and recruited. Patients were included in the study if they met the following requirements: Diagnosed with type 2 DM and were receiving sulfonylurea or a sulfonylurea and metformin combination treatment; baseline postprandial blood glucose records were available; had the ability to understand the study rationale and were willing to sign a written consent for enrolment at the screening visit before any protocol specific procedure was performed; and did not have any co-morbidities or concomitant medications prescribed that would affect the CYP2C9 activity. Type 2 DM patients who were: Seriously ill or hospitalised; received antidiabetic medication other than sulfonylurea or a sulfonylurea and metformin combination; had a recent history of renal or hepatic disease or insufficiency; whose baseline diagnostic haematological record were unavailable; were unwilling to undergo genetic analysis; or were receiving any concomitant medications that might induce (amiodarone, cimetidine, cotrimoxazole, disulfiram, fluvastatin, fluvoxamin, fluconazole, isoniazid, ketoconazole, metronidazole, sulfinpyrazole, ticlopidine, zafirlukast) or inhibit (barbiturates, carbamazipine, phenobarbital, phenytoin primidone, rifampin) CYP2C9 activity were excluded from the study.
The treatment regimen was subjective according to the patient’s glycaemic status and physician’s decision. The patient treatment groups were classified based on the type of sulfonylurea prescribed to them. A 5 ml venous blood sample was collected using ethylenediamine tetraacetic acid (EDTA) as an anticoagulant from each patient while undergoing haematological testing for blood glucose as prescribed by the physician during their routine follow-up visit and was used for the genotypic analysis. Baseline and posttreatment postprandial blood glucose records for each patient were recorded. The patients were genotyped for CYP2C9*2 and CYP2C9*3 alleles using the multiplex polymerase chain reaction (PCR) technique [32,33].
Postprandial blood glucose concentration was selected as the phenotypic marker. In case of already diabetic patients (more than 1 year of diabetes) the blood glucose levels before and after a constant drug dosage interval were selected. The change in postprandial blood glucose concentration over the treatment period was transformed into glucose drop per milligram (DPM) of daily drug dosage value for each treatment group and was used as an index for the genotype– phenotype correlation. DPM values were arrived at using the formula, DPM value = (pretreatment glucose concentration–posttreatment glucose concentration)/ daily dose of drug in milligram. Statistical analysis was performed considering P<0.05 to be statistically significant. Observed frequencies for the diabetic study population were compared with the predicted frequencies according to Hardy Weinberg law using the chi-square test {χ2(3, 0.05) = 7.815 >5.2035(χ2 cal)}. Differences in genotypic frequencies among the diabetic and healthy Gujarat populations were also assessed using chi-square test (2×4 RC Contingency table) {χ2(3, 0.05) = 7.815 >2.9377 (χ2 cal)}. The patients within each treatment group were classified into their respective genotypic classes, as evident from the genotyping results and their mean glucose (DPM) values were compared statistically using analysis of variance (ANOVA) followed by Tukey’s test. Results are expressed as mean glucose DPM values±SEM. Statistical analysis was performed using SPSS version 16.
Results and Discussion
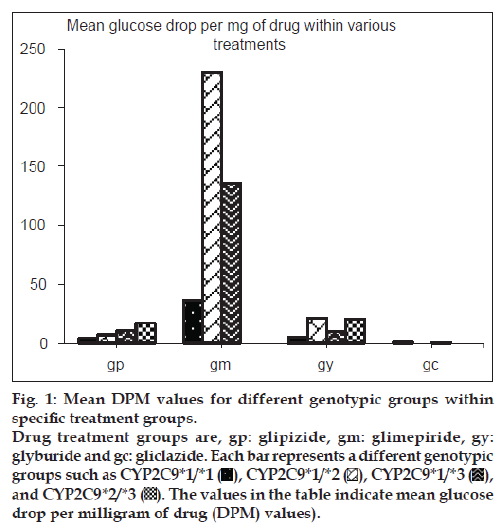
The allelic and genotype frequencies observed in this study are in accordance to the predicted frequencies calculated by the Hardy–Weinberg law. Out of 109 patient genotypes that were confirmed, the distribution of various polymorphic genotypes within the specific treatment groups is shown in fig. 1. Of the 109 patients, 80 (73.4%) had CYP2C9*1/*1 [wild-type] genotype, 9 (8.3%) had CYP2C9*1/*2 [heterozygous mutant] genotype, 14 (12.9%) had CYP2C9*1/*3 [heterozygous mutant] genotype and 6 (5.5%) had CYP2C9*2/*3 [mixed homozygous mutant] genotype. No patient homozygous for the same mutant allele was identified.
