- *Corresponding Author:
- Xiaotong Zhang
Department of Computer Science and Technology, University of Science and Technology Beijing, Beijing 100083, China
E-mail: zxt@ustb.edu.cn
| This article was originally published in a special issue, “Integrative Approaches in Biomedical Sciences for Drug Discovery and Development” |
| Indian J Pharm Sci 2024:86(6) Spl Issue “9-18” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study proposes a novel approach to drug response prediction by incorporating pharmacological mechanisms into graph convolutional networks. The developed model, termed pharmacological knowledge graph convolutional network, leverages prior knowledge of drug-target interactions, pharmacokinetics, and pharmacodynamics to improve both the accuracy and interpretability of predictions. By integrating comprehensive pharmacological datasets such as DrugBank and the Cancer Genome Atlas, the pharmacological knowledge graph convolutional network model demonstrates enhanced predictive capabilities, outperforming baseline models that lack prior knowledge integration. Experimental results highlight the potential of this approach in advancing personalized medicine and drug repurposing strategies by offering deeper insights into drug mechanisms across diverse biological contexts. Pharmacological knowledge graph convolutional network’s ability to predict drug responses accurately underscores its utility in the development of tailored therapeutic interventions and the acceleration of new drug discoveries.
Keywords
Autism spectrum disorder, graph neural networks, drug-target interactions, pharmacokinetics, magnetic resonance imaging
The era of precision medicine has ushered in the need to unravel the complex dynamics of drug responses, which are characterized by significant inter-individual variability. The heterogeneity in patient responses to therapeutics is driven by an intricate array of factors, including genetic polymorphisms, environmental exposures, and the multifaceted nature of drug-drug and drug-disease interactions. The complexity of these determinants has rendered conventional approaches to predicting drug responses inadequate, necessitating a more sophisticated model that integrates pharmacological knowledge[1].
In recent years, Graph Convolutional Networks (GCNs) have demonstrated substantial potential in handling non-Euclidean data structures, particularly in the context of biological networks. By leveraging the graph-based representation of drug-target interactions, pharmacokinetics (absorption, distribution, metabolism, and excretion), and pharmacodynamics (mechanisms of action and dose-response relationships), GCNs provide a robust framework for predicting drug efficacy and toxicity[2,3].
This study introduces the Pharmacological Knowledge Graph Convolutional Network (PKGCN), a novel model that incorporates rich pharmacological prior knowledge into the GCN architecture to enhance drug response prediction. The PKGCN model leverages known drug-target interactions, pharmacokinetic profiles, and pharmacodynamics effects to improve predictive accuracy and interpretability. This approach allows for a more informed understanding of drug actions across various biological contexts, paving the way for applications in personalized medicine and drug repurposing[4,5]. In addition to applying PKGCN to pharmacological datasets such as DrugBank and The Cancer Genome Atlas (TCGA), this study assesses the model’s performance across different therapeutic classes. By integrating these diverse datasets, PKGCN demonstrates its potential to generalize across various pharmacological scenarios, thereby offering significant improvements in the prediction of drug responses and enabling more personalized and targeted therapeutic strategies[6].
As the volume and complexity of biological data increase, traditional machine learning approaches struggle to capture the intricate relationships inherent in drug responses. Machine learning algorithms such as Support Vector Machines (SVM) and Random Forests (RF) have demonstrated some utility in this field but are often limited by their inability to effectively model non-linear, graph-based relationships present in biological networks[7,8]. Deep learning and specifically GCNs, offers a solution by enabling the integration of both graph-structured and feature-based data, leading to improved performance in drug response prediction tasks[9].
Previous studies have highlighted the efficacy of GCNs in various biomedical applications, including disease classification and biomarker discovery. However, these models often overlook critical pharmacological information such as drug-target interactions, metabolic pathways, and dose-response relationships, which are essential for accurate drug response predictions[10]. The incorporation of pharmacological knowledge into GCNs could significantly enhance the model’s ability to predict drug efficacy and toxicity, making it a valuable tool in the development of personalized treatments[11,12].
In this study, we propose the development of the PKGCN to address the limitations of existing models. The PKGCN model integrates prior pharmacological knowledge at multiple levels, utilizing drug-target interactions, pharmacokinetic data, and pharmacodynamics mechanisms to improve prediction accuracy. By incorporating this knowledge into the graph convolutional layers and pooling mechanisms, the PKGCN model can better capture the complex interplay between drugs and biological systems, leading to more accurate and interpretable predictions[13-19].
Materials and Methods
The functional Magnetic Resonance Imaging (fMRI) datasets:
fMRI is utilized as a cerebral imaging method for analysing brain functions. Within fMRI datasets, brain volume is segmented into tiny cubic units referred to as voxels. A time series is extracted from each voxel by monitoring its activity over time, constituting the Blood Oxygen-Level Dependent (BOLD) signal. The application of fMRI technology during the performance of a specific task is termed task-fMRI, while imaging during resting periods is referred to as resting-state fMRI (rs-fMRI). Both task-fMRI and rs-fMRI are widely used to analyse brain disorders.
