- *Corresponding Author:
- Jing Yuan
School of Basic Medical Sciences, Peking University Health Science Center, Haidian 100191, China
E-mail: yuanjing6212@163.com
| This article was originally published in a special issue, “Integrative Approaches in Biomedical Sciences for Drug Discovery and Development” |
| Indian J Pharm Sci 2024:86(6) Spl Issue “58-70” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
With ongoing progression of severe acute respiratory syndrome coronavirus-2 induced coronavirus disease-19, the post-acute coronavirus disease-19 syndrome (long coronavirus disease) has garnered increasing wide attention as a novel multi system disorder. Epilepsy is one of neurological manifestations associated with long coronavirus disease. However, the underlying common pathogenic mechanisms responsible for epilepsy induced by coronavirus disease-19 remain elusive. This research focused on identifying and elucidating the common underlying molecular mechanisms and biological processes underlying coronavirus disease-19 related epilepsy through bioinformatic methods. Gene expression omnibus and array express databases provided gene expression profiles associated with coronavirus disease-19 and epilepsy. Weighted gene co-expression network analysis revealed the major modules and the shared genes between epilepsy and coronavirus disease-19. A protein-protein interaction network was established using these common genes, and relevant hub genes were recognized through Cytoscape. Additionally, gene ontology and Kyoto encyclopedia of genes and genomes enrichment analyses revealed the shared pathogenic molecular mechanism between epilepsy and coronavirus disease-19. 373 common genes shared by the two diseases were chosen for further analysis. Cytoscape identified 20 hub genes related to epilepsy and coronavirus disease-19. The enrichment analysis indicated that activation of the immune response might be a potential biological process involved in coronavirus disease-19 related epilepsy. The immune cell infiltration analysis showed that dendritic cells after being activated were positively correlated with both the two diseases. This research expounded the common pathogenesis of epilepsy and coronavirus disease-19, offering potential novel insights into diagnostic and clinical management strategies for coronavirus disease-19 related epilepsy.
Keywords
Coronavirus disease-19, epilepsy, weighted gene co-expression network analysis, Hub gene, proteinprotein interaction
Since initial identification of Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) infection in December 2019, its rapid global spread has led to significant consequences. Coronavirus Disease (COVID)-19 was declared a pandemic by the World Health Organization (WHO) on March 11th 2020, signifying the most severe public health emergency of the 21st century[1]. Initially, COVID-19 shows extra pulmonary manifestations[2]. However, with the pandemic progressed, a considerable number of recovered people after COVID-19 began experiencing persistent complications, collectively referred to as long COVID[3,4]. Long COVID is commonly defined as the post COVID-19 condition, which is observed in people who had a confirmed or probable SARSCoV- 2 infection, typically manifesting 3 mo after the onset of COVID-19 symptoms and lasting for over 2 mo, without an alternative explanation for the symptoms[5]. Statistically, incidence of long COVID is estimated to range 10 %-30 % among nonhospitalized cases[6], 50 %-70 % among hospitalized cases[7], and 10 %-20 % among vaccinated cases[8]. The clinical presentations of long COVID exhibit considerable variation in terms of symptoms, severity and duration. Multiple systems, such as respiratory, cardiovascular, digestive, nervous, and hematological, are usually impaired by the condition[9].
Epilepsy, a prevalent neurological disorder, presents challenges in achieving optimal seizure control through pharmacotherapy, as over 30 % of patients experience inadequate seizure control despite drug treatment[10,11]. Patients with intractable epilepsy usually undergo surgery to mitigate their mortality risks[12,13]. Notably, the most frequently reported neurological conditions linked to COVID-19 are acute symptomatic epileptic seizures and Status Epilepticus (SE) with a high rate of mortality (5 %-39 %)[14]. A comprehensive analysis conducted in October 2022, based on a retrospective cohort encompassing approximately 1.5 million patients diagnosed with COVID-19, revealed an increased risk of new epilepsy or seizure diagnosis during the first 6 mo as well as 2 y following the initial COVID-19 diagnosis, when compared to adequately matched patients with other respiratory infections[15]. These compelling clinical and epidemiological findings all revealed the link between epilepsy and COVID-19.
COVID-19 related epilepsy can arise from a range of pathophysiological mechanisms. Seizures commonly manifest as initial symptoms in viral infections of Central Nervous System (CNS)[16]. Notably, brain tissues of some individuals with COVID-19 have exhibited neuropathological changes, along with observed SARS-CoV-2 RNA and viral proteins[17]. Based on current evidence, SARS-CoV-2 can access the CNS via two possible routes; the olfactory route and the hematogenous pathway. The virus can access the CNS through the olfactory route by infecting the olfactory neurons in the nasal cavity. In the hematogenous pathway, viruses can enter the CNS through the infiltration of infected blood cells or the direct infection of endothelial cells at the Blood-Brain Barrier (BBB). This invasion can subsequently result in encephalitis, which may be accompanied by the occurrence of seizures[18,19]. Additionally, systemic inflammation induced by viral infection, even without neuroinvasion or true encephalitis, can contribute to seizures. Previously, important role of pro-inflammatory cytokines in initiation and persistence of epilepsy has been focused on[20]. SARS-CoV-2 infection is typically accompanied by fever and elevated circulating inflammatory cytokines, which can disrupt the BBB integrity and potentially facilitate the transfer of inflammatory cytokines into the CNS[21]. Moreover, systemic inflammatory cytokines could also enter the CNS through the vagal nerve in the gut-brain axis[22]. Furthermore, following SARS-CoV-2 infection, immune cells often become over-activated, leading to increased infiltration of immune cells from the bloodstream into the brain. These immune cells may contribute to neurotoxicity, synaptic dysregulation, and ictogenesis. Additionally, immune cells are usually over activated after SARS-CoV-2 infection, which may increase the infiltration of immune cells from circulation to brain. These cells may contribute to neurotoxicity, synaptic dysregulation, and ictogenesis[23,24]. Nonetheless, the precise underlying molecular and biological mechanisms that link epilepsy with COVID-19 remain fully elucidated. The Ribonucleic Acid (RNA)-seq and microarray technologies were utilized to explore the common pathogenesis and interaction between different diseases. This research aimed to recognize the hub genes related to the common pathogenesis between epilepsy and COVID-19 through Weighted Gene Co-Expression Network Analysis (WGCNA). 373 common genes were identified between epilepsy and COVID-19. Next, the Protein-Protein Interaction (PPI) network was constructed to elucidate the relationships among these proteins. In addition, using the Cytohubba analysis, the top 20 genes with the most active interaction were designated as hub genes and primarily enriched in immune response activation, suggesting a potential common mechanism for epilepsy and COVID-19. The combination of Cluster of Differentiation (CD) 38 and Protein Kinase C Alpha (PRKCA) was identified as the shared diagnostic genes between epilepsy and COVID-19 by employing LASSO regression, and this finding was validated in additional datasets. Moreover, immune cell infiltration analysis revealed the involvement of activated Dendritic Cells (DCs) and CD8 T cells in pathogenic processes of the two diseases. Finally, the interplay among hub genes, Transcription Factors (TFs), and microRNA (miRNA) were forecasted based on different databases. Overall, this research elucidated the molecular mechanisms underlying the association between epilepsy and COVID-19. Meanwhile, the identified hold promises as potential targets for diagnosing and intervening COVID-19 related epilepsy.
