- *Corresponding Author:
- R. K. Kancha
Molecular Medicine and Therapeutics Laboratory,
Centre for Plant Molecular Biology,
Osmania University,
Hyderabad 500007,
India
E-mail: ickrishna@gmail.com
| Date of Received | 12 November 2020 |
| Date of Revision | 14 June 2021 |
| Date of Acceptance | 01 September 2021 |
| Indian J Pharm Sci 2021;83(5):871-885 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The coronavirus disease 2019 is a disease caused by severe acute respiratory syndrome coronavirus 2 and continues to pose a global threat resulting in thousands of deaths. In the absence of effective therapeutics, implementation of non-pharmaceutical interventions still remains an important prevention strategy, while various vaccine candidates are either approved or in clinical trials, several drugs are also being tested in pre-clinical studies for their potential for repurposing to treat the coronavirus disease 2019. The mechanistic details underlying disease pathogenesis and therapeutic efficacy of various drugs are rapidly emerging, which will be useful for fast-tracking pharmaceutical leads to clinical utility. The current review details the mechanistic understanding of viral pathogenesis including host cell interactions, viral replication, altered host cell processes, cytokine release syndrome and the role of coagulation pathway, renin angiotensin system and kallikrein-bradykinin pathways in acute respiratory distress syndrome. Further, a detailed overview of therapeutic candidates including antibodies, targeted enzyme inhibitors, drugs that showed clinical efficacy and drugs with anti-severe acute respiratory syndrome coronavirus 2 activities in pre-clinical models are discussed. Additionally, the approval and availability of various vaccines and their mechanism of action are also detailed. Importantly, emerging clinical evidence regarding immune biomarkers that help in patient stratification into moderate or severe cases are highlighted that may aid in reducing mortality by choosing appropriate treatment strategies. Taken together, the report gives up to date mechanistic information on disease pathogenesis and potential therapeutics.
Keywords
Coronavirus disease 2019, severe acute respiratory syndrome coronavirus 2, cytokine storm, acute respiratory distress syndrome, vaccine
The pandemic coronavirus disease 2019 (COVID-19) has caused immense loss of lives and global burden on economic and social fronts. The only currently available option to limit the COVID-19 pandemic is classic epidemic control that involves case isolation, contact-tracing, quarantine, physical distancing and hygiene measures. A report that studied measures taken in Wuhan city and Hubei province concluded that the National emergency response (NER), which included public health intervention (i.e. non-pharmaceutical) resulted in slowing down of the epidemicn [1]. In addition, an estimated reduction of R0 from 3.54 to 0.28 in Wuhan was attributed to strict adherence of multiple non-pharmaceutical interventionsn [2]. Furthermore, COVID-19 displays interesting transmission dynamics wherein index cases transmit the disease at higher rates before or immediately after the initial onset of symptomsn [3]. Among health care workers, nearly 3 % were tested positive to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) despite being asymptomatic suggesting a need to screen widely in populations to contain spread of the disease. Taken together, non-pharmaceutical interventions were very successful in reducing the spread of COVID-19 pandemic and continuation of these measures was recommended to achieve an Rt value below 1 in the absence of an available treatment option.
Clinical Symptoms and the Evolution of Virus
Based on the data from 72 314 cases as on 11 February 2020, 3 % of patients were older than 80 y, 87 % were aged between 30 and 79 y, 1 % were aged 10 to 19 y and 1 % were aged 9 y or youngern [4]. Nearly 81 % of reported cases were mild, 14 % were severe and 5 % were criticaln [4]. The overall case-fatality rate (CFR) was 2.3 % but increased significantly to 8 % in the age group of 70 to 79 y and 14.8 % in those aged 80 y or aboven [4]. The estimated infection-to-fatality ratio (IFR) increases with age, with an IFR ratio of 0.001 % in 5-9 y olds and 8.29 % in individuals above 80 yn [5]. Most of the SARS-CoV-2 infected patients showed following symptoms (as on 15 June 2020): fever (70-90 %), cough (60-86 %), dyspnoea (53-80 %), myalgia (15-44 %), fatigue (38 %), nausea/vomiting or diarrhoea (15-39 %), headache, weakness (25 %) and rhinorrhoea (7 %)n [6]. Higher viral loads and long viral shedding periods are associated with severe clinical symptoms and thus may serve as prognostic markersn [7].
Individuals with comorbidities were reported to be at higher risk of SARS-CoV-2 infection (as per the data on 9 February 2020): acute respiratory distress syndrome (ARDS) (67 %), hyperglycaemia (35 %), acute kidney injury (29 %), liver dysfunction (29 %), cardiac injury (23 %), hospital acquired pneumonia (11.5 %), gastrointestinal haemorrhage (4 %), pneumothorax (2 %), bacteraemia (2 %) and urinary tract infection (2 %)n [8]. In addition, a high mortality rate due to COVID-19 was observed in cancer patients, especially high in thoracic cancer patientsn [9]. A spectrum of neurological symptoms were reported in less severe patients while disorders of consciousness occurred commonly (19.6 %) in older patients and in severe casesn [10]. Furthermore, geographic location specific (nearly 65 % in Europeans and 5 % in Chinese) dysfunction of olfactory and gustatory senses were reported in COVID-19 patientsn [11].
Close resemblance between SARS-CoV-2 and other animal coronaviruses indicates possible zoonotic transmission by acquiring key functional regions within the spike protein that enhances its binding to human angiotensin converting enzyme 2 (hACE2) receptorn [12]. Interestingly, SARS-CoV-2 has been suggested to have evolved from sarbecoviruses that may have been circulating among bats for several decades before their recent zoonotic transmission to humansn [13]. Phylogenetic analysis of SARS-CoV-2 genomes revealed three central variants A (ancestral type), B and C; while B is the most common type in east Asia, A and C types are present mostly in Europe and American [14]. Interestingly, most of the recurrent mutations identified in SARS-CoV-2 genome are missense mutations indicating a possible ongoing functionally relevant adaptationn [15]. For example, emergence and dominance of SARS-CoV-2 strain that contains D614G mutation in the spike protein having enhanced infectivity and higher viral loads in upper respiratory tract indicates the fitness advantage of this mutant over the wild type strainn [16]. Surprisingly, the D614G mutation didn’t alter the efficacy of therapeutic antibodiesn [16]. Notably, a 382-nucleotide deletion in the open reading frame 8 (ORF8) was shown to be associated with milder symptoms than the wild-type virus despite unaltered replicative fitness and viral loadn [17]. However, not all mutations are created equal: certain mutants were shown to be less infectious indicating the functional significance of those mutated residues, while some mutations were shown to alter sensitivity towards neutralizing antibodiesn [18].