The fMRI data utilized in this paper was sourced from the publicly available Autism Brain Imaging Data Exchange (ABIDE) dataset. The ABIDE initiative comprises of two extensive collections; ABIDE I and ABIDE II. Our study focused on modelling and used the pre-processed ABIDE I dataset[20]. The ABIDE I dataset consists of 1112 samples, including 539 individuals with Autism Spectrum Disorder (ASD) and 573 typical controls from 16 international imaging sites. The pre-processing of the dataset was conducted using the Configurable Pipeline for the Analysis of Connectomes (CPAC), which includes various pre-processing steps such as motion correction, anatomical/functional coregistration, spatial normalization, spatial and temporal filtering, tissue segmentation, slice-timing correction, several variations of nuisance signal removal, and volume censoring (motion "scrubbing"). After pre-processing, the effective sample size was reduced to 871. The dataset includes structural and resting-state functional MRI data, along with a comprehensive array of phenotypic information, as illustrated in Table 1.
| Data sources | Samples (ASD/control) | Age | Gender (male/female) | FIQ | VIQ | PIQ |
|---|---|---|---|---|---|---|
| CALTECH | 19/19 | 17.0-56.2 | 30/8 | 93.0-134.0 | 80.0-135.0 | 84.0-129.0 |
| CMU | 14/13 | 19.0-40.0 | 21/6 | 95.0-134.0 | 89.0-132.0 | 92.0-129.0 |
| KKI | 22/33 | 8.1-12.8 | 42/13 | 69.0-131.0 | / | / |
| LEUVEN | 29/35 | 12.1-32.0 | 56/8 | 89.0-146.0 | 50.0-136.0 | 74.0-155.0 |
| MAX_MUN | 24/33 | 7.0-58.0 | 50/7 | 79.0-133.0 | / | 83.0-126.0 |
| NYU | 79/105 | 6.5-39.1 | 147/37 | 76.0-148.0 | 73.0-143.0 | 67.0-149.0 |
| OHSU | 13/15 | 8.0-15.2 | 28/0 | 69.6-132.0 | / | / |
| OLIN | 20/16 | 10.0-24.0 | 31/5 | 71.0-135.0 | / | / |
| PITT | 30/27 | 9.3-35.2 | 49/8 | 81.0-131.0 | 81.0-132.0 | 83.0-128.0 |
| SBL | 15/15 | 20.0-64.0 | 30/0 | 95.0-125.0 | 93.0-133.0 | 84.0-135.0 |
| SDSU | 14/22 | 8.7-17.2 | 29/7 | 81.0-141.0 | 83.0-147.0 | 81.0-140.0 |
| STANFORD | 20/20 | 7.5-12.9 | 32/8 | 78.0-148.0 | 67.0-149.0 | 81.0-157.0 |
| TRINITY | 24/25 | 12.0-25.9 | 49/0 | 72.0-135.0 | 81.0-137.0 | 63.0-132.0 |
| UCLA | 62/47 | 8.4-17.9 | 96/13 | 64.0-132.0 | 59.0-132.0 | 72.0-132.0 |
| UM | 68/77 | 8.2-28.8 | 117/28 | 76.0-147.5 | 75.0-180.0 | 59.0-148.0 |
| USM | 58/43 | 8.8-50.2 | 101/0 | 65.0-148.0 | 55.0-140.0 | 72.0-155.0 |
| YALE | 28/28 | 7.0-17.8 | 40/16 | 41.0-141.0 | 42.0-143.0 | 37.0-139.0 |
Table 1: Demographic information of ABIDE I dataset.
Prior knowledge:
Prior knowledge related to fMRI and mental disorders encompasses various aspects, including changes in brain tissues, alterations in the activation of specific brain regions, disruptions in connectivity between brain regions, and other symptoms attributed to the disorder. Our emphasis is on the disorder of Functional Connectivity (FC) and the involvement of Reactive Oxygen Intermediates (ROIs), as increasing evidence in indicates that ASD is characterized by connectivity abnormalities[21]. Taking into account the information presented above, prior knowledge related to ASD can be broadly categorized into ROI activation alterations in connectivity patterns between ROIs. Through a comprehensive review of existing research findings on ASD, we aim to synthesize and consolidate prior knowledge pertaining to this disorder (Table 2).
| SVM | RF | CNN | MLP | GCN | GAT | GraphSAGE | PKGCN | |
|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | 54.86 | 58.29 | 65.41 | 63.38 | 67.92 | 76.10 | 78.50 | 90.36 |
| F1 (%) | 53.89 | 56.29 | 68.36 | 76.54 | 74.92 | 76.02 | 75.56 | 85.85 |
| Precision (%) | 58.44 | 57.32 | 75.00 | 86.11 | 69.06 | 75.34 | 74.84 | 89.11 |
| Recall (%) | 50.00 | 55.56 | 61.93 | 68.89 | 81.90 | 78.73 | 78.31 | 88.18 |
Table 2: Comparison of the classification performance.