Materials and Methods
Datasets collection and preprocessing:
Fig. 1, depicts operations for data collection and analysis. The COVID-19 dataset (GSE196822) and epilepsy dataset (E-MTAB-3123) were obtained from the publicly available Gene Expression Omnibus (GEO) and Array Express public database, respectively. The GSE196822 dataset comprised 34 patients with COVID-19 and 9 non COVID-19 controls. This dataset was generated using the GPL20301 Illumina HiSeq 4000 (Homo sapiens) platform. On the other hand, the E-MTAB-3123 dataset consisted of 24 patients with temporal lobe epilepsy and 23 post-mortem controls without epilepsy. This dataset was generated based on the Agilent-039906 custom chip platform.
WGCNA:
WGCNA is a powerful analytical technique utilized to examine the gene expression patterns across multiple samples. This method identifies gene clusters with similar expression patterns and explores the correlations between modules and specific traits or phenotypes[25]. The WGCNA R package enabled the construction of the gene co-expression modules of COVID-19 and epilepsy. Firstly, the pickSoftThreshold function was employed to utilize and define the optimal soft threshold power (β) for constructing the adjacency matrix construction based on the scale-free topology criterion. Subsequently, the TOM similarity algorithm converted the adjacency matrix into a topological overlap matrix. Hierarchical clustering analysis of the topological overlap matrix, along with the employment of the Dynamic Branch-Cut method, was performed to recognize co-expression modules. Module Eigengene (ME) was computed using the ME algorithm. Spearman correlation analysis was adopted to determine the link of ME to clinical traits, with p<0.05 and absolute correlation coefficient (r)>0.3 were identified statistically significant. Modules with high trait correlation were considered as crucial modules, and genes within those modules were selected for further analyses.
Identification of common and hub genes in COVID-19 and epilepsy:
COVID-19 related and epilepsy-relevant modules were intersected to confirm their common genes. In this research, STRING (https://cn.string-db. org/) was used to contribute a PPI network of the common genes with a score (combined score) >0.7. This network was subjected to visualization through Cytoscape (version 3.9.1). The hub genes (top 20) with the strongest interactions were screened by Cytohubba plug-in in Cytoscape through degree topological analysis method.
Functional enrichment and pathway analysis of hub genes:
The clue Gene Ontology (GO) plug-in in Cytoscape and the clusterProfiler package performed GO and KEGG enrichment analyses on those common genes and hub genes to understand the shared underlying pathogenesis between COVID-19 and epilepsy. The biological functions of the hub genes were described by the GO analysis from Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). Employing clue GO allowed for the reduction of redundant GO terms while preserving more representative parent or child terms[26]. KEGG analysis elucidated the pathways in which the hub genes were involved. p<0.05 was considered as the threshold of significant enrichment.
Immune cell infiltration analyses:
Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) enabled the characterization of immune cell infiltration in epilepsy and control (http://CIBERSORT.stanford. edu/). CIBERSORT can transform the expression data of genes into composition of infiltrating immune cells[27]. By employing the LM22 gene file of CIBERSORT, the components and percentages of 22 infiltrating immune cells were identified. The Wilcoxon test was performed to identify immune cell populations that exhibited significant differences between epilepsy and control. Visualization of the immune cell infiltration results was achieved using the ggplots and pheatmap R packages. Additionally, Spearman’s rank correlation test with p<0.05 was utilized to explore the link between hub genes and the infiltrating immune cells.
Diagnostic biomarker selection based on machine learning:
The LASSO is a popular regression method that aids in variable selection and enhances predictive accuracy[28]. A glmnet R package was adopted for performing the LASSO regression, aiming to identify the optimal diagnostic biomarkers of epilepsy and COVID-19 in the hub genes.
Diagnostic value evaluation:
The two candidate biomarkers were externally validated based on the Receiver Operating Characteristic (ROC) curve analysis in separate datasets for epilepsy and COVID-19. The GSE177477 dataset contained 11 COVID-19 patients and 18 controls. The 9 epilepsy samples and 50 controls in GSE143272 dataset were selected.
Prediction of TFs and miRNAs:
TFs corresponding to hub genes were forecasted by five databases (ChEA, ENCODE, hTFtarget, TRANSFAC, and TRRUST). TFs which displayed interactions in at least three databases were selected for further analysis. Additionally, five bioinformatics tools (miRMap, miRanda, miRDB, TargetScan, and miTarBase) were applied to forecast the interplays between miRNA and hub genes[29-33]. Only the miRNAs which were forecasted by all five tools were selected. The TF-miRNA-mRNA regulatory network was established with Cytoscape (version 3.9.1).