Cellular and Molecular Pathogenesis
Virus-host cell interaction:
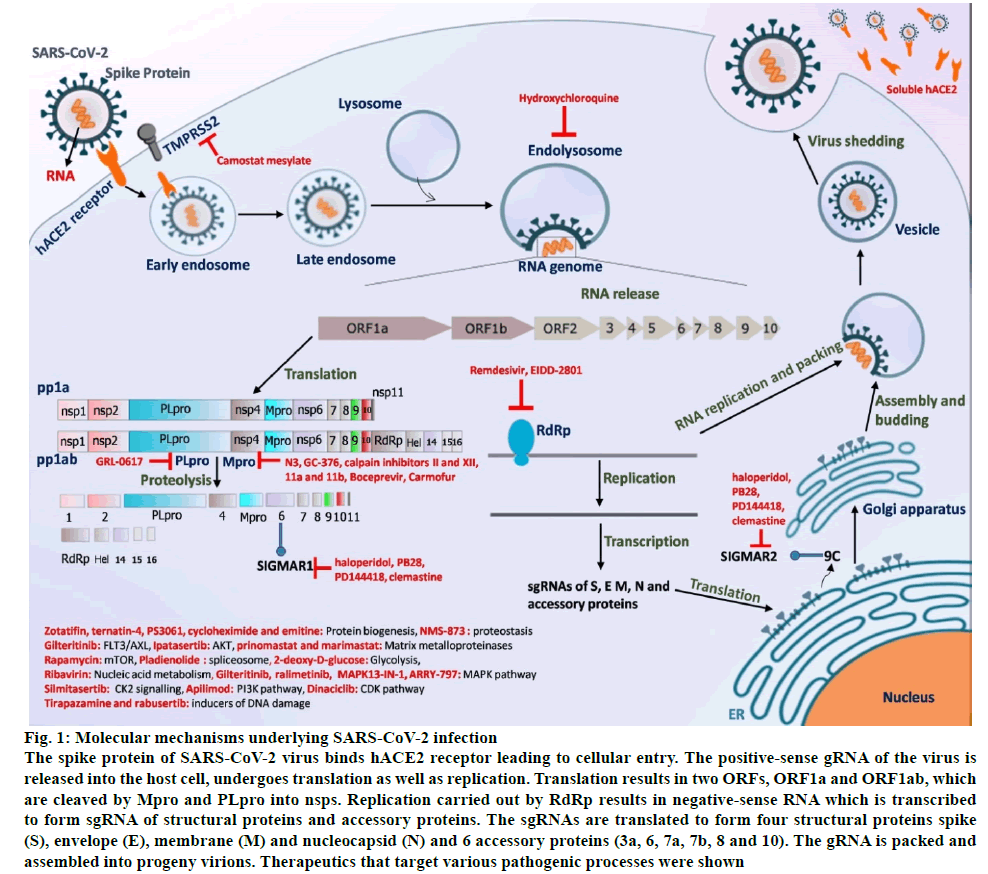
The spike (S) protein of SARS-CoV-2 forms trimern [19] and binds to the N-terminal peptidase domain of hACE2 receptorn [20] leading to the entry of virus into the cell (fig. 1)n [21]. Comprehensive analysis of tissue-specific angiotensin converting enzyme 2 (ACE2) expression revealed higher to moderate expression in enterocytes, renal tubules, gallbladder, cardiomyocytes, male reproductive cells, placental trophoblasts, ductal cells, eye and vasculaturen [22]. ACE2 gene expression is very low in the nasal epithelium of children and increases with age, which explains the lower susceptibility of younger children to SARS-CoV-2 infection than adultsn [23]. Furthermore, ACE2 receptor along with the transmembrane serine protease 2 (TMPRSS2) was shown to be expressed in nasal goblet and ciliated cellsn [24]. Expression of ACE2 receptor is higher in the epithelium of upper respiratory tract than that of lower respiratory tractn [25]. In lungs, ACE2 receptor is expressed predominantly in type II pneumocytesn [24] and transient secretory cells of the subsegmental bronchial branchesn [26]. Interestingly, ACE2 is an interferon (INF) regulated genen [24] and is upregulated by inflammatory stimuli as well as cigarette smoking consequently increasing the susceptibility to SARSCoV- 2 infectionn [27].
Figure 1: Molecular mechanisms underlying SARS-CoV-2 infection
The spike protein of SARS-CoV-2 virus binds hACE2 receptor leading to cellular entry. The positive-sense gRNA of the virus is released into the host cell, undergoes translation as well as replication. Translation results in two ORFs, ORF1a and ORF1ab, which are cleaved by Mpro and PLpro into nsps. Replication carried out by RdRp results in negative-sense RNA which is transcribed to form sgRNA of structural proteins and accessory proteins. The sgRNAs are translated to form four structural proteins spike (S), envelope (E), membrane (M) and nucleocapsid (N) and 6 accessory proteins (3a, 6, 7a, 7b, 8 and 10). The gRNA is packed and assembled into progeny virions. Therapeutics that target various pathogenic processes were shown
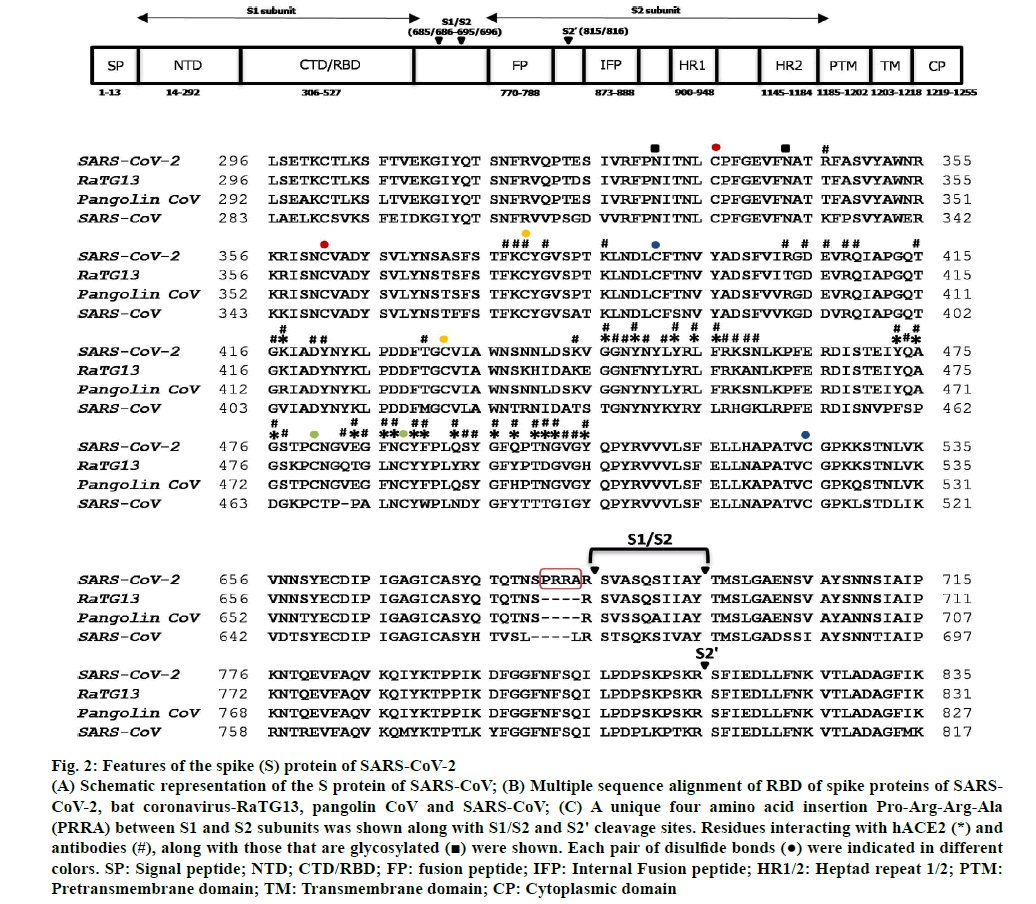
The spike (S) protein is cleaved by host cell proteases such as TMPRSS2 and cathepsin B/L into S1 and S2 subunitsn [21]. The S1 subunit is comprised of an N-terminal domain (NTD), C-terminal domain (CTD) and also receptor binding domain (RBD) both of which are involved in receptor binding while S2 is involved in membrane fusion (fig. 2)n [28]. The S1 subunit undergoes hinge-like movement resulting in its existence either in “up” conformation (receptor-accessible) or in “down” conformation (receptor-inaccessible)n [19]. A unique feature that distinguishes the spike protein of SARS-CoV-2 from that of other corona viruses is the presence of a four amino acid residue insertion (P681-RR-A684) between S1 and S2 subunits, which creates a recognition site for cleavage by pro-protein convertase furin (fig. 2)n [21]. However, the recognition sequence of an additional cleavage site (S2’) within S2 domain is conserved between SARS-CoV and SARSCoV- 2 (fig. 2)n [21]. An extended insertion within the RBD forms a receptor binding motif (RBM), which contains most of the residues that contact the hACE2 (fig. 2)n [20]. Despite structural similarities in receptor binding, SARS-CoV-2-CTD binds hACE2 with higher affinity when compared to that of SARS-CoV-RBD possibly due to larger binding receptor interface, few additional interactions and altered surface electrostatic potentialn [19,20,28]. Interestingly, nearly 90 % of receptor interacting residues of SARS-CoV-2-Spike-RBD are conserved in the spike protein of pangolin-CoV (fig. 2). Several antibodies were found to interact with SARSCoV- 2-Spike-RBD residues that are critical for hACE2 receptor binding (fig. 2). Notably, each one among the 21 hACE2 interacting residues was found to interact with at least one of the several antibodies (fig. 2). Binding of linoleic acid to SARS-CoV-2 spike protein reduces its interaction with ACE2 implicating its role in pathogenesisn [29].