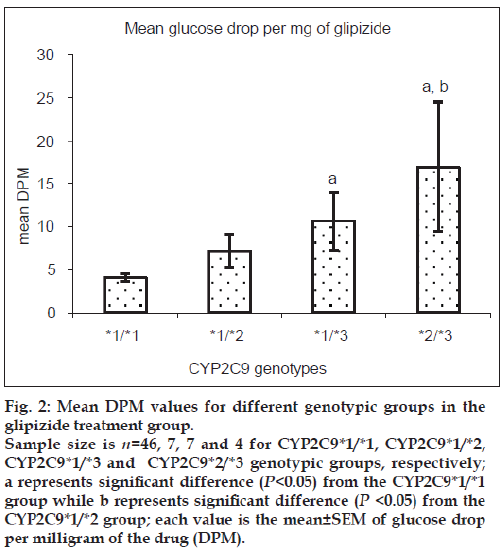
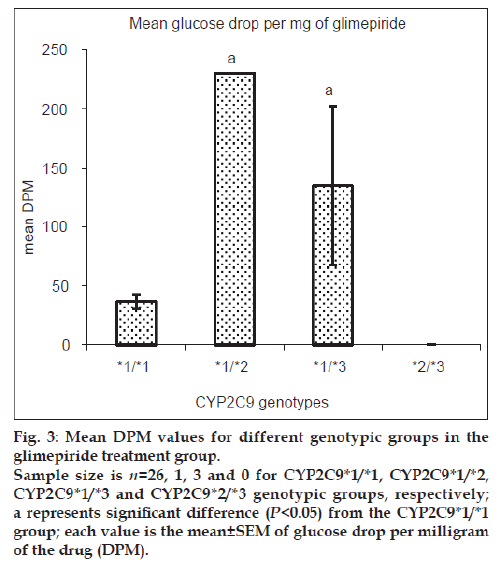
Patients heterozygous for CYP2C9*3 mutant allele showed greater mean glucose DPM values than homozygous wild-type patients for both glipizide and glimepiride treatment groups. Patients with mixed homozygous mutant genotype (CYP2C9*2/*3) showed greater mean glucose DPM values than the homozygous wild-type patients in the glipizide treatment group. Patients heterozygous mutant for CYP2C9*2 allele did not show any significant difference in the mean glucose DPM values when compared with homozygous wild-type patients in the glipizide treatment group while the number of patients in the glimepiride treatment group was inadequate for this comparison. The comparison for glyburide and gliclazide groups was not possible due to insufficient number of patients available in respective genotypic groups. Figs. 2 and 3 show the mean glucose DPM value comparison for the glipizide and glimepiride treatment groups, respectively.
Figure 2: Mean DPM values for different genotypic groups in the
glipizide treatment group.
Sample size is n=46, 7, 7 and 4 for CYP2C9*1/*1, CYP2C9*1/*2,
CYP2C9*1/*3 and CYP2C9*2/*3 genotypic groups, respectively;
a represents significant difference (P<0.05) from the CYP2C9*1/*1
group while b represents significant difference (P <0.05) from the
CYP2C9*1/*2 group; each value is the mean±SEM of glucose drop
per milligram of the drug (DPM).
Figure 3: Mean DPM values for different genotypic groups in the
glimepiride treatment group.
Sample size is n=26, 1, 3 and 0 for CYP2C9*1/*1, CYP2C9*1/*2,
CYP2C9*1/*3 and CYP2C9*2/*3 genotypic groups, respectively;
a represents significant difference (P<0.05) from the CYP2C9*1/*1
group; each value is the mean±SEM of glucose drop per milligram
of the drug (DPM).
This is the first study to establish the CYP2C9 allelic and genotypic frequencies in diabetic population of Gujarat state. However, the genotypic analysis of healthy population of the same region has been studied already [30]. The study showed that polymorphic genotypes have a significant effect on plasma glucose concentration in patients with compromised glycaemic control. The results support the rational for conducting a study in diabetes patients whose glucose metabolism is impaired unlike those performed in healthy subjects with normal intact homeostatic mechanisms.
Niemi et al. showed that the exposure to glyburide and glimepiride was markedly greater in healthy subjects heterozygous for CYP2C9*3 allele than in homozygous wild-type individuals [14]. Results of the present study support the findings by Neimi et al. that the CYP2C9*3 allele significantly affects glimepiride pharmacokinetics substantiated by the pharmacodynamic effects on plasma glucose concentration as evident in the present study. Since only one patient was found with CYP2C9*1/*2 genotype on glimepiride treatment, the effect of CYP2C9*2 allele on plasma glucose concentration is inconclusive. If the effects of CYP2C9*2 and CYP2C9*3 alleles on the pharmacokinetics of sulfonylureas like glyburide and glimepiride are extended for glipizide, the major treatment group in the present study, results are analogous to the ones reported by Neimi et al., that is, CYP2C9*2 allele did not have significant effect on plasma glucose concentration while presence of CYP2C9*3 allele caused significant drop in plasma glucose levels as compared with the wild-type patients.