Results and Discussion
The research procedure was illustrated in fig. 1. Four datasets were selected, and their key information is summarized in Table 1. The E-MTAB-3123 and GSE196822 datasets were undertaken as the discovery cohorts for conducting the WGCNA analysis. Subsequently, the GSE143272 and GSE177477 datasets were based to assess the diagnostic value of the shared diagnostic biomarkers.
| Disease | Dataset | Platform | Samples | Tissue |
|---|---|---|---|---|
| COVID-19 | GSE196822 | GPL20301 | 34 patients and 9 controls | Peripheral blood |
| Epilepsy | E-MTAB-3123 | Agilent SurePrint G3 | 24 patients and 23 controls | Hippocampal tissue |
| COVID-19 | GSE177477 | GPL23159 | 11 patients and 18 controls | Peripheral blood |
| Epilepsy | GSE143272 | GPL10558 | 9 patients and 50 controls | Peripheral blood |
Table 1: Information of Datasets Containing the Epilepsy and Covid-19 Patients
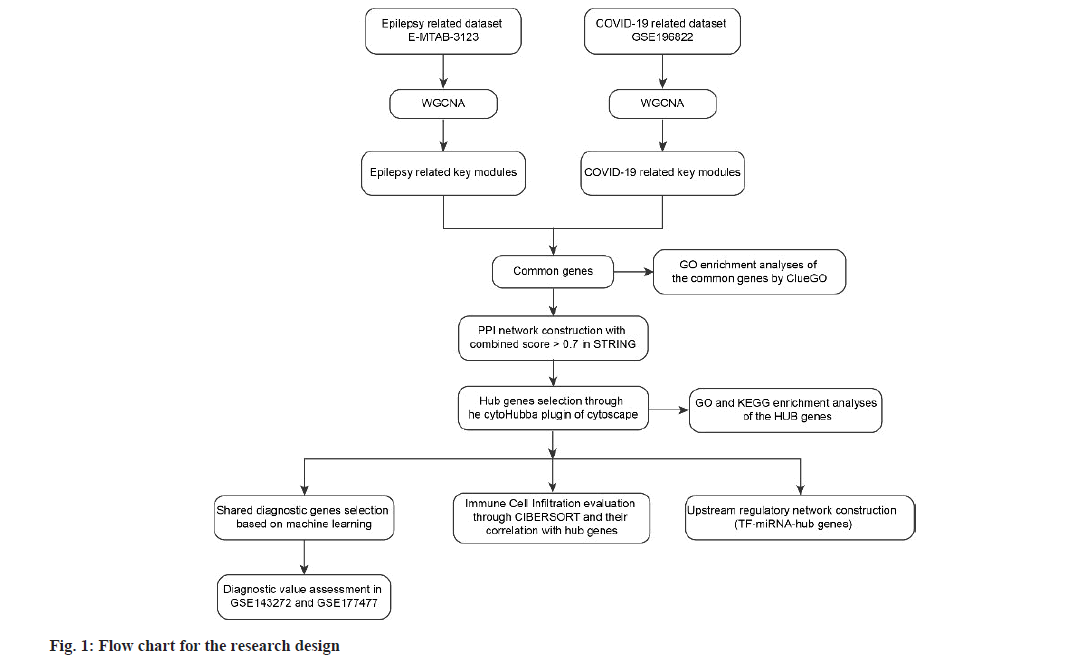
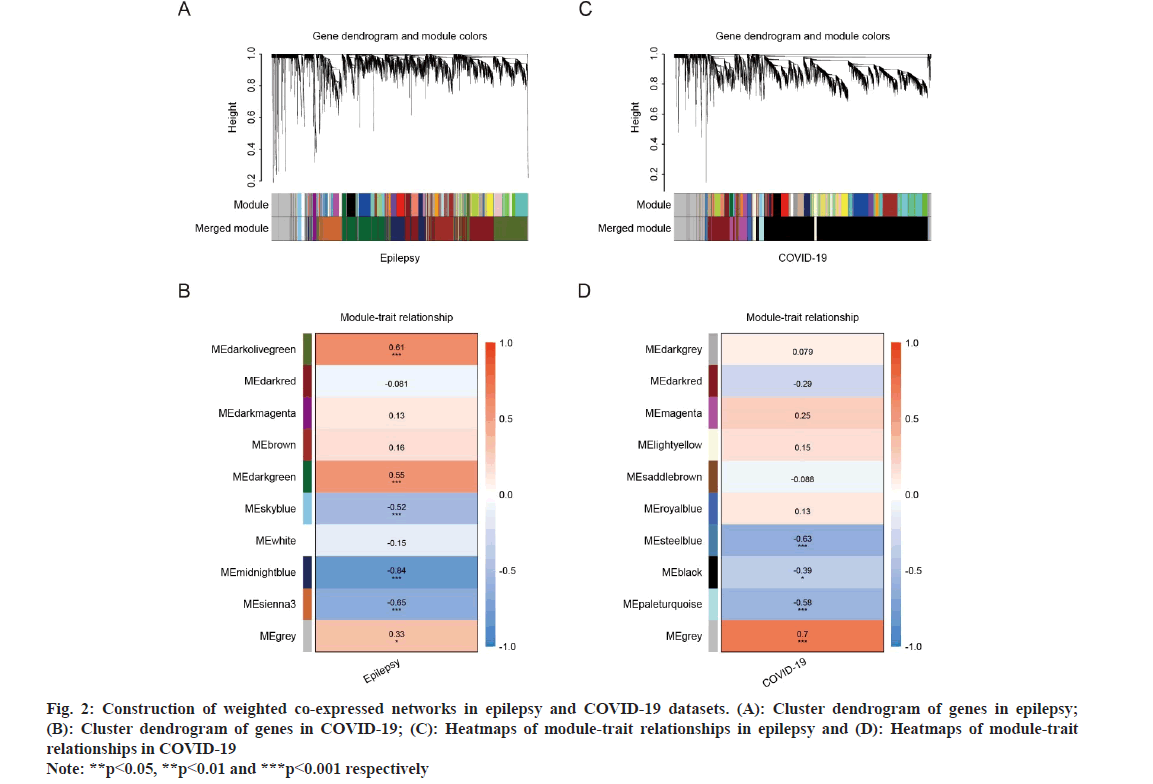
WGCNA explored the co-expressed gene modules in epilepsy and COVID-19 related datasets. For the E-MTAB-3123 dataset, a soft threshold power (β) of 18 was determined, forming a scale-free topological index (R2) of 0.815 (fig. 2A). The network exhibited a scale-free topology distribution, in line with the corresponding model and mean connectivity. The association between gene modules and clinical traits (diseases and health states) was calculated based on Spearman correlation analyses (fig. 2A and fig. 2B). Five modules were found to be greatly associated with epilepsy and were designated as epilepsy-related key modules (the dark olive green module r=0.61, p=5e-6; the dark green module r=0.55, p=7e-5; the sky blue r=-0.52, p=2e-4; the mid night blue module r=-0.84, p=2e-13 and the sienna3 module r=-0.65, p=9e-7). Similarly, the β of 28 and R2 of 0.749 were set in GSE196822 dataset. Among the ten identified modules, the steel blue module (r=-0.63, p=4e-5), the pale turquoise module (r=-0.58, p=3e-4), and the black module (r=-0.39, p=0.02) exhibited strong correlations with COVID-19 (fig. 2C and fig. 2D). The common genes were identified by intersecting the epilepsy and COVID-19-related modules, which were considered to be extremely related to the pathogenesis of epilepsy and COVID-19 (fig. 3A).