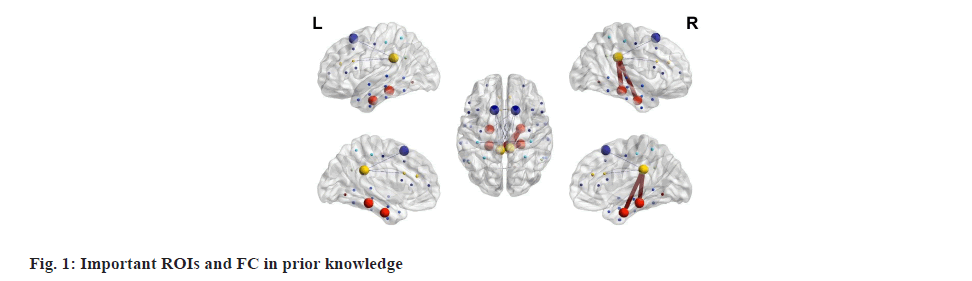
Before delving into the details, it is crucial to acknowledge that the information presented below pertains to the most common diagnostic features or biomarkers of ASD as delineated in numerous papers. However, these findings may not be universally applicable to individuals with more specific or rare symptoms. In fig. 1, different colours of nodes represent distinct functional brain regions, with the size of the nodes indicating the degree of association with ASD. Thin lines denote under-connectivity between brain regions, while thick lines signify over-connectivity. The figure illustrates that individuals with ASD often exhibit heightened activity in the default network, encompassing the medial Prefrontal Cortex (mPFC), Posterior Cingulate Cortex (PCC)/Precuneus (PCu), inferior parietal lobe, lateral temporal cortex, and hippocampus, even during periods of rest[22].
Many of these areas also exhibit abnormal connectivity with one another. Compared to the control group, the ASD group tends to have weaker FC between regions such as the PCC and superior frontal gyrus, the frontal lobe and the parietal lobe, the anterior cingulate gyrus region and the posterior cingulate gyrus, and the left para hippocampal gyrus and middle prefrontal cortex. Discovered that the ASD group exhibits stronger connectivity between regions such as the PCC and the right temporal lobe, the PCC and right para hippocampal gyrus, the right parietal lobe and the prefrontal region, posterior occipital and temporal cortices[23].
GCNs:
The graph neural network was introduced as a solution for analysing non-Euclidean data samples. Traditional machine learning and artificial neural networks such as Convolutional Neural Network (CNN)[24] or Recurrent Neural Network (RNN) are effective in Euclidean spaces like language and images but not suitable for graph data. Graph neural networks have emerged as a promising solution for processing non-Euclidean data, including knowledge graphs, social networks, biological networks, chemical structures, etc. These networks are quickly gaining widespread usage and effectiveness.
Methods:
Previous studies analysing fMRI data for brain disorder classification using GCN treated all nodes as identical, assigning them the same embedding algorithm. This approach proved to be problematic with the human brain, where nodes can vary significantly. Even with Graph Attention Networks (GATs) highlighting node differences, the model’s theoretical interpretability needed verification. In contrast, our proposed GCN framework includes a prior knowledge attention graph convolutional layer and a Prior Knowledge Aware pooling layer (PKA pooling), emphasizing node differences and prior knowledge to enhance the model’s results. This approach differs from previous work and shows promising potential for improving brain disorder classification.
Brain representation in graph:
In graph theory, a graph is defined as a collection of vertices and edges. To apply this concept to brain connectivity, we identify regions of interest (ROIs) of the brain as vertices in the graph. The quantity of ROIs is n and varies depending on the brain atlas and parcellation method used, which can be considered as vertices. For example, the Harvard-Oxford atlas from FSL contains 97 ROIs[25], while the Multi-Subject Dictionary Learning (MSDL) brain Probabilistic atlas provided by has 39 ROIs[26]. The edges in the graph represent connections between ROIs and are assigned an edge weight, if there is an edge from vertex Vi to vertex Vj. The graph is characterized by two matrices; the adjacency matrix Eadj, which displays the network structure, and the edge weight matrix Ew, which shows connectivity strength. FC between ROIs can be calculated with different methods, such as covariance, correlation, partial correlation, tangent or precision[27-30].
In GCNs, each node has its own node features. For brain networks, the BOLD time series signal data ST can be the node features. In brain applications, the node feature could be the maximum, minimum, mean, variance, standard deviation, mode, median and any other evaluation indicators that can reveal the characteristics of BOLD signal of Vi. Assuming that every node Vi has m features, except the statistical indicators of distribution, then the node features of Vi. To unify node feature space, assume that the whole time period of Vi BOLD signal is t, we set an adaptive time window to achieve this.
Prior knowledge modelling:
Given the considerable number of studies about ASD biomarkers, two types of prior knowledge, abnormal brain areas and brain connectivity, can be abstracted as nodes and edges in a graph. For convenience, abnormal brain areas can be named as hv, which is a set of k elements. Similarly, abnormal brain connectivity can be expressed as a set with size m, denoted as he.