Figure 2: Features of the spike (S) protein of SARS-CoV-2
(A) Schematic representation of the S protein of SARS-CoV; (B) Multiple sequence alignment of RBD of spike proteins of SARSCoV-2, bat coronavirus-RaTG13, pangolin CoV and SARS-CoV; (C) A unique four amino acid insertion Pro-Arg-Arg-Ala(PRRA) between S1 and S2 subunits was shown along with S1/S2 and S2' cleavage sites. Residues interacting with hACE2 (*) andantibodies (#), along with those that are glycosylated  were shown. Each pair of disulfide bonds
were shown. Each pair of disulfide bonds  were indicated in different colors. SP: Signal peptide; NTD; CTD/RBD; FP: fusion peptide; IFP: Internal Fusion peptide; HR1/2: Heptad repeat 1/2; PTM: Pretransmembrane domain; TM: Transmembrane domain; CP: Cytoplasmic domain
were indicated in different colors. SP: Signal peptide; NTD; CTD/RBD; FP: fusion peptide; IFP: Internal Fusion peptide; HR1/2: Heptad repeat 1/2; PTM: Pretransmembrane domain; TM: Transmembrane domain; CP: Cytoplasmic domain
In addition to ACE2 receptor, the SARS-CoV-2 spike protein interacts with neurophilin-1 (NRP1), which was shown to be blocked by monoclonal antibodies and NRP1 antagonistn [30]. The spike protein is glycosylated; a total of 22 N-linked glycans were observed that may serve to camouflage protein epitopes and consequently immune evasionn [31]. The importance of glycans on spike protein is exemplified by their effect on antigenicity towards cross-reactive antibodies: a single amino acid residue change resulting in the loss of a glycan modification contributed to the reduced binding affinity of the CR3022 antibody towards spike protein of SARSCoV- 2 when compared to that of the SARS-CoVn n [32]. In addition to the hACE2 binding domain, certain regions that are conserved between SARS-CoV and SARSCoV- 2 may serve as good epitopes to be neutralized by cross-reactive antibodies generated against the SARSCoVn n [ 32]. For example, the CR3022 antibody that was derived from convalescent SARS-CoV patient binds the spike protein on SARS-CoV-2 albeit with less affinity as compared to that of the SARS-CoVn n [32]. It may thus be important to produce monoclonal antibodies with high cross-reactivity that helps to combat multiple corona viral infections as well as potential future novel coronaviruses.
Replication of virus:
SARS-CoV-2 has a positive-sense, single-stranded ribonucleic acid (RNA) genome of approximately 30kb in size (fig. 1)n n [33]. An RNA dependent RNA polymerase (RdRp/non-structural proteins (nsp12)) performs replication and transcription using viral genome as a template (fig. 1)n n [33]. Negative-sense RNA intermediates are generated, which serve as templates for the synthesis of positive-sense genomic RNA (gRNA) and sub-genomic RNAs (sgRNAs) (fig. 1)n n [33]. The 2’-O-methylation on the first nucleotide of SARS-CoV-2 messenger RNAs (mRNAs) by nsp10/nsp16 heterodimer results in mimicking of host cell mRNA thus protecting the virus from host innate immune restrictionn n [34]. Inside the host cell, the gRNA is translated from two open reading frames (ORFs), ORF1a and ORF1b (fig. 1)n n [33]. The ORF1a produces polypeptide 1a (pp1a, 440-500 kDa) and ORF1b produces a comparatively larger polypeptide 1ab (pp1ab, 740-810 kDa) (fig. 1)n n [33]. 1a and 1ab are further cleaved into 11 and 15 nsps respectively by viral proteases nsp3 (papain-like protease (PLpro) domain) and nsp5 (3C-like protease (3CLpro) domain) (fig. 1)n n [33]. Interestingly, the C-terminus of viral protein nsp1 blocks host cell translation by binding to the 40S subunit of ribosome potentially leading to the blunted innate immune responsen [35]. Further, the NSP16 binds U1/U2 mRNA recognition domains while NSP8 and NSP9 binds 7SL RNA causing suppression of mRNA splicing and protein traffickingn [35]. Shorter sgRNAs encode conserved structural proteins (spike protein n [S], envelope protein n [E], membrane protein n [M] and nucleocapsid protein n [N]) and about 6 accessory proteins (3a, 6, 7a, 7b, 8 and 10) (fig. 1)n [33]. The gRNA is packaged by structural proteins to assemble progeny virions (fig. 1)n [33]. Progeny virions leave the host cells via lysosomal trafficking, which can be blocked by CID1067700, a Ras-related protein (Rab7) guanosine triphosphatase (GTPase) competitive inhibitor (fig. 1)n [36].
Effect of infection on host cell processes:
The SARS-CoV-2 infection leads to perturbation of multiple processes of host cell such as transcription, translation, RNA splicing, carbon metabolism, nucleic acid metabolism and proteostasisn [33]. Proteomic analysis of virus-host, protein-protein interactions revealed dysregulation of processes such as nuclear transport, vesicular trafficking pathway, ribosome biogenesis, translation, ubiquitination, lipoprotein metabolism and innate immune signaling pathwayn [37]. Interestingly, most of the host proteins that interacted with viral proteins were enriched in lung tissue indicating it as a primary target organ for pathogenesisn [37]. Analysis of transcriptome, proteome, ubiquitinome and phosphoproteome of A549 cells upon SARS-CoV-2 infection revealed perturbation of pathways such as stress and deoxyribonucleic acid (DNA) damage response, transcription regulation and cell junction organizationn [38]. A significant increase in phosphorylation was observed both for viral and host cell proteins indicating this posttranslational modification as a major cellular response to infectionn [39]. This resulted in a strong activation of p38/mitogen-activated protein kinase (MAPK), protein kinase B (AKT) and extracellular regulated kinase (ERK), Rho-GTPase cytoskeleton and casein kinase 2 (CK2) cytoskeletal related target pathways along with dysregulated cell cyclen [39]. Proteomic and metabolomic profiling of the sera from COVID-19 patients revealed disease-specific differential expression of 105 proteins and 373 metabolites, of which 93 proteins and 204 metabolites correlated with disease severityn [40]. In addition to the proposition of potential diagnostic markers, this study also demonstrated the dysregulation of immune and metabolic pathways that can act as potential therapeutic targetsn [40]. Further, using genomescale clustered regularly interspaced short palindromic repeats (CRISPR) loss of function screen in human alveolar epithelial cells revealed transcriptional changes in cholesterol biosynthesis pathwayn [41].