Fig. 2: Construction of weighted co-expressed networks in epilepsy and COVID-19 datasets. (A): Cluster dendrogram of genes in epilepsy; (B): Cluster dendrogram of genes in COVID-19; (C): Heatmaps of module-trait relationships in epilepsy and (D): Heatmaps of module-trait relationships in COVID-19 Note: **p<0.05, **p<0.01 and ***p<0.001 respectively
To understand the potential biological functions of the 373 shared genes between epilepsy and COVID-19, GO enrichment analysis was conducted using clue GO (fig. 3B). The results revealed that they were mainly enriched in various biological activities, including T cell activation, lymphocyte activation, regulation of cytokine production, peptide antigen assembly with Major Histocompatibility Complex (MHC) class II protein complex, cellular response to chemical stress, and leukocyte mediated cytotoxicity. Notably, T cell activation accounted for about 50 % of total GO terms (fig. 2A). These results indicated that these common genes may collectively participate in the progression of both epilepsy and COVID-19 through immune activation related biological functions or signaling pathways.
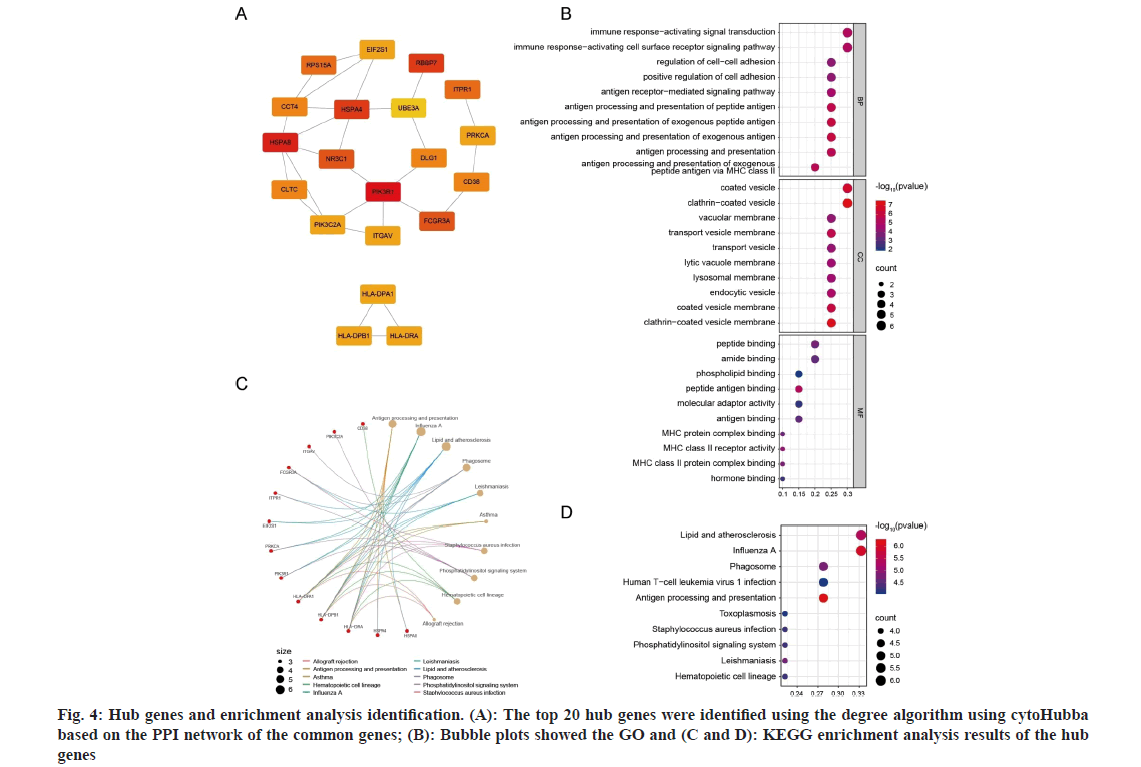
To understand the inter correlations among the common genes, a PPI network was proposed based on the STRING database. This network allowed to identify the key proteins that play essential biological roles and reveal the underlying mechanisms[34]. This network for the common genes contained 185 nodes and 247 interaction pairs (fig. 3A). The CytoHubba plug-in in Cytoscape analyzed the PPI network based on the degree algorithms (fig. 4A). The top 20 genes were selected as hub genes, because they had the strongest interactions, which were Phosphoinositide- 3-Kinase Regulatory 1 (PIK3R1), Heat Shock Protein Alpha 8 (HSPA8), Retinoblastoma-Binding Protein 7 (RBBP7), Heat Shock 70 kDa Protein 4 (HSPA4), NR3C1, Fc Gamma Receptor 3A (FCGR3A), Ribosomal Protein S15A (RPS15A), Inositol 1,4,5-Trisphosphate Receptor Type 1 (ITPR1), Discs Large MAGUK Scaffold Protein 1 (DLG1), Clathrin Heavy Chain (CLTC), CD38, Chaperonin Containing TCP1 4 (CCT4), PRKCA, PIK3C2A, Integrin Subunit Alpha V (ITGAV), Histocompatibility Complex, Class II, DR Alpha (HLA-DRA), Histocompatibility Complex, Class II, DP Beta 1 (HLA-DPB1), HLA-DPA1, Eukaryotic Translation Initiation Factor 2 Subunit Alpha (EIF2S1), and Ubiquitin Protein Ligase E3A (UBE3A). The strongest interactions among these genes suggested their potential importance in the pathogenic mechanisms of both epilepsy and COVID-19.