PKGCN model:
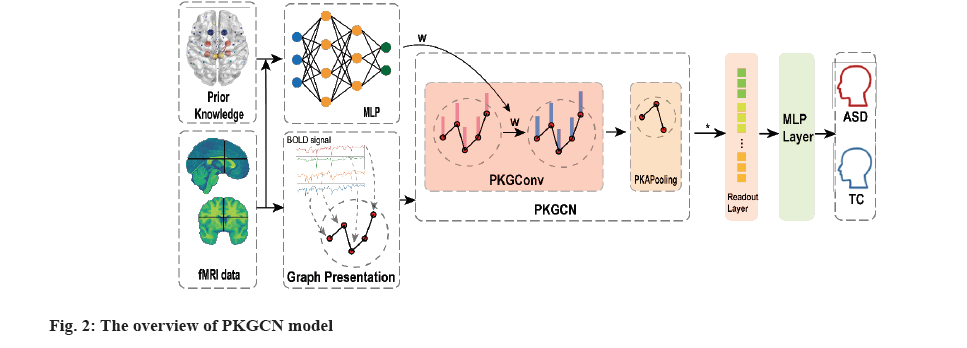
In contrast to the existing GCN framework mentioned above, the PKGCN implements different weights for nodes and incorporates edge weights by employing a Multi-Layer Perceptron (MLP). These weights are then utilized in graph convolutional layers during training. In the pooling layers, TopK pooling is applied to consider both node features and graph topology. Finally, a readout layer is used to integrate the embedding node features into a form that can be processed by a graph classifier. The complete architecture of the PKGCN model is illustrated in fig. 2-fig. 4.
The inputs include the prior knowledge matrix, and the graph data, such as graph structure, node features and edge weights. The MLP computes the node weights as shown in box labelled ‘MLP’, and these weights will train in PKGConv as shown in pink box. In yellow box, it is the modified TopK Pooling layer and the readout layer is in orange box. Notably, the (*) symbol on the arrow signifies the repetition of graph convolutional and pooling layers.
The classification on graphs depends on the three layers in PKGCN; graph convolutional layers, node pooling layers and a readout layer. The graph convolutional layer, PKGConv, probes the graph structure using edge features, the pooling layer reduces the nodes, and the readout layer summarizes the features. The node features can be embedded into a low-dimensional space by the weight factor (w) computed by the input and prior knowledge with MLP during this process. Finally, the summarized vector is fed into an MLP to achieve the final classification result.
PKA pooling:
Due to the multi dimensions of node features, pooling layers will implement features dimensionality reduction to reduce training complexity. Moreover, pooling layers optimize graph representation through selection, reduction and connection steps for downstream tasks such as graph classification.
As previously discussed, certain brain regions are more indicative of brain disorder diagnosis. Thus, these ROIs should be retained in the subsequent steps even after the pooling layers. In other words, these ROIs will act as super nodes and will occupy a larger proportion in subsequent training. Following the design of TopK Pooling, the preserved nodes after the PKA Pooling layer should meet the conditions that the scores of these nodes should be ranked high. In essence, the learnable vector w of lth layer can project the node features matrix to a normalized One-Dimensional (1D) representation for each node, resulting in the score vectors. After this, we obtain a ranking node list, and the top nodes from this list can be sampled to implement graph node reduction.
Readout layer:
At last, a readout layer is needed to turn the preserved information into an appropriate presentation that can be processed by MLP layers for the final prediction result.
Loss function:
The learnable vector p, which is associated with the prior knowledge and incorporated into the graph representation at the PKGconv layer, necessitates a redesign of the loss function. This modification aims to regulate the training process and mitigate the introduction of ambiguous values. The detailed elements of the loss function definition are expounded below.
Cross entropy loss:
The cross-entropy loss serves as the primary loss function for training the model. However, given the additional factors introduced into the model, an adaptation of the loss function is essential for optimal results. The designed loss function consists of several components, each contributing to the overall objective with varying weights.
Unit loss:
As mentioned earlier, the factor influenced by prior knowledge, w(l) is calculated by MLP, and the pooling score was computed . This calculation involves multiple parameters, which may lead to an ambiguous situation where inputs from different samples have the same value of w(l).To avoid this situation, a unit loss is employed based on the idea of a unit vector.
Prior Knowledge Prefer Loss (PKP-loss):
The PKP-loss is introduced to impose constraints on different samples, serving two objectives. Firstly, it encourages the selection of similar ROIs, especially those outlined in prior knowledge, across various samples. Secondly, it regulates the loss of the ROIs specified in the prior knowledge. If, after the first pooling layer, the selected ROIs in different samples have no duplicate elements, the model’s design becomes futile. Consequently, in the PKGCN model, the PKP-loss is designed to compel the model to choose similar ROIs, particularly those in the prior knowledge, after the first Top-K pooling layer.