Holstein et al. suggested that genotyping might be a tool for the better prediction of adverse effects caused by oral hypoglycaemic agents [7]. In accordance to this, the present study provides quantitative evidence for the incidence of hypoglycaemia. The evidence of the differences in the blood glucose control between various genotypic classes advocates a need for further controlled studies targeting specific populations on larger prospective cohorts in order to establish perfect genotype–phenotype correlation.
Plasma glucose concentration is a complex physiological marker affected by multiple factors involved in body’s homeostatic mechanisms. Unless the variables like food intake, physical activity and compliant drug regimen are controlled, plasma glucose concentration cannot be used as a robust phenotypic marker, which may have contributed to the nonuniformity of data in the present study. Although glycated haemoglobin (HbA1c) does not reflect transient periods of hypoglycaemia, it can nullify the effect of the aforementioned confounding effects and may serve as a robust phenotypic marker. Better genotype– phenotype correlations are more likely be achieved if a controlled, in-patient study, where all the abovementioned confounding factors are minimised and blood glucose or HbA1c is used as phenotypic marker. Another reason for the lack of strong genotype– phenotype correlation may be due to fewer numbers of patients on particular treatment with different polymorphic genotypes, which made their comparison less affirmative. Thus, future studies recruiting larger patient pools to ensure adequate sample size in the polymorphic genotype groups is required.
Understanding the role of genetic polymorphisms in drug responses helps ensuring adequate drug efficacy and decrease in the incidence of adverse drug effects by tailoring medications according to the patients’ genetic profile. Advances in this area have important implications in the design of drug dose regimens and rational drug prescriptions. Use of pharmacogenetic knowledge during the drug discovery and developmental process can accelerate the improvement of targeted therapeutic interventions where the pharmacophore is explicitly designed for particular responder patient groups. Such genetic stratification of patients especially in clinical trials can enhance the statistical power of the study and reduce the number of subjects required; leading to significant time and resource savings in drug development by reducing drug trial failure rates. Therefore, advanced technologies that identify genetic polymorphisms rapidly, accurately and economically will be of significant value to pharmaceutical research and development [34].
In the practice of medicine, a major problem with the pharmacotherapy of type 2 DM patients taking sulfonylureas or any other oral hypoglycaemic agent is uncontrolled hyperglycaemia, while hypoglycaemic events account for very low percentages. In fact, polymorphic patients expressing polymorphic CYP2C9 alleles could be better treated with lower doses of sulfonylureas in order to achieve tightly controlled plasma glucose levels. The clinical situations become more complex when multiple CYP2C9 substrates like losartan, diclofenac, imipramine, phenytoin or CYP2C9 inducing drugs like cimetidine, ketoconazole, metronidazole or inhibitors like carbamazipine, barbiturates are coprescribed for the management of coexisting disorders. The current study thus opens new avenues for greater and serious investigations on coprescriptions in DM in the light of coexistence of polymorphisms in order to handle clinical situations systematically and attain better patient care.
Acknowledgements
The authors are grateful to Dr. Chitra Joshi, Dr. B. D. Makad, Dr. A. N. Shah for their cooperation during the collection of samples from Civil hospital, Ahmedabad, India. A special thanks to Dr. Bansi Saboo for his help and cooperation during the study.
References
- American Diabetes Association: Diagnosis and classification of diabetes mellitus. Diabetes Care 2004;27Suppl 1:S5-10.
- King H, Aubert RE, Herman WH. Global burden of diabetes, 1995-2025: Prevalence, numerical estimates, and projections. Diabetes Care 1998;21:1414-31.
- Ramachandran A, Snehalatha C, Kapur A, Vijay V, Mohan V, Das AK, et al. High prevalence of diabetes and impaired glucose tolerance in India: National Urban Diabetes Survey. Diabetologia 2001;44:1094-101.
- Beck-Nielsen H, Groop LC. Metabolic and genetic characterization of prediabetic states.Sequence of events leading to non-insulin-dependent diabetes mellitus. J Clin Invest 1994;94:1714-21.
- Hansen T. Genetics of type-2 diabetes. Curr Sci 2002;83:1477-82.
- Holstein A, Egberts EH. Risk of hypoglycaemia with oral antidiabetic agents in patients with Type 2 diabetes.Exp Clin Endocrinol Diabetes 2003;111:405-14.