To explore the shared regulatory pathways and biological functions of the hub genes, the GO and KEGG analyses were implemented. The GO terms of BPs uncovered that the hub genes were significantly enriched in immune response-activating, antigen processing and presentation, and regulation of cellcell adhesion. In terms of CCs, these genes were primarily associated with coated vesicle, clathrincoated vesicle, transport vesicle, endocytic vesicle, and their membranes, as well as lysosomal membrane. For MFs, the genes were predominantly involved in amide binding, peptide binding, phospholipid binding, antigen binding, and binding to MHC proteins (fig. 4B). Moreover, the KEGG analysis demonstrated that the hub genes were primarily associated with pathways related to infectious diseases, immune response, and antigen processing and presentation (fig. 4C and fig. 4D). Collectively, the hub genes may contribute to the progression of epilepsy and COVID-19 primarily through immune response-related biological functions or signaling pathways.
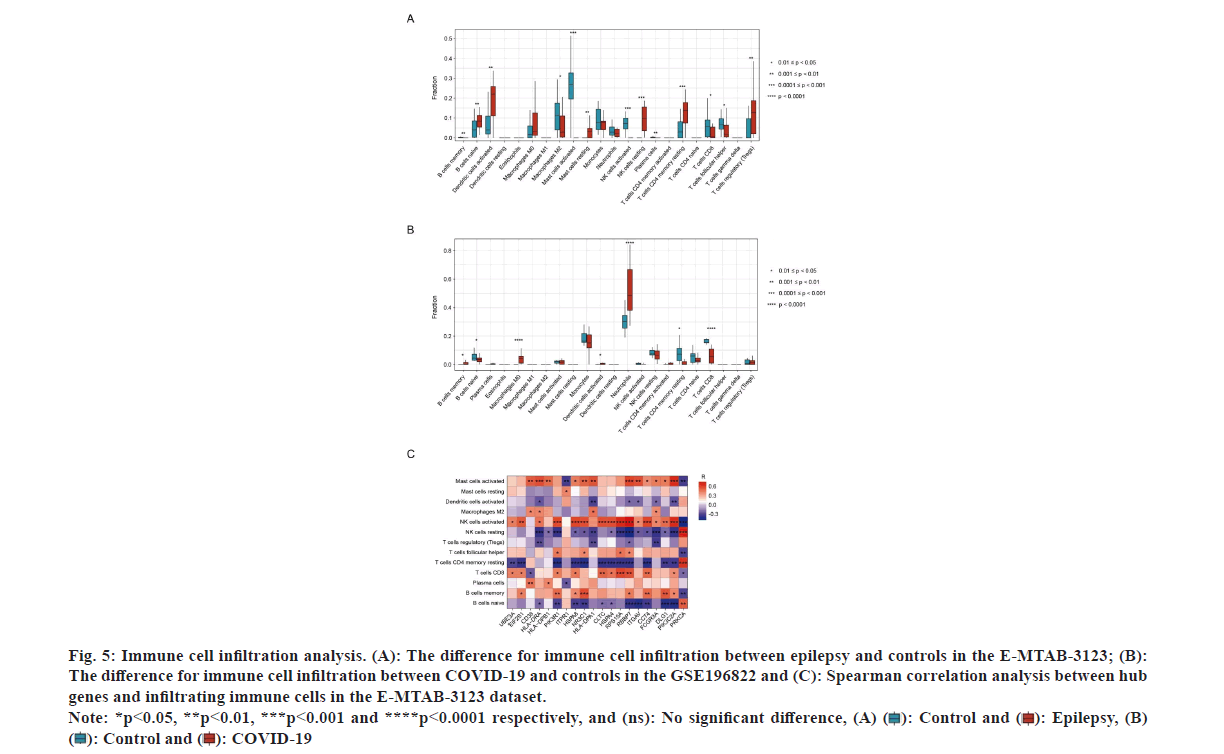
Given that the above analysis of the common and hub genes has revealed the important role for immune responses in the progression of epilepsy and COVID-19, the 22 immune cells in two groups were investigated through CIBERSORT analyses to explore their potential immune mechanisms. The analysis indicated that the epilepsy samples were positively linked to naive B cell, activated DCs, mast cells in a resting state, NK cells in an inactive state, CD4 memory T cells with a naive phenotype, and T cells with a regulatory function. While, negative links were observed with memory B cells, M2 macrophages, activated mast and NK cells, plasma cells, CD8 T cells, and follicular helper T cells (fig. 5A). For COVID-19 samples, positive correlations were observed with memory B cells, M0 macrophages, activated DCs, neutrophils, and CD8 T cells, while negative relationships were noted with naive B cells, activated CD4 memory T cells, and CD8 T cells (fig. 5B). Notably, epilepsy and COVID-19 patients exhibited increased activated DCs and reduced CD8 T cells compared to controls. These results suggested the potential significance of activated DCs and CD8 T cells in COVID-19 related epilepsy.