Results and Discussion
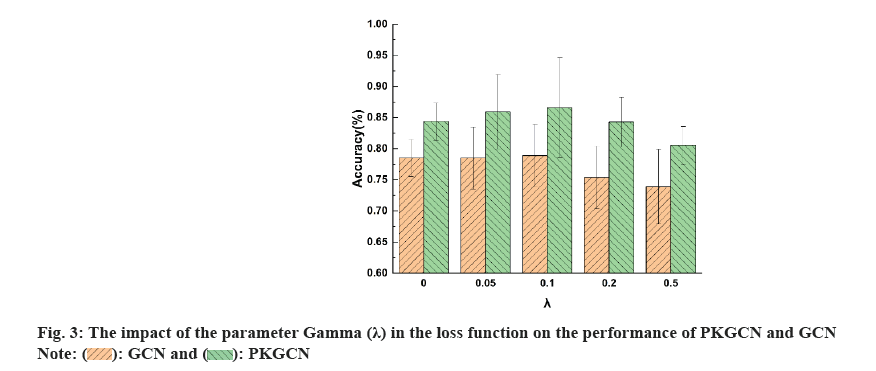
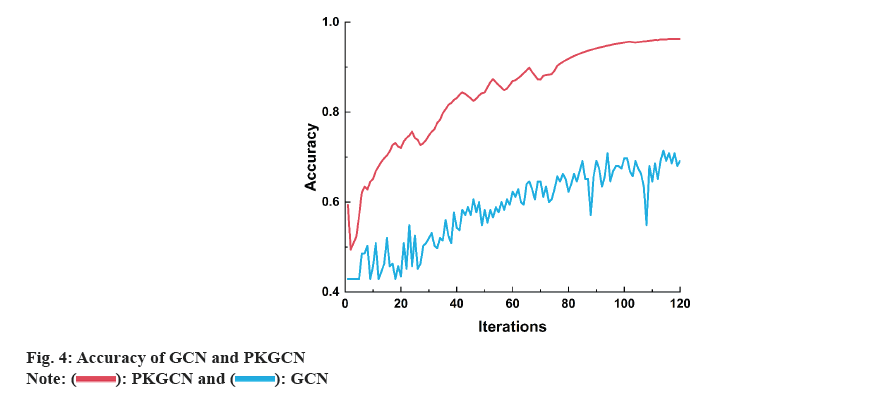
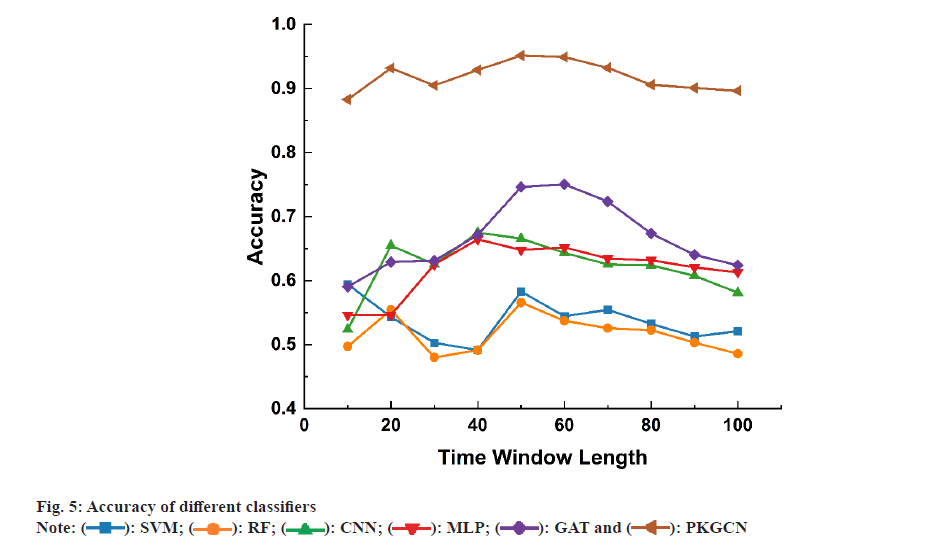
To systematically evaluate the performance of the PKGCN model in predicting drug responses, we conducted a series of experiments. Our experiments were designed to compare the predictive accuracy of PKGCN with established baseline models, including traditional machine learning algorithms and other graph-based neural network approaches. We selected these baseline models for their prevalence in the field and their ability to serve as a benchmark for the innovative aspects of PKGCN. The dataset was split into training, validation, and test sets to ensure the robustness of our findings. We meticulously trained the PKGCN model, tuning its parameters to optimize performance while avoiding overfitting. Throughout the training process, we monitored the model’s convergence and predictive accuracy on the validation set (fig. 5).