- Holstein A, Plaschke A, Ptak M, Egberts EH, El -Din J, Brockmoller J, et al. Association between CYP2C9 slow metabolizer genotypes and severe hypoglycaemia on medication with sulphonylurea hypoglycaemic agents. Br J Clin Pharmacol 2005;60:103-6.
- Lapple F, von Richter O, Fromm MF, Richter T, Thon KP, Wisser H, et al. Differential expression and function of CYP2C isoforms in human intestine and liver. Pharmacogenetics 2003;13:565-75.
- Lasker JM, Wester MR, Aramsombatdee E, Raucy JL. Characterization of CYP2C19 and CYP2C9 from human liver: Respective roles in microsomal tolbutamide, S- mephenytoin, and omeprazole hydroxylations. Arch Biochem Biophys 1998;353:16-28.
- Kirchheiner J, Bauer S, Meineke I, Rohde W, Prang V, Meisel C, et al. Impact of CYP2C9 and CYP2C19 polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics 2002;12:101-9.
- Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. ClinPharmacolTher 2005;77:1-16.
- Kirchheiner J, Brockmoller J, Meineke I, Bauer S, Rohde W, Meisel C, et al. Impact of CYP2C9 amino acid polymorphisms on glyburide kinetics and on the insulin and glucose response in healthy volunteers. Clin Pharmacol Ther 2002;71:286-96.
- Kirchheiner J, Meineke I, Muller G, Bauer S, Rohde W, Meisel C, et al. Influence of CYP2C9 and CYP2D6 polymorphisms on the pharmacokinetics of nateglinide in genotyped healthy volunteers. Clin Pharmacokinet 2004;43:267-78.
- Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivisto KT. Glyburide and glimepiride pharmacokinetics in subjects with different CYP2C9 genotypes. Clin Pharmacol Ther 2002;72:326-32.
- Grant RW, Wexler DJ. Loss-of-function CYP2C9 variants: Finding the correct clinical role for Type 2 diabetes pharmacogenetic testing. Expert Rev Cardiovasc Ther 2010;8:339-43.
- Surendiran A, Pradhan SC, Agrawal A, Subrahmanyam DK, Rajan S, Anichavezhi D, et al. Influence of CYP2C9 gene polymorphisms on response to glibenclamide in type 2 diabetes mellitus patients. Eur J Clin Pharmacol 2011;67:797-801.
- Homepage of the human cytochrome P450 (CYP) allele nomenclature committee. Available from: http://www.imm.ki.se/cypalleles/CYP2C9.htm. [Updated Mar 2006; accessed 2013 Nov 28].
- Rettie AE, Haining RL, Bajpai M, Levy RH. A common genetic basis for idiosyncratic toxicity of warfarin and phenytoin. Epilepsy Res 1999;35:253-5.
- Yamazaki H, Inoue K, Chiba K, Ozawa N, Kawai T, Suzuki Y, et al. Comparative studies on the catalytic roles of cytochrome P450 2C9 and its Cys- and Leu-variants in the oxidation of warfarin, flurbiprofen, and diclofenac by human liver microsomes. Biochem Pharmacol 1998;56:243-51.
- Yasar U, Tybring G, Hidestrand M, Oscarson M, Ingelman-Sundberg M, Dahl ML, et al. Role of CYP2C9 polymorphism in losartan oxidation. Drug MetabDispos 2001;29:1051-6.
- Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999;353:717-9.
- Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH: Cytochrome P450 oxidoreductase. Pharmacogenetics 1997;7:203-10.
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994;4:39-42.
- Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 2000;96:1816-9.
- Chauhan N, Shubha R, Padh H. Pharmacogenetics: Basis for rational drug therapy. Indian J Pharm Sci 2007;69:180-9.
- Daly AK. Molecular basis of polymorphic drug metabolism. J Mol Med 1995;73:539-53.
- Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin.ThrombHaemost 2004;91:87-94.
- Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287:1690-8.
- Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics 2004;14:539-47.
- Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 2009;19:170-9.
- Rosemary J, Adithan C, Soya SS, Nathalie G, Shashindran C, Benny KA, et al. CYP2C9 and CYP2C19 genetic polymorphisms: Frequencies in the sounth Indian population. Fund Clin Pharmacol 2005;19:101-5.
- Chauhan N. Inter-individual variability of cytochromoe P450 and pharmacokinetics in Indian population. PhD Thesis, 2007.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory Manual. New York: Cold Spring Harbour Laboratory Press; 1989.
- Padh H. Human genome project and pharmacogenomics: Leading towards individualized medication. Indian Drugs 2001;38:160-3.