Fig. 5: Immune cell infiltration analysis. (A): The difference for immune cell infiltration between epilepsy and controls in the E-MTAB-3123; (B):
The difference for immune cell infiltration between COVID-19 and controls in the GSE196822 and (C): Spearman correlation analysis between hub
genes and infiltrating immune cells in the E-MTAB-3123 dataset. Note: *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001 respectively, and (ns): No significant difference, (A) (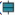 ): Control and (
): Control and ( ): Epilepsy, (B)
(
): Epilepsy, (B)
( ): Control and (
): Control and ( ): COVID-19
): COVID-19
Subsequent to that, the association of hub genes to the significantly differential immune cells in epilepsy samples was assessed. The findings demonstrated predominantly positive correlations between most of the hub genes and activated mast cells, activated NK cells, CD8 T cells, and memory B cells. On the other hand, they were negatively linked to resting NK and CD4 memory T cells, and naïve B cells (fig. 5C). As a result, these genes may participate in the pathogenesis of epilepsy together with immune cells.
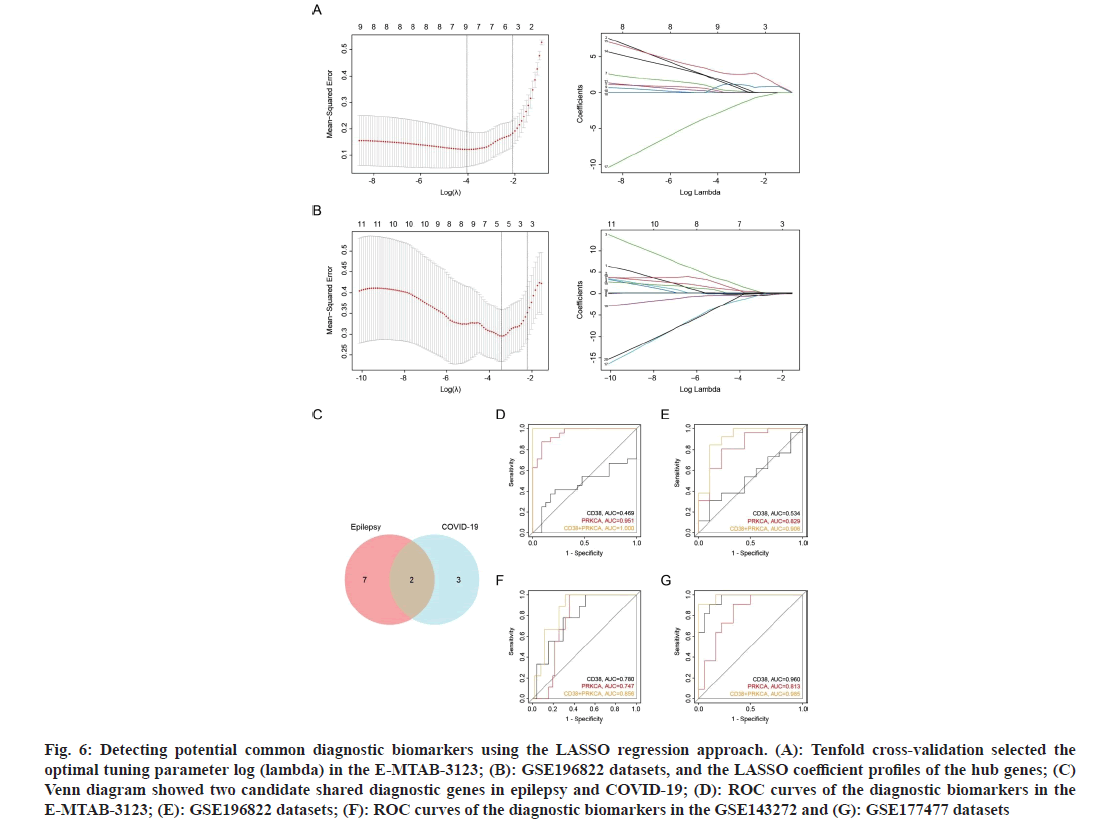
Given the importance of hub genes in the pathogenic mechanisms of epilepsy and COVID-19, the LASSO algorithm was employed to construct a diagnostic model and identify the potential shared diagnostic genes of the two diseases. Nine genes in E-MTAB-3123 and five genes in GSE196822 were identified as potential candidates of epilepsy and COVID-19, respectively (fig. 6A and fig. 6B). Besides, CD38 and PRKCA were found to be overlapping and designated as the common diagnostic biomarkers for epilepsy and COVID-19 (fig. 6C).
Fig. 6: Detecting potential common diagnostic biomarkers using the LASSO regression approach. (A): Tenfold cross-validation selected the optimal tuning parameter log (lambda) in the E-MTAB-3123; (B): GSE196822 datasets, and the LASSO coefficient profiles of the hub genes; (C) Venn diagram showed two candidate shared diagnostic genes in epilepsy and COVID-19; (D): ROC curves of the diagnostic biomarkers in the E-MTAB-3123; (E): GSE196822 datasets; (F): ROC curves of the diagnostic biomarkers in the GSE143272 and (G): GSE177477 datasets
To evaluate the diagnostic performance of the biomarkers, a ROC curve was constructed to determine the diagnostic specificity and sensitivity of CD38 and PRKCA. In the E-MTAB-3123 dataset, PRKCA exhibited a nearly perfect diagnostic value (Area Under the Curve (AUC)=0.951) for epilepsy (fig. 6D). In the GSE196822 dataset, PRKCA demonstrated a good diagnostic value (AUC=0.829) for COVID-19 (fig. 6E). Notably, the combination of CD38 and PRKCA showed the highest diagnostic values in both datasets. To validate these findings, the diagnostic efficacy was further evaluated using two external datasets. CD38 and PRKCA exhibited strong predictive performance in both the epilepsy dataset GSE143272 and COVID-19 dataset GSE177477 (fig. 6F and fig. 6G). Similarly, the combination of CD38 and PRKCA demonstrated the highest predictive performance. Given these results, the combination of CD38 and PRKCA had a powerful discriminatory capability, which may be a potential prevention and diagnostic target for COVID-19 related epilepsy patients.