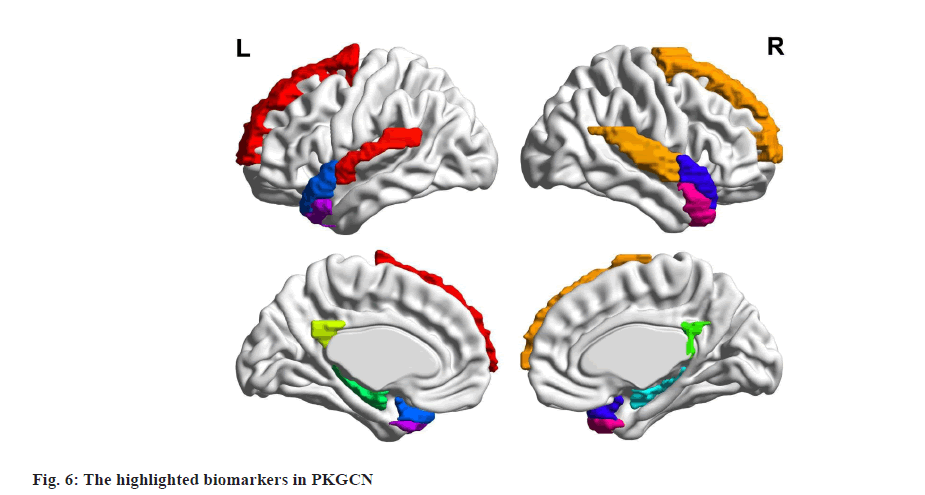
Post-training, we analysed the predictions made by the PKGCN model and correlated them with existing pharmacological data. This analysis was pivotal in understanding the model’s predictive capabilities and its alignment with known biological mechanisms of drug action. Our results revealed that the PKGCN model demonstrated a significant enhancement in predictive accuracy compared to the baseline models. The incorporation of pharmacological prior knowledge into the model’s architecture allowed for a more nuanced understanding of drug responses, leading to improved predictions. Furthermore, the analysis highlighted the discovery of biomarkers and pathways that were significantly associated with specific drug responses. These findings were in concordance with established pharmacological knowledge, thereby validating the biological relevance of the model’s predictions. The identification of these biomarkers and pathways also provided novel insights into the mechanisms of drug action, offering potential avenues for drug repurposing and the development of targeted therapies (fig. 6).
An important aspect of PKGCN is the integration of drug mechanisms, particularly through the incorporation of drug-target interactions and pharmacokinetics/pharmacodynamics data. By embedding these interactions into the graph convolutional layers, the model captures the biological pathways that drugs engage with, optimizing predictive performance. For example, in the case of anti-cancer drugs with complex mechanisms, the PKGCN model considers how the drugs interact with specific biological pathways, influencing key targets like enzymes or receptors. This enhances the model's ability to align its predictions with actual experimental data, offering improved accuracy in forecasting drug responses in a clinical setting.
We applied PKGCN to several anti-cancer drugs, which involve intricate mechanisms such as multi-target interactions or modulation of immune responses. Through case studies, we observed that PKGCN was able to predict responses more accurately than traditional models. For instance, when predicting the response to kinase inhibitors, the model successfully aligned predicted outcomes with known experimental data. This demonstrates the model’s effectiveness in dealing with complex pharmacological profiles, reinforcing its practical applications in oncology.
PKGCN’s performance was benchmarked against other well-established pharmacological models, such as those available in DeepChem. By integrating pharmacological prior knowledge, PKGCN consistently outperformed traditional GCN models and other machine learning approaches. Specifically, we compared PKGCN with existing models on DrugBank and TCGA datasets, where PKGCN demonstrated superior accuracy and robustness in predicting drug responses. This further highlights PKGCN’s advantage in modelling drug mechanisms and applying them to real-world pharmacological data.
The potential of PKGCN extends to personalized medicine, where individual patient profiles, including genetic data and drug interactions, can be leveraged to tailor therapies. PKGCN facilitates this by predicting how a specific drug will behave in a patient based on their unique biological makeup. Additionally, the model shows promise in drug repurposing, where it can identify new therapeutic uses for existing drugs by analysing their effects across different biological networks. This capability can significantly accelerate the process of drug discovery and optimization of treatment strategies.
Bringing PKGCN into clinical practice introduces certain challenges, including the need for comprehensive and high-quality pharmacological datasets. The model’s success is dependent on the accuracy and completeness of input data, especially regarding drug-target interactions. In clinical settings, the integration of patient-specific data, such as genetic profiles, will be crucial for optimizing the model’s predictions. Moreover, there is a need for user-friendly interfaces to facilitate the adoption of PKGCN by clinicians. Developing tools that can seamlessly integrate with existing clinical systems will be key to translating PKGCN into everyday medical practice, ultimately enhancing decision-making in personalized therapy and accelerating the drug development process.
While PKGCN demonstrates significant promise, it has some limitations. The model’s reliance on the availability and completeness of pharmacological data can impact its accuracy, especially in cases where drug-target interactions or metabolic pathways are under-characterized. Additionally, the complexity of the PKGCN model presents challenges in terms of computational efficiency, particularly when scaling to large datasets. To address these limitations, future research could focus on optimizing the model’s architecture for greater scalability while maintaining high predictive accuracy.
Moreover, expanding the model’s application beyond pharmacology to include other therapeutic areas and integrating more diverse data types, such as patient-specific genetic and environmental factors, would further enhance its utility. As PKGCN evolves, future work should also explore novel optimization techniques and regularization strategies to prevent overfitting, particularly with complex, large-scale data. Another promising direction is the development of user-friendly interfaces that allow clinicians to interact with the model more intuitively, facilitating its integration into clinical workflows.