Cytokine release syndrome:
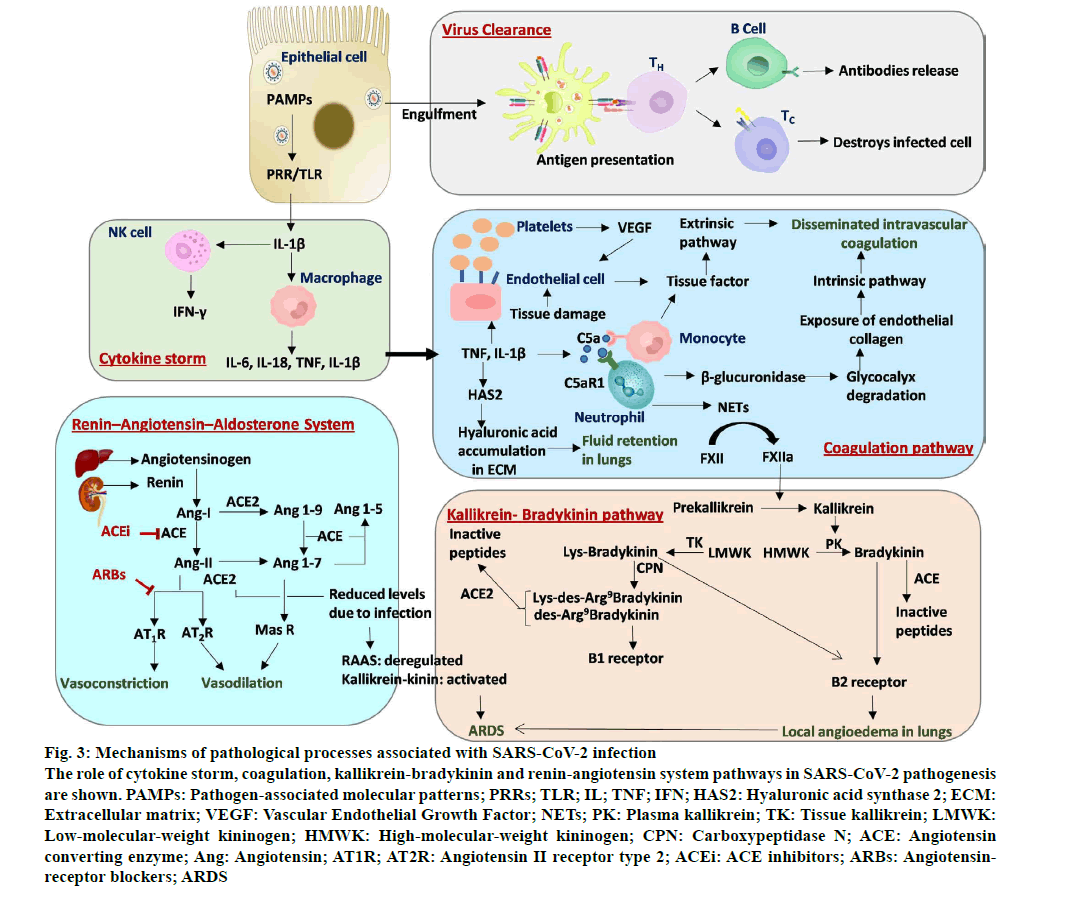
SARS-CoV-2 is a cytopathic virus that causes pyroptosis of infected cells resulting in the release of interleukin 1 beta (IL-1β), which will lead to a localized inflammatory response (fig. 3)n [42]. Here, genomic single stranded RNA (ssRNA) and double-stranded RNA (dsRNA) intermediately act as pathogen associated molecular patterns (PAMPs) that are recognized by pattern recognition receptors (PRRs) resulting in an increased production of IL-1β (fig. 3)n [43]. The IL-1β activates macrophages to release IL-6, IL-18, tumor necrosis factor (TNF) and IL-1β, as well as natural killer cells to release interferon gamma (IFN-γ) (fig. 3)n [43]. Infiltration of T-lymphocytes and monocytes from blood into the lungs lead to clearing of infection from the lungs and subsequent recovery of patient from the disease (fig. 3)n [42]. But, in patients with dysfunctional immune system, a cytokine storm ensues that includes excessive infiltration of immune cells into the lung and overproduction of inflammatory cytokines (fig. 3)n [42]. The cytokine storm combined with reduced levels of oxygen results in multi-organ failure in infected persons. Several recent studies report observations that support the induction of cytokine storm and disease severity in COVID-19 patients.
Heterogeneity in immune response was observed in hospitalized COVID-19 patients and has been classified broadly into 3 groups: Immunotype 1 is associated with severe disease and is characterized by robust cluster of differentiation 4 (CD4+) T-cell activation and exhaustion of CD8+ T-cells, immunotype 2 is characterized by less CD4+ T-cell activation and more CD8+ cells and immunotype 3 represents failure of immune response as exemplified by the lack of activation of lymphocytesn [44]. Furthermore, reduced levels of circulating CD4+ and CD8+ T-cells combined with elevated levels of IL-6 and IL-8 were shown to be indicators for disease progressionn [45]. In addition, differential subsets of circulating monocytes and neutrophils were reported in mild and severe COVID-19 patientsn [46]. Moreover, differential immune cell type profiles were observed also in bronchoalveolar lavage fluid thus distinguishing severe (monocytederived macrophages and neutrophils) from moderate (CD8+ T lymphocytes) COVID-19 patientsn [47]. Analysis of postmortem spleen and lymph nodes of COVID-19 patients revealed reduction in B-cell lymphoma 6 (Bcl- 6+) germinal center B cellsn [48] and profiling of humoral immune response in COVID-19 patients with varying degrees of severity revealed robust immunoglobulin M (IgM) and immunoglobulin A (IgA) development in both severe COVID-19 survivors and non-survivors. However, survivors class-switched to immunoglobulin G (IgG) while non-survivors showed attenuated IgG responsen [49]. Importantly, patients with sustained antibody response quickly recovered from the disease and harbored high somatic mutations in virus specific memory B cellsn [50]. Elevated levels of inflammatory cytokines in bronchoalveolar lavage fluid of severe COVID-19 patients suggest their possible role in the disruption of lung epithelial barrier and consequent bacterial infectionn [47]. In addition, increased levels of proinflammatory cytokines and chemokines (C-X-C motif chemokine ligand 8 (CXCL8), CXCL9, CXCL16, C-C motif chemokine ligand 2 (CCL2) and CCL8 were detected in serum samples of COVID-19 patientsn [51]. Further, the shift from mild to moderate disease is marked by an enhanced inflammatory signalling along with loss of lipid, amino acid and xenobiotic metabolisms possibly due to hepatic dysfunctionn [52]. Furthermore, high levels of soluble complement component 5a (C5a)n [53] as well as calprotectin, CXCL- 8, CXCL-12 and IL-6n [46] were observed in severe COVID-19 patients. Moreover, time-dependent elevated levels of inflammatory cytokines in the blood samples were observed in intensive care unit (ICU) admitted COVID-19 patients: all 7 d (TNF, granzyme B, heat shock protein 70 and IL-18), 2 to 3 d (IFN-γ inducible protein 10) and 2 to 7 d (elastase 2)n [54]. A “core COVID-19 signature” comprising of elevated inflammatory cytokines interleukin 1 alpha (IL-1α), IL- 1β, IL-17A, IL-12p70 and interferon alpha (IFN-α) was observed in all COVID-19 patientsn [55]. Notably, higher inflammatory cytokines thyroperoxidase (TPO), IL-33, IL-16, IL-21, IL-23, interferon lambda (IFN-λ), eotaxin and eotaxin-3 were observed in patients with severe COVID-19 diseasen [55]. Importantly, core COVID-19 signature declined after 10 d of disease onset in moderate patients but maintained further in severe patientsn [55]. Further, based on four defined immune signatures, Lucas et al.. stratified patients into three clusters that are predictive of disease prognosisn [55].
Coagulation pathway:.
The mobilized immune cells and inflammatory cytokines along with vasoactive molecules increase the endothelial cell contractility and loosens the interendothelial junctions resulting in creating gaps at these junctionsn [56]. Endothelial cells that are activated by IL- 1β and TNF express P-selectin, von Willebrand factor and fibrinogen resulting in the binding of platelets (fig. 3)n [56]. The infiltrated activated neutrophils release β-glucuronidases that degrade the glycocalyx, a protective layer with anti-coagulative properties (fig. 3). Additionally, the cytokines IL-1β and TNF upregulates the expression of hyaluronic acid synthase 2 resulting in the production of hyaluronic acid in the extra cellular matrix causing fluid retention in lungs (fig. 3)n [56]. Thus, altogether these events lead to increased vascular permeability and vascular leakage.
Several parallel pathways act and converge to activate the coagulation pathway (fig. 3). The exposure of endothelial collagen due to the loss of vascular integrity and endothelial dysfunction, results in the activation of intrinsic pathway where factor XIIa acts downstream converting the factor X to factor Xa (fig. 3). Cytokines released by endothelial cells increases the production of platelets that releases vascular endothelial growth factor (VEGF) which in turn drives endothelial cells to express tissue factor (fig. 3). In addition, endothelial cell or tissue damage also leads to the expression of tissue factor which activates extrinsic pathway where factor VIIa converts factor X to factor Xa (fig. 3). Activated factor Xa by extrinsic and intrinsic pathways initiate coagulation pathway resulting in the formation of fibrin clots (fig. 3). Recently, Carvelli et al.. reported the activation of coagulation pathway via complement component C5a (C5a)-complement C5a receptor 1 (C5aR1) in COVID-19 patients. Here, binding of soluble C5a to its receptor C5aR1 present on circulating neutrophils and monocytes results in recruitment, activation and adherence of neutrophils to the pulmonary endothelium (fig. 3)n [53]. The activated monocytes and neutrophils further express tissue factor and neutrophil extracellular traps (NETs) respectively (fig. 3). These NETs expressed by neutrophils activate factor XII, a zymogen form into factor XIIa, a coagulation factor (fig. 3). Finally, these events result in disseminated intravascular coagulation (DIC) (fig. 3)n [56]. Taken together, endothelial dysfunction, vascular leakage, DIC, inflammation and higher levels of cytokines result in alveolar dysfunction, ARDS with respiratory failure leading to multi organ failuren [56]. Considering the role of coagulation in COVID-19 disease, the potential of blood thinners and other anti-coagulants were tested. Early initiation of anti-coagulation therapy involving either heparin or enoxaparin was shown to be associated with a decrease in 30 d mortalityn [57] indicating its promise in the treatment of COVID-19 patients.