To identify the TFs and miRNAs which regulating the hub genes, a TF-miRNA-hub gene regulatory network was constructed (fig. 7). Using five databases, including ChEA, ENCODE, hTFtarget, TRANSFAC, and TRRUST, 35 TFs that may regulate the hub genes were identified. Noteworthy, CREB1 and TCF3 emerged as important TFs with the highest number of connections to the hub genes. Similarly, other databases were used and the interactions of 16 miRNAs with the hub genes were found. Five essential miRNAs were revealed, such as hsa-miR- 106b-5p, hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR- 20b-5p, and hsa-miR-93-5p.
Ever since COVID-19 emerged in 2019, more and more cases of seizures in patients infected with SARS-CoV-2 have been reported[35,36]. A case report in May 2020 reported the occurrence of focal SE as the first symptom of a patient infected with SARSCoV- 2[37]. Another report in October 2020 described a patient with refractory SE 6 w after SARS-CoV-2[38]. As the epidemic progresses, numerous studies have confirmed the strong association between epilepsy and COVID-19[39]. Several theoretical mechanisms have been proposed to explain this correlation, such as fever during infection, neuroinvasive of SARSCoV- 2, elevated circulating cytokines, destruction of BBB, and hyper activated immune cells[40-43]. However, the intricate molecular mechanisms underlying the interaction between epilepsy and COVID-19 still have not been completely elucidated. This research investigated the common genes and shared signatures of epilepsy and COVID-19 using bioinformatics methods, so as to facilitate early detection and improve therapeutic strategies for COVID-19-related epilepsy.
In this research, 373 common genes between epilepsy and COVID-19 were identified using the WGCNA. The enrichment analysis disclosed that the identified genes were dominantly enriched in immune response-related pathways and functions, such as antigen presentation, lymphocyte activation, and regulation of cytokine production. These results highlight the crucial role of the immune response in the common pathogenesis of epilepsy and COVID-19. SARS-CoV-2 infection may impact the CNS either by directly invading neurons or by causing immunemediated nervous damage[44]. However, detecting SARS-CoV-2 RNA in the cerebrospinal fluid of COVID-19 patients with neurological symptoms is relatively rare, which suggested that direct neuronal invasion may not be the main mechanism. Frequently, COVID-19 is commonly accompanied by massive release of various cytokines and hyper activation of immune response[45]. Some of the circulating cytokines could increase BBB permeability, which allows immune cells to infiltrate the CNS and attack neurons[46]. Previous reports have indicated that certain respiratory viruses can trigger an inflammatory cascade during infections, resulting in the release of cytokines that activate glutamate receptors, thus leading to neuronal hyperexcitation and the onset of acute symptomatic seizures[47]. Consequently, it was speculated that the elevated cytokines and immune activation may be one of the pathogenesis of COVID-19 related epilepsy.
PPI analysis further identified 20 hub genes which were closely related to epilepsy and COVID-19, including PIK3R1, HSPA8, RBBP7, HSPA4, NR3C1, FCGR3A, RPS15A, ITPR1, DLG1, CLTC, CD38, CCT4, PRKCA, PIK3C2A, ITGAV, HLA-DRA, HLA-DPB1, HLA-DPA1, EIF2S1, and UBE3A. As well known, PIK3R1 and PIK3C2A are classical members of PI3Ks to activate the PI3K/Akt/mTOR pathway, which is crucial in the regulation of various cell functions[48]. Meanwhile, it has been confirmed to be activated in patients with COVID-19, and its inhibitors could exert antiviral effects[47]. Moreover, it may be increased in brain tissue of people with epilepsy[49]. Inhibiting the mTOR signaling pathway could effectively reduce seizure frequency[50]. HAPA4 and HSPA8 encoded heat-shock proteins that participated in antigen presentation and functioned as cytokines, which stimulated the production of proinflammatory cytokines and enhanced DC maturation[51]. CLTC encodes clathrin heavy chain 1, which was a component of clathrin. Clathrin could form a curved envelope and cover the vesicle cytoplasmic face which was coated by clathrin[52]. These special organelles are linked to intracellular trafficking of receptors and endocytosis of various macromolecules[53]. A previous study indicated that the CLTC variants were associated with epilepsy phenotypes[54]. The disruption of UBE3A causes Angelman Syndrome (AS), which featured with delayed development, intellectual disability, and seizures[55]. In mouse models of AS, UBE3A has been demonstrated to degrade ubiquitinmediated big potassium channels to inhibit neuronal hyperexcitability, thereby improving susceptibility to epilepsy[56]. It was interesting that three of the hub genes encode human MHC, also known as human leukocyte antigens, including HLA-DRA, HLADPB1, and HLA-DPA1. Previous studies had shown that HLA antigens and haplotypes, such as HLADR4, HLA-DR7, and HLA-DQ2, are in connection with different epilepsies[57]. They may play a role in different types of epilepsies. HLA molecules were also critical for antigen presentation to T cells and mediating immune responses during SARS-CoV-2 infection[58]. Briefly, these hub genes identified in this study may play important roles in the pathogenesis of COVID-19 related epilepsy.