Our research presents the PKGCN model, an innovative framework that integrates rich pharmacological prior knowledge to predict drug responses with enhanced accuracy and interpretability. This model represents a significant advancement in the field of pharmacology by providing a nuanced understanding of drug actions and interactions. The key findings of our study underscore the importance of incorporating drug-target interactions, pharmacokinetics, and pharmacodynamics into predictive models. The PKGCN model's ability to leverage this knowledge results in a substantial improvement in predictive performance, offering a more precise tool for drug response prediction.
The novel aspects of the PKGCN model, including the use of a multilayer graph convolutional network with attention coefficients derived from prior knowledge, a prior knowledge-aware pooling layer, and an adjusted loss function, have collectively contributed to its success. These features have been instrumental in refining the model's output and ensuring that it aligns with established pharmacological principles.
Looking ahead, the potential applications of the PKGCN model are vast and promising, particularly in the realms of drug discovery and personalized medicine. The model’s capacity to predict individualized drug responses can facilitate the development of tailored treatment plans, thereby personalizing healthcare to an unprecedented degree. Furthermore, its ability to identify significant biomarkers and pathways may accelerate the identification of new drug targets and the repurposing of existing drugs for novel therapeutic indications. As we move forward, the PKGCN model’s application to more complex datasets and diverse therapeutic areas will be crucial. We anticipate extending its use beyond the current scope to encompass a broader range of pharmacological data. This includes integrating patient-specific genetic information and environmental factors, which could further enhance the model’s predictive capabilities and applicability in real-world clinical settings. In conclusion, the PKGCN model represents a transformative tool in pharmacology, with the potential to reshape drug response prediction and contribute to the advancement of personalized medicine. Its successful integration of prior knowledge and GCNs marks a significant step towards more accurate, efficient, and patient-centred therapeutic strategies.
Acknowledgements:
This work was supported by the National Key R&D Program of China (No: 2023YFB3812901).
Conflict of interests:
The authors declared no conflict of interests.
References
- Vahia VN. Diagnostic and statistical manual of mental disorders 5: A quick glance. Indian J Psychiatry 2013;55(3):220-3.
[Crossref] [Google Scholar] [PubMed]
- Yingling ME. Participation in part C early intervention: One key to an earlier diagnosis of autism spectrum disorder? J Pediatr 2019;215:238-43.
[Crossref] [Google Scholar] [PubMed]
- Demirci O, Clark VP, Magnotta VA, Andreasen NC, Lauriello J, Kiehl KA, et al. A review of challenges in the use of fMRI for disease classification/characterization and a projection pursuit application from a multi-site fMRI schizophrenia study. Brain Imaging Behav 2008;2(3):207-26.
[Crossref] [Google Scholar] [PubMed]
- Orban P, Dansereau C, Desbois L, Mongeau-Perusse V, Giguere CE, Nguyen H, et al. Multisite generalizability of schizophrenia diagnosis classification based on functional brain connectivity. Schizophrenia Res 2018;192:167-71.
[Crossref] [Google Scholar] [PubMed]
- Pflanz CP, Egle MS, O'Brien JT, Morris RG, Barrick TR, Blamire AM, et al. Association of blood pressure lowering intensity with white matter network integrity in patients with cerebral small vessel disease. Neurology 2022;99(17):e1945-53.
[Crossref] [Google Scholar] [PubMed]
- Chyzhyk D, Savio A, Graña M. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Networks 2015;68:23-33.
[Crossref] [Google Scholar] [PubMed]
- Zheng J, Wei X, Wang J, Lin H, Pan H, Shi Y. Diagnosis of schizophrenia based on deep learning using fMRI. Comput Math Methods Med 2021;2021(1):8437260.
[Crossref] [Google Scholar] [PubMed]
- Zhao J, Huang J, Zhi D, Yan W, Ma X, Yang X, et al. Functional Network Connectivity (FNC)-based Generative Adversarial Network (GAN) and its applications in classification of mental disorders. J Neurosci Methods 2020;341:108756.
[Crossref] [Google Scholar] [PubMed]
- Yan W, Calhoun V, Song M, Cui Y, Yan H, Liu S, et al. Discriminating schizophrenia using recurrent neural network applied on time courses of multi-site fMRI data. EBioMedicine 2019;47:543-52.
[Crossref] [Google Scholar] [PubMed]
- Amsalem D, Jankowski SE, Pagdon S, Smith S, Yang LH, Valeri L, et al. “It’s tough to be a black man with schizophrenia”: Randomized controlled trial of a brief video intervention to reduce public stigma. Schizophrenia Bull 2024;50(3):695-704.
[Crossref] [Google Scholar] [PubMed]
- Lin QH, Niu YW, Sui J, Zhao WD, Zhuo C, Calhoun VD. SSPNet: An interpretable 3D-CNN for classification of schizophrenia using phase maps of resting-state complex-valued fMRI data. Med Image Anal 2022;79:102430.