ARDS:
Damage of lung is caused both by the virus and secreted proteases. Compromised functions of type I pneumocytes that are involved in gas exchange as well as alveolar surfactant producing type II pneumocytes leads to dyspnea and low blood oxygen levels apart from vulnerability to secondary bacterial infections. SARSCoV- 2 binds to and downregulates ACE2 receptor leading to the activation of Kallikrein-bradykinin pathway and deregulation of renin- angiotensin aldosterone system (RAAS) pathway resulting in ARDS (fig. 3)n [56].
Kallikrein-bradykinin pathway:
Kallikreins are a group of serine proteases which are broadly classified into two types: Plasma kallikreins and tissue kallikreins (fig. 3)n [58]. Plasma kallikrein is synthesised in the liver, gets activated by coagulation factor XIIa, which then process high molecular weight kininogens (HMWK) to kinins such as bradykinin (fig. 3)n [58]. On the other hand, tissue kallikreins process low molecular weight kininogen (LMWK) to lys-bradykinin (fig. 3)n [58]. Both bradykinin and lys-bradykinin act as ligands for bradykinin receptor B2 (B2R) (fig. 3)n [58]. Further, bradykinin and lys-bradykinin are processed into des-Arg9-bradykinin and Lys-des-Arg9-bradykinin by carboxypeptidases M and N, respectively (fig. 3)n [58]. These des forms of bradykinin act as ligands for bradykinin receptor B1 (B1R) (fig. 3)n [58]. Binding of bradykinins to B1R and B2R on endothelial cells results in vascular permeability and vasodilation leading to local angioedema in lungs (fig. 3)n [58]. Importantly, ACE receptor inactivates the B2R ligands such as bradykinin that is linked to systemic acquired angioedema and ACE2 receptor inactivates B1R ligands such as des-Arg9-bradykinin and Lys-des-Arg9- bradykinin that is linked to pulmonary edema (fig. 3)n [58]. Reduced levels of ACE2 due to SARS-CoV-2 binding results in the activation of B1R by active des- Arg9-bradykinin and Lys-des-Arg9-bradykinin leading to lung injury (fig. 3).
RAAS pathway:
The RAAS pathway also plays an important role in regulating lung health. Angiotensinogen, which is produced by the liver, is converted into angiotensin I (Ang-I) by renin that is secreted by the kidney (fig. 3) n [59]. Ang-I is converted either to angiotensin II (Ang- II) by ACE or to angiotensin 1-9 (Ang 1-9) by ACE2 receptors (fig. 3)n [59]. Ang-II and Ang 1-9 are further converted to angiotensin 1-7 (Ang 1-7) by ACE2 and ACE enzymes, respectively (fig. 3)n [59]. Ang-II interacts with angiotensin II type I receptor (AT1R) that results in vasoconstriction (fig. 3)n [59]. Ang 1-7 either interacts with MAS1 oncogene (MAS receptor) leading to vasodilation or is converted to angiotensin 1-5 (Ang 1-5) by the ACE enzyme (fig. 3)n [59]. Upon SARSCoV- 2 infection, ACE2 levels are decreased leading to the upregulation of (Ang-II)-AT1R signaling as well as downregulation of (Ang 1-7) MAS signaling and higher binding of Ang-II to AT1R leading to pulmonary edema and consequently lung injury.
Therapeutics
Antibodies and vaccines:
Identification of viral protein specific circulating CD4+ and CD8+ T-lymphocytesn [60] as well as antibodies (spike- RBD-specific B38, H4, REGN10987, REGN10933, BD-368-2, CC6.33, CoV-2-2196 and CoV-2-2381 and spike-NTD-specific 4A8) in the plasma of convalescent individuals suggests the possibility of induction of a natural immune response by potential vaccines alson [61– 70]. Structural analysis of human neutralizing antibodies in complex with SARS-CoV-2 spike or RBD revealed distinct interaction modes, with the antibodies binding to RBD’s conformations: ‘’up’’ RBDs; ”up” and “down” RBDs that can contact adjacent RBDs and outside the ACE2 and can recognize “up” and “down” RBDsn [67]. Notably, a significant difference in the presence of neutralizing antibodies was observed between asymptomatic (81.1 %) and symptomatic (62.2 %) SARS-CoV-2 infected patientsn [68]. SARS-CoV-2-Spike- RBD specific monoclonal antibodies (P2B-2F6, P2C- 1F11, P2C-1A3, CA1 and CB6) failed to cross react with spike proteins of either SARS-CoV or middle east respiratory syndrome (MERS-CoV) indicating epitopic differences specifically in the RBDn [71–73]. But, antibodies from the plasma of severely infected individuals with SARS-CoV-2 showed cross reactivity towards spike protein of SARS-CoV and MERS-CoVn [71]. Similarly, cross neutralization of SARS-CoV-2 by the sera from SARS-CoV convalescent patients was demonstrated in vitron [21]. In addition, monoclonal antibodies (S309, ADI-55689 and ADI-56046) derived from memory B cells of a SARS-CoV patient showed significant cross-reactivity against SARS-CoV-2n [74,75]. However, it is important to note that the neutralizing activity of S309 is not due to the blocking of receptor binding because the epitope for this antibody is not the RBD of spike proteinn [74]. The possibility of emergence of escape mutants towards specific monoclonal antibodies was demonstrated recentlyn [76]. Therefore, a cocktail of antibodies with non-overlapping epitopes such as REGN10987-REGN10933n [76], B38-H4n [61] or CoV2-2196-CoV2-2130 pairsn [70] was suggested for an increased efficacy for COVID-19 treatment. An engineered bivalent VHH (single domain antibody, VHH-72-Fc) displayed cross neutralization activity against both SARS-CoV and SARS-CoV-2n [77].
Additionally, nanobodies that are derived upon immunization of camelids, showed good anti-SARSCoV- 2 activity, with increased activity in its multivalent formn [78]. Taken together, these observations indicate the possibility of development of vaccines that elicit immune response as well as application of therapeutic antibodies against SARS-CoV-2 infection. In addition, a human monoclonal antibody (47D11) produced in transgenic H2L2 mice was shown to neutralize both SARS-Co-V and SARS-CoV-2 indicating its utility in both diagnostic serological tests as well as therapeutic applicationsn [79].
Surprisingly, SARS-CoV-2 reactive T cells were detected in a significant fraction (20-60 %) of uninfected healthy individuals suggesting immune cross-reactivity that develops from the past infections caused by common cold coronavirusesn [60]. Further, anti-SARSCoV- 2 antibodies, predominantly of IgG class that bind S2 subunit were detected in SARS-CoV-2 uninfected individuals, which are able to neutralize SARS-CoV-2 and SARS-CoV pseudotypesn [80]. An inactivated SARSCoV- 2 vaccine candidate Sinopharm COVID-19 vaccine or BIBP vaccine (BBIBP-CorV) and purified inactivated SARS-CoV-2 vaccine (PiCoVacc) not only induced immunogenicity in diverse animal species such as mice, rats, guinea pigs and rabbits, but also efficiently blocked the SARS-CoV-2 infection in a monkey model of COVID-19 diseasen [81,82]. Further, a recombinant adenovirus-based vaccine that expressed SARSCoV- 2 spike protein showed successful antigenicity combined with tolerability and good safety profilen [83]. Additionally, a DNA-based vaccine (INO-4800) that expressed SARS-CoV-2-spike protein induced immune response in mice and guinea pigsn [84]. The resulting antibodies not only displayed limited cross-reactivity towards SARS-CoV spike protein but also competed with ACE2 receptor for binding to the SARS-CoV-2 spike proteinn [84]. Moreover, an alpha-virus-derived replicon RNA (repRNA) and lipid nanoparticle encapsulated mRNA (ARCoV) that expressed the spike protein of SARS-CoV-2 elicited long-term immune response both in mice and non-human primatesn [85,86]. Both mRNA vaccines (mRNA-1273 and BNT162b1) and adenoviral based vaccines (ChAdOx1, rAd26 and rAd5) displayed robust anti-SARS-CoV-2 immune response pre-clinicallyn [87-89] and clinicallyn [90-93] and displayed favorable safety profiles in phase 1/2. As on 7 December 2020, the adenoviral vaccine AZD1222 (ChAdOx1, from AstraZeneca) displayed 82 % efficacy with a two-dose regimenn [94] and the World Health Organization (WHO’s) Strategic Advisory Group of Experts (SAGE) has issued interim recommendations for its use. On 11 December 2020 the United States Food and Drug Administration (FDA) has issued an emergency use authorization for the mRNA-based vaccine BNT162b2 (Pfizer-BioNTech) that showed a 95 % efficacy in a phase 2/3 trialn [95]. On 18 December 2020, the US FDA has issued an emergency use authorization for the vaccine mRNA-1273 (Moderna), which demonstrated an efficacy of 94.1 % in a phase 3 studyn [96]. It is important to note that recent studies indicated the efficacy of vaccines (BNT162b2) against the mutated strain such as B.1.1.7 also thus indicating their potential to control the pandemicn [97,98].