In this research, hub genes are primarily involved in the progressions of both epilepsy and COVID-19 through immune-related biological pathways and functions. Our results showed that activated DCs were positively associated with both the two diseases, suggesting an important role of DCs in the pathogenesis of epilepsy and COVID-19. DCs are the most efficient antigen-presenting cells and are crucial in generating immune responses, creating connections between innate and adaptive immunity[59]. During SARS-CoV-2 infection, when the DCs were activated by the pathogen, they secreted proinflammatory cytokines and expressed surface molecules, such as MHC and costimulatory molecules, and induced innate and adaptive immune responses. It had been identified that the number of activated DCs was positively correlated with the severity of COVID-19[60]. DCs also present in the brain, which respond to pathogens such as viruses or bacteria, and to interferons or cytokines that were produced during inflammation or injury[61]. Activated DCs mobilized effector T-cells to provide protection. However, the T cells that enter the CNS sometimes could also cause tissue damage under certain conditions[62].
Further exploration of the hub genes identified the combination of CD38 and PRKCA as potential diagnostic genes of epilepsy and COVID-19. This result was validated in two validated cohorts. The deficiency of Nicotinamide Adenine Dinucleotide (NAD+) is an important pathological factor in various diseases related to CNS[63]. CD38 is a key regulator of degrading NAD+ in hippocampus during epileptogenesis. The production of CD38 had been identified to be increased in the hippocampus of epilepsy rats, while CD38 inhibitor could reduce the tonic-clonic seizure severity and seizure duration[64]. A previous study also suggested that CD38 could involve in the progress of COVID-19 from several aspects, such as viral invasion, viral evasion from innate and adaptive immune responses, and hyper inflammation associated to metabolic conditions lymphopenia[65]. Although the biological functions of PRKCA in the pathogenesis of COVID-19, which encoding protein kinase C had not been fully discussed, network pharmacology had revealed it as therapeutic target for COVID-19[66].
This research aimed to investigate the common pathogenesis between epilepsy and COVID-19. However, it is essential to acknowledge the limitations of our study. First, the specificity of epilepsy patients prevented us from including more samples in our work. Second, the diagnostic value of the combination of CD38 and PRKCA needs to be further verified in future clinical trials. In conclusion, this research proposed the potential common pathogenetic mechanisms between epilepsy and COVID-19. We identified the common genes and BP of these two diseases, which help to better understand their connections. The hub genes, especially CD38 and PRKCA, may be the promising prevention and diagnostic targets for COVID-19-related epilepsy.
Funding:
This study was supported by the Postdoctoral Research Fund of Chaoyang District, Beijing, China in 2022. Natural Science Foundation for Key Programs of China Grants (No: 82130065), National Natural Science Foundation of China (No: 82002330), and FENG foundation (No: FFBR 202103).
Acknowledgments:
The authors acknowledge all the contributors for publicly sharing the gene expression profiling datasets used in this study.
Author’s contributions:
Conceptualization by YL and CJ; transcriptomic analysis by YL; investigation by YL; methodology by YL; software by YL; supervision by YL and CJ; writing original draft by YL and writing-review and editing by YL and CJ. All authors have read and agreed to the published version of the manuscript.
Conflict of interests:
The authors declared no conflict of interests.
References
- Koc HC, Xiao J, Liu W, Li Y, Chen G. Long COVID and its management. Int J Biol Sci 2022;18(12):4768-80.
- Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 2021;19(3):141-54.
[Crossref] [Google Scholar] [PubMed]
- Chen C, Haupert SR, Zimmermann L, Shi X, Fritsche LG, Mukherjee B. Global prevalence of post-Coronavirus Disease-2019 (COVID-19) condition or long COVID: A meta-analysis and systematic review. J Infect Dis 2022;226(9):1593-607.
[Crossref] [Google Scholar] [PubMed]
- Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, et al. 6-month consequences of COVID-19 in patients discharged from hospital: A cohort study. Lancet 2021;397(10270):220-32.
[Crossref] [Google Scholar] [PubMed]
- Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis 2022;22(4):e102-7.
[Crossref] [Google Scholar] [PubMed]
- Davis HE, McCorkell L, Vogel JM, Topol EJ. Long COVID: Major findings, mechanisms and recommendations. Nat Rev Microbiol 2023;21(3):133-46.
[Crossref] [Google Scholar] [PubMed]
- Ceban F, Ling S, Lui LM, Lee Y, Gill H, Teopiz KM, et al. Fatigue and cognitive impairment in post-COVID-19 syndrome: A systematic review and meta-analysis. Brain Beha Immun 2022;101:93-135.
[Crossref] [Google Scholar] [PubMed]
- Al-Aly Z, Bowe B, Xie Y. Long COVID after breakthrough SARS-CoV-2 infection. Nat Med 2022;28(7):1461-7.
[Crossref] [Google Scholar] [PubMed]
- Zawilska JB, Kuczyńska K. Psychiatric and neurological complications of long COVID. J Psychiatr Res 2022;156:349-60.
[Crossref] [Google Scholar] [PubMed]
- Pfisterer U, Petukhov V, Demharter S, Meichsner J, Thompson JJ, Batiuk MY, et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun 2020;11(1):5038.
[Crossref] [Google Scholar] [PubMed]
- Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, et al. Definition of drug resistant epilepsy: Consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia 2010;51(6):1069-77.
[Crossref] [Google Scholar] [PubMed]
- Engel J, McDermott MP, Wiebe S, Langfitt JT, Stern JM, Dewar S, et al. Early surgical therapy for drug-resistant temporal lobe epilepsy: A randomized trial. JAMA 2012;307(9):922-30.
[Crossref] [Google Scholar] [PubMed]
- Schramm J, Lehmann TN, Zentner J, Mueller CA, Scorzin J, Fimmers R, et al. Randomized controlled trial of 2.5-cm vs. 3.5-cm mesial temporal resection in temporal lobe epilepsy-part 1: Intent-to-treat analysis. Acta Neurochir 2011;153:209-19.
[Crossref] [Google Scholar] [PubMed]
- Evangelista G