[Crossref] [Google Scholar] [PubMed]
- Zhuang H, Liu R, Wu C, Meng Z, Wang D, Liu D, et al. Multimodal classification of drug-naïve first-episode schizophrenia combining anatomical, diffusion and resting state functional resonance imaging. Neurosci Lett 2019;705:87-93.
[Crossref] [Google Scholar] [PubMed]
- Fang S, Liu Z, Qiu Q, Tang Z, Yang Y, Kuang Z, et al. Diagnosing and grading gastric atrophy and intestinal metaplasia using semi-supervised deep learning on pathological images: Development and validation study. Gastr Cancer 2024;27(2):343-54.
[Crossref] [Google Scholar] [PubMed]
- Onicas AI, Deighton S, Yeates KO, Bray S, Graff K, Abdeen N, et al. Longitudinal functional connectome in pediatric concussion: An advancing concussion assessment in pediatrics study. J Neurotrauma 2024;41(5-6):587-603.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Gong Z, Wang W, Wang C, Xu Z, Lv J, et al. Adversarial caching training: Unsupervised inductive network representation learning on large-scale graphs. IEEE Trans Neural Networks Learn Syst 2022;33(12):7079-90.
- Jorge-Hernandez F, Garcia Chimeno Y, Garcia-Zapirain B, Cabrera Zubizarreta A, Gomez Beldarrain MA, Fernandez-Ruanova B. Graph theory for feature extraction and classification: A migraine pathology case study. Biomed Mater Eng 2014;24(6):2979-86.
[Crossref] [Google Scholar] [PubMed]
- Lei D, Qin K, Pinaya WH, Young J, van Amelsvoort T, Marcelis M, et al. Graph convolutional networks reveal network-level functional dysconnectivity in schizophrenia. Schizophrenia Bull 2022;48(4):881-92.
[Crossref] [Google Scholar] [PubMed]
- Yang C, Wang P, Tan J, Liu Q, Li X. Autism spectrum disorder diagnosis using graph attention network based on spatial-constrained sparse functional brain networks. Comput Biol Med 2021;139:104963.
[Crossref] [Google Scholar] [PubMed]
- Li X, Zhou Y, Dvornek N, Zhang M, Gao S, Zhuang J, et al. Braingnn: Interpretable brain graph neural network for fMRI analysis. Med Image Anal 2021;74:102233.
[Crossref] [Google Scholar] [PubMed]
- Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED, et al. Multisite functional connectivity MRI classification of autism: ABIDE results. Front Hum Neurosci 2013;7:599.
[Crossref] [Google Scholar] [PubMed]
- Maximo JO, Cadena EJ, Kana RK. The implications of brain connectivity in the neuropsychology of autism. Neuropsychol Rev 2014;24:16-31.
[Crossref] [Google Scholar] [PubMed]
- Monk CS, Peltier SJ, Wiggins JL, Weng SJ, Carrasco M, Risi S, et al. Abnormalities of intrinsic functional connectivity in autism spectrum disorders. Neuroimage 2009;47(2):764-72.
[Crossref] [Google Scholar] [PubMed]
- Spreng RN, Stevens WD, Chamberlain JP, Gilmore AW, Schacter DL. Default network activity, coupled with the frontoparietal control network, supports goal-directed cognition. Neuroimage 2010;53(1):303-17.
[Crossref] [Google Scholar] [PubMed]
- Kourounis G, Elmahmudi AA, Thomson B, Hunter J, Ugail H, Wilson C. Computer image analysis with artificial intelligence: A practical introduction to convolutional neural networks for medical professionals. Postgraduate Med J 2023;99(1178):1287-94.
- Craddock RC, James GA, Holtzheimer III PE, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp 2012;33(8):1914-28.
[Crossref] [Google Scholar] [PubMed]
- Varoquaux G, Gramfort A, Pedregosa F, Michel V, Thirion B. Multi-subject dictionary learning to segment an atlas of brain spontaneous activity. Info Process Med Imag 2011;22:562-73.
[Crossref] [Google Scholar] [PubMed]
- Varoquaux G, Baronnet F, Kleinschmidt A, Fillard P, Thirion B. Detection of brain functional-connectivity difference in post-stroke patients using group-level covariance modeling. Med Image Comput Assist Interv 2010;13(1):200-8.
[Crossref] [Google Scholar] [PubMed]
- Xia M, Wang J, He Y. BrainNet viewer: A network visualization tool for human brain connectomics. PloS One 2013;8(7):e68910.
[Crossref] [Google Scholar] [PubMed]
- Steardo Jr L, Carbone EA, de Filippis R, Pisanu C, Segura-Garcia C, Squassina A, et al. Application of support vector machine on fMRI data as biomarkers in schizophrenia diagnosis: A systematic review. Front Psychiatry 2020;11:588.
[Crossref] [Google Scholar] [PubMed]
- Dvornek NC, Ventola P, Pelphrey KA, Duncan JS. Identifying autism from resting-state fMRI using long short-term memory networks. Mach Learn Med Imag 2017:10541:362-370.
[Crossref] [Google Scholar] [PubMed]
 .
.
 .
.
 .
.