Chemical inhibitors targeting viral enzymes:
The RdRp or nsp12 catalyzes RNA synthesis, assisted by cofactors nsp7 and nsp8, along with the helicase nsp13 and thus is an important therapeutic targetn [99,100]. The RdRp consists of a nidovirus unique N-terminal extension domain (NiRAN) and a right-hand polymerase domain, which are connected by an interface domainn [99]. The polymerase domain further consists of a fingers subdomain (motifs F and G), a palm subdomain (motifs A, B, C and D) and a thumb subdomainn [99]. The in vitro catalytic activity of RdRp was shown to be inhibited effectively by the active form remdesivir triphosphate (RTP) but not either by the pro-drug remdesivir or by the inactive remdesivir monophosphate (RMP)n [101]. The incorporated RMP interacts with K545 and R555 of RdRp, upstream base of the primer strand and uridine base of the template strandn [101]. Even though remdesivir was approved by FDA following preliminary results from the phase III trialsn [102], no statistically significant clinical benefit was observed with remdesivir treatment in adults with severe COVID-19n [103]. However, further studies involving strategies such as higher dose regimen or combination with other antivirals or neutralizing antibodies was suggested to enhance the efficacy of remdesivirn [103].
The main protease (Mpro), also known as 3CLpro, together with papain-like proteases (PLPs) cleaves polyproteins into 16 nsps that plays an important role in viral life cycle (fig. 1). A high degree of similarity of Mpro between related coronaviruses combined with the absence of a human homolog makes it an ideal therapeutic target. The Mpro functions as a dimer with each protomer (monomer) comprising three domains (I, II and III)n [104]. The substrate-binding site is present in the cleft formed between domains I and II and contains four subsites (S1, S2, S4 and S1'), while His41 and Cys145 forms a catalytic dyadn [104]. Investigational drugs (peptidomimetic aldehydes 6e, 11a and 11b, calpain inhibitors II and XII, GC-376 and N3) displayed Mpro inhibitory activity (fig. 1)n [104-106]. In addition, anti-viral drugs (boceprevir) and anti-neoplastic drugs (carmofur) showed activity against Mpro suggesting the possibility of repurposing of these drugs for COVID-19 treatment (fig. 1)n [104,106]. Inhibitor interacting residues of Mpro enzyme are fully conserved between SARS-CoV-2 and SARS-CoV but only 60 % of residues are conserved between SARS-CoV-2 and MERS-CoV (Data not shown). Interestingly, blocking of another protease (PLpro) by a naphthalene-based inhibitor GRL-0617 not only blunted viral replication in infected cells but also promoted anti-viral immunity (fig. 1)n [107]. These results indicate the promise of inhibition of proteases either alone or in combinations to effectively combat SARS-CoV-2 infectionn [107].
Figure 3:Mechanisms of pathological processes associated with SARS-CoV-2 infection
The role of cytokine storm, coagulation, kallikrein-bradykinin and renin-angiotensin system pathways in SARS-CoV-2 pathogenesis are shown. PAMPs: Pathogen-associated molecular patterns; PRRs; TLR; IL; TNF; IFN; HAS2: Hyaluronic acid synthase 2; ECM:Extracellular matrix; VEGF: Vascular Endothelial Growth Factor; NETs; PK: Plasma kallikrein; TK: Tissue kallikrein; LMWK: Low-molecular-weight kininogen; HMWK: High-molecular-weight kininogen; CPN: Carboxypeptidase N; ACE: Angiotensin converting enzyme; Ang: Angiotensin; AT1R; AT2R: Angiotensin II receptor type 2; ACEi: ACE inhibitors; ARBs: Angiotensinreceptor blockers; ARDS
Drugs with clinical efficacy:
In hospitalized severe COVID-19 patients, treatment with dexamethasone, an anti-inflammatory drug approved for rheumatoid arthritis, resulted in reduced duration of mechanical ventilation and mortalityn [108]. Further, meta-analysis of clinical trials with dexamethasone, hydrocortisone or methylprednisolone treatment revealed that these corticosteroids are associated with lower mortality rate when compared to usual care or placebon [109]. Furthermore, tocilizumab, an anti-IL-6R, showed effective clinical outcomes with reduced requirement of oxygen therapy in a small cohort of severe COVID-19 patientsn [110]. Phosphorylation of Bruton tyrosine kinase (BTK) was significantly elevated in peripheral blood monocytes of COVID-19 patients as compared to healthy volunteersn [111]. Offlabel treatment with BTK inhibitor acalabrutinib has improved the condition of severely ill COVID-19 patients by normalizing inflammatory markersn [111]. Ruxolitinib, a Janus kinase (JAK) inhibitors, in combination with standard of care agent showed rapid clinical improvement in severe COVID-19 patientsn [112]. Another JAK inhibitor baricitinib, in combination with lopinavir/ritonavir, showed promise with a lower fatality rate, decreased ICU admission and a higher discharge rate when compared with standard of care compounds (hydroxychloroquine with lopinavir/ ritonavir) in moderate COVID-19 patientsn [113]. Further, in SARS-CoV-2 infected rhesus macaques, it limited the lung infiltration of inflammatory cells and restored innate anti-SARS-CoV-2 T-cell responsen [114]. Self-administration of famotidine, a histamine type 2 receptor antagonist (approved for treating ulcers), by non-hospitalized symptomatic COVID-19 patients was found to be effective in reducing the symptoms such as cough, shortness of breath, fatigue, headache and anosmia and improved patient reported outcomesn [115]. In a phase 2 clinical trial of combined interferon beta 1b (IFN β-1b), lopinavir-ritonavir and ribavirin in COVID-19 patients, reduced the viral shedding period and hospital stay when compared with control group (lopinavir-ritonavir) with favorable safety profilen [116]. However, a combination of antiviral protease inhibitors lopinavir and ritonavir showed no benefit in adult COVID-19 patientsn [117].
Drugs with pre-clinical activity:
Based on affinity purification mass spectrometry (AP-MS), a total of 332 high-confidence interactions were observed between 26 SARS-CoV-2 proteins and host cell proteins resulting in the identification of 66 druggable human proteins that can be targeted by 69 compoundsn [37]. Inhibitors of protein biogenesis (zotatifin, ternatin-4 and PS3061) and Sigma R1/R2 (haloperidol, PB28, PD144418, hydroxychloroquine, clemastine, cloperastine and progesterone) showed significant anti-SARS-CoV-2 activity (fig. 1)n [37]. Similarly, AP-MS analysis identified 1484 interactions between 24 SARS-CoV-2 and 27 SARS-CoV proteins and 1086 host cell proteins of which majority interactions were found to be common between SARS-CoV and SARS-CoV-2n [38]. Potent anti-SARSCoV- 2 activity was displayed by inhibitors of fms-like tyrosine kinase 3 (FLT3)/AXL receptor tyrosine kinase (AXL) (gilteritinib), AKT (Ipatasertib) and matrix metalloproteinases (prinomastat and marimastat)n [38]. In addition, inducers of DNA damage (tirapazamine, rabusertib) or the mammalian target of rapamycin (mTOR) inhibitor (rapamycin) suppressed SARSCoV- 2 growthn [38]. Based on phosphoproteomic analysis, inhibitors that target upregulated signaling pathways showed significant anti-viral activity: CK2 signaling (silmitasertib), MAPK pathway (gilteritinib, ralimetinib, MAPK13-IN-1, ARRY-797), phosphatidylinositol 3?kinase (PI3K) pathway (apilimod) and cyclindependent protein kinase (CDK) signaling pathway (dinaciclib)n [39]. Furthermore, screening of a library with about 12 000 molecules identified clinical stage drugs such as FYVE finger-containing phosphoinositide kinase (PIKfyve) inhibitor apilimod and several cysteine protease inhibitors (MDL-28170, ZLVG CHN2, VBY- 825 and ONO 5334) that displayed anti-SARS-CoV-2 activity at therapeutically achievable dosesn [118].
Using cell culture based infection model, perturbation of key metabolic pathways was observed and further demonstrated the anti-COVID-19 efficacy of inhibitors of translation (cycloheximide and emitine), spliceosome (pladienolide), glycolysis (2-deoxy-D-glucose), nucleic acid metabolism (ribavirin) and proteostasis (NMS-873) n [119]. Notably, engineered soluble hACE2 was shown to efficiently reduce SARS-CoV-2 entry into host cell indicating its potential therapeutic value (fig. 1)n [120]. An inhibitor of serine protease TMPRSS2, camostat mesylate efficiently blocked the viral entry into the host cell and may thus be repurposed for COVID-19 treatment (fig. 1)n [21]. Teicoplanin, a glycopeptide antibiotic, inhibited the activity of cathepsin L towards spike protein of SARS-CoV and MERS-CoVn [121]. Therefore, teicoplanin may thus be used as one of the potential treatment options for COVID-19 patients as the cathepsin L activity is conserved among SARSCoV- 2, MERS-CoV and SARS-CoV virusesn [122]. Ivermectin, an anti-parasitic agent, displayed in vitro anti-viral action against SARS-CoV-2 possibly via inhibiting importin α/β1 (IMP-α/β1) mediated nuclear transport of viral proteinsn [123]. It was hypothesized that 4-hydroxychloroquine, an anti-malarial medication, has anti-SARS-CoV-2 effect due to its ability to increase the pH of lysosome (fig. 1). Despite pre-clinical activity in vitron [124], 4-hydroxychloroquine failed to display anti-COVID-19 activity in patients either alone or in combination with azithromycinn [125]. In human-C5aR1 knock in mice, anti-C5aR1 monoclonal antibodies inhibited acute lung injury by preventing infiltration of monocyte/macrophage and neutrophils, indicating its therapeutic potentialn [53].
Conclusion
In the absence of effective therapeutics, nonpharmaceutical interventions still remains an important preventive option. Thus, there is an urgent need either to develop a novel antiviral drug or identify existing clinically approved therapeutics that has potential to reduce mortality by interfering with pathological processes such as cytokine storm, ARDS or coagulation. Therefore, it is important to fast-track the trials by repurposing drugs that have shown proof of concept anti-SARS-CoV-2 activity in pre-clinical COVID-19 disease models. Recent studies have proposed a number of host genetic factors that determine the infectivity and pathogenicity of SARS-CoV-2. For example, a gene cluster of Neanderthal origin on chromosome 3 spanning approximately 50 kb of the genome was associated with susceptibility to COVID-19n [126]. In addition, a positive correlation between rs2285666 polymorphism of ACE2 with reduced infectivity and CFR was observed in Indian populationn [127]. Further, in 3.5 % of COVID-19 patients with life-threatening pneumonia, genetic defects at 8 of the 13 loci involved in the toll-like receptor-3 (TLR-3) and interferon regulatory factor 7 (IRF7) dependent induction and amplification of type I IFNs was observedn [128]. The genes that cluster into pathways including vacuolar adenosine triphosphatase (ATPase) proton pump, retromer, commander complexes and SWItch/Sucrose non-fermentable (SWI/SNF) chromatin remodeling complex were identified as host factors for SARSCoV- 2 infectionn [41]. Interestingly, loss of function variant of X-chromosomal TLR7 gene were shown to be associated with decreased production of IFN-γ, which is a crucial mediator of immune response in COVID-19 patients indicating an important role of host factors in disease pathogenesisn [129]. Therefore, stratification of patients based on immune biomarkers and host genetic factors may help in designing patient specific treatment strategies.
Author’s contributions:
Shravanthi Ravula and Sai Charitha Mullaguri contributed equally to this work.
Acknowledgements:
Rama Krishna Kancha acknowledges funding from University Grants Commission-Faculty Recharge Programme (UGC-FRP) (No. F.4-5(136- FRP)/2014(BSR)). Sravani Akula and Shravanthi Ravula acknowledge fellowships from the Indian Council of Medical Research (ICMR) (SRF: 2019-6060/CMSBMS) and Council of Scientific & Industrial Research (CSIR) (JRF: 09/132(0884)/2019-EMR-I), respectively. The authors also acknowledge the participation of Ms. Manasa Desai, Ms. Veena Akula, Ms. Salwa Hareem, Mr. Vigneshwar Reddy Ashireddygari and Mr. Mohd Adnan in scientific discussions.
Conflicts of interest:
The authors declared no conflict of interest.
References
- Tian H, Liu Y, Li Y, Wu CH, Chen B, Kraemer MU, et al.. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science 2020;368(6491):638-42.
- Hao X, Cheng S, Wu D, Wu T, Lin X, Wang C. Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature 2020;584(7821):420-4.
- Cheng HY, Jian SW, Liu DP, Ng TC, Huang WT, Lin HH. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med 2020;180(9):1156-63.
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020;323(13):1239-42.
- O’Driscoll M, Dos Santos GR, Wang L, Cummings DA, Azman AS, Paireau J, et al.. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 2021;590(7844):140-5.
- Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA 2020;324(8):782-93.
- Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al.. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020;20(6):656-7.
- Yang X, Yu Y, Xu J, Shu H, Liu H, Wu Y, et al.. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020;8(5):475-81.
- Mehta V, Goel S, Kabarriti R, Cole D, Goldfinger M, Acuna-Villaorduna A, et al.. Case fatality rate of cancer patients with COVID-19 in a New York hospital system. Cancer Discov 2020;10(7):935-41.
- Romero-Sanchez CM, Diaz-Maroto I, Fernandez-Diaz E, Sanchez-Larsen A, Layos-Romero A, Garcia-Garcia J, et al.. Neurologic manifestations in hospitalized patients with COVID-19: the ALBACOVID registry. Neurology 2020;95(8):e1060-70.
- Spinato G, Fabbris C, Polesel J, Cazzador D, Borsetto D, Hopkins C, et al.. Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection. JAMA 2020;323(20):2089-90.
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al.. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 2020;30(11):2196-203.
- Boni MF, Lemey P, Jiang X, Lam TT, Perry BW, Castoe TA, et al.. Evolutionary origins of the SARS-CoV-2 Sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 2020;5(11):1408-17.
- Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA 2020;117(17):9241-3.
- van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, et al.. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 2020;83:104351.
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021;592(7852):116-21.
- Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al.. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet 2020;396:603-11.
- Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al.. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020;182(5):1284-94.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al.. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367(6483):1260-3.
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al.. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581(7807):215-20.
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al.. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271-80.
- Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 2020;16(7):e9610.
- Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA 2020;323(23):2427-9.
- Ziegler CG, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181(5):1016-35.
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon III KH, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 2020;182(2):429-46.
- Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS?CoV?2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39(10):e105114.
- Smith JC, Sausville EL, Girish V, Yuan ML, Vasudevan A, John KM, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell 2020;53(5):514-29.
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al.. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020;181(4):894-904.
- Toelzer C, Gupta K, Yadav SK, Borucu U, Davidson AD, Williamson MK, et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020;370(6517):725-30.
- Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Anton-Plagaro C, et al.. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020;370(6518):861-5.
- Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020;369(6501):330-3.
- Yuan M, Wu NC, Zhu X, Lee CC, So RT, Lv H, et al.. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020;368(6491):630-3.
- Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell 2020;181(4):914-21.
- Viswanathan T, Arya S, Chan SH, Qi S, Dai N, Misra A, et al.. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun 2020;11(1):1-7.
- Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 2020;183(5):1325-39.
- Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 2020;183(6):1520-35.
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al.. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583(7816):459-68.
- Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021;594(7862):246-52.
- Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Marrero MC, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020;182(3):685-712.
- Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al.. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020;182(1):59-72.
- Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, et al.. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 2021;184(1):92-105.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al.. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497-506.
- Cardone M, Yano M, Rosenberg AS, Puig M. Lessons learned to date on COVID-19 hyperinflammatory syndrome: considerations for interventions to mitigate SARS-CoV-2 viral infection and detrimental hyperinflammation. Front Immunol 2020;11:1131.
- Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al.. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020;369(6508).
- Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, et al.. Viral and host factors related to the clinical outcome of COVID-19. Nature 2020;583(7816):437-40.
- Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al.. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 2020;182(6):1401-18.
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al.. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26(6):842-4.
- Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al.. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 2020;183(1):143-57.
- Zohar T, Loos C, Fischinger S, Atyeo C, Wang C, Slein MD, et al.. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 2020;183(6):1508-19.
- Chen Y, Zuiani A, Fischinger S, Mullur J, Atyeo C, Travers M, et al.. Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell 2020;183(6):1496-507.
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al.. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181(5):1036-45.
- Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, et al.. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 2020;183(6):1479-95.
- Carvelli J, Demaria O, Vely F, Batista L, Benmansour NC, Fares J, et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 2020;588(7836):146-50.
- Fraser DD, Cepinskas G, Slessarev M, Martin C, Daley M, Miller MR, et al. Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit Care Explor 2020;2(6):e0144.
- Lucas C, Wong P, Klein J, Castro TB, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020;584(7821):463-9.
- Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol 2020;20(7):389-91.
- Rentsch CT, Beckman JA, Tomlinson L, Gellad WF, Alcorn C, Kidwai-Khan F, et al.. Early initiation of prophylactic anticoagulation for prevention of coronavirus disease 2019 mortality in patients admitted to hospital in the United States: cohort study. BMJ 2021 11;372.
- van de Veerdonk FL, Netea MG, van Deuren M, van der Meer JW, de Mast Q, Bruggemann RJ, et al.. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife 2020;9:e57555.
- Lubel J, Garg M. Renin-angiotensin-aldosterone system inhibitors in Covid-19. N Engl J Med 2020;382:e92.
- Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NM, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020;5(48).
- Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, et al.. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020;368(6496):1274-8.
- Robbiani DF, Gaebler C, Muecksch F, Lorenzi JC, Wang Z, Cho A, et al.. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020;584(7821):437-42.
- Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, et al.. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020;369(6506):1010-4.
- Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann AJ, et al.. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020;5(48):eabc8413.
- Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al.. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 2020;182(1):73-84.
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al.. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020;369(6504):650-5.
- Barnes CO, West Jr AP, Huey-Tubman KE, Hoffmann MA, Sharaf NG, Hoffman PR, et al.. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 2020;182(4):828-42.
- Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al.. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26(6):845-8.
- Le Bert N, Tan AT, Kunasegaran K, Tham CY, Hafezi M, Chia A, et al.. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020;584(7821):457-62.
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020;584(7821):443-9.
- Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al.. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020;584(7819):115-9.
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, et al.. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020;584(7819):120-4.
- Brouwer PJ, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al.. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020;369(6504):643-50.
- Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020;583(7815):290-5.
- Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020;369(6504):731-6.
- Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al.. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020;369(6506):1014-8.
- Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, et al.. Structural basis for potent neutralization of Betacoronaviruses by single-domain camelid antibodies. Cell 2020;181(5):1004-15.
- Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, et al.. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 2020;370(6523):1473-9.
- Wang C, Li W, Drabek D, Okba NM, van Haperen R, Osterhaus AD, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 2020;11(1):1-6.
- Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, et al.. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020;370(6522):1339-43.
- Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, et al.. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 2020;182(3):713-21.
- Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, et al.. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020;369(6499):77-81.
- Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, et al.. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020;395(10240):1845-54.
- Smith TR, Patel A, Ramos S, Elwood D, Zhu X, Yan J, et al.. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 2020;11(1):1-3.
- Erasmus JH, Khandhar AP, O’Connor MA, Walls AC, Hemann EA, Murapa P, et al.. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med 2020;12(555).
- Zhang NN, Li XF, Deng YQ, Zhao H, Huang YJ, Yang G, et al.. A thermostable mRNA vaccine against COVID-19. Cell 2020;182(5):1271-83.
- Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al.. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med 2020;383(16):1544-55.
- van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al.. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020;586(7830):578-82.
- Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, et al.. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell 2020;183(1):169-84.
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al.. An mRNA vaccine against SARS-CoV-2-preliminary report. N Engl J Med 2020;383(20):1920-31.
- Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020;586(7830):589-93.
- Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al.. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020;396(10249):467-78.
- Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, et al.. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet 2020;396(10255):887-97.
- Voysey M, Clemens SA, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021;397(10269):99-111.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020;383(27):2603-15.
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al.. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021;384(5):403-16.
- Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al.. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 2021;27(4):620-1.
- Muik A, Wallisch AK, Sanger B, Swanson KA, Muhl J, Chen W, et al.. Neutralization of SARS-CoV-2 lineage B. 1.1. 7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 2021;371(6534):1152-3.
- Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al.. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020;368(6492):779-82.
- Chen J, Malone B, Llewellyn E, Grasso M, Shelton PM, Olinares PD, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020;182(6):1560-73.
- Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, et al.. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020;368(6498):1499-504.
- Rubin D, Chan-Tack K, Farley J, Sherwat A. FDA approval of remdesivir-a step in the right direction. N Engl J Med 2020;383(27):2598-600.
- Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al.. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020;395(10236):1569-78.
- Jin Z, Zhao Y, Sun Y, Zhang B, Wang H, Wu Y, et al.. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol 2020;27(6):529-32.
- Rathnayake AD, Zheng J, Kim Y, Perera KD, Mackin S, Meyerholz DK, et al. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci Transl Med 2020;12(557).
- Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, et al.. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 2020;30(8):678-92.
- Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020;587(7835):657-62.
- Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al.. Dexamethasone in Hospitalized Patients with Covid-19-Preliminary Report. N Engl J Med 2021;384(8):693-704.
- Sterne JA, Murthy S, Diaz JV, Slutsky AS, Villar J, Angus DC, et al.. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: A meta-analysis. JAMA 2020;324(13):1330-41.
- Xu X, Han M, Li T, Sun W, Wang D, Fu B, et al.. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA 2020;117(20):10970-5.
- Roschewski M, Lionakis MS, Sharman JP, Roswarski J, Goy A, Monticelli MA, et al.. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci Immunol 2020;5(48).
- Cao Y, Wei J, Zou L, Jiang T, Wang G, Chen L, et al.. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): A multicenter, single-blind, randomized controlled trial. J Allergy Clin Immunol 2020;146(1):137-46.
- Cantini F, Niccoli L, Nannini C, Matarrese D, Di Natale ME, Lotti P, et al.. Beneficial impact of Baricitinib in COVID-19 moderate pneumonia; multicentre study. J Infect 2020;81(4):647-79.
- Hoang TN, Pino M, Boddapati AK, Viox EG, Starke CE, Upadhyay AA, et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell 2021;184(2):460-75.
- Janowitz T, Gablenz E, Pattinson D, Wang TC, Conigliaro J, Tracey K, et al.. Famotidine use and quantitative symptom tracking for COVID-19 in non-hospitalised patients: a case series. Gut 2020;69(9):1592-7.
- Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al.. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020;395(10238):1695-704.
- Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al.. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 2020;382(19):1787-99.
- Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al.. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020;586(7827):113-9.
- Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al.. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020;583(7816):469-72.
- Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, et al.. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020;181(4):905-13.
- Zhou N, Pan T, Zhang J, Li Q, Zhang X, Bai C, et al.. Glycopeptide antibiotics potently inhibit cathepsin l in the late endosome/lysosome and block the entry of ebola virus, middle east respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J Biol Chem 2016;291(17):9218-32.
- Baron SA, Devaux C, Colson P, Raoult D, Rolain J-M. Teicoplanin: an alternative drug for the treatment of COVID-19? Int J Antimicrob Agents 2020;55(4):105944.
- Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 2020;178:104787.
- Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, et al.. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 2020;6(1):1-4.
- Cavalcanti AB, Zampieri FG, Rosa RG, Azevedo LC, Veiga VC, Avezum A, et al.. Hydroxychloroquine with or without azithromycin in mild-to-moderate Covid-19. N Engl J Med 2020;383(21):2041-52.
- Zeberg H, Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 2020;587(7835):610-2.
- Srivastava A, Bandopadhyay A, Das D, Pandey RK, Singh V, Khanam N, et al.. Genetic association of ACE2 rs2285666 polymorphism with COVID-19 spatial distribution in India. Front Genet 2020;11:1163.
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al.. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020;370(6515).
- Van Der Made CI, Simons A, Schuurs-Hoeijmakers J, Van Den Heuvel G, Mantere T, Kersten S, et al.. Presence of genetic variants among young men with severe COVID-19. JAMA 2020;324(7):663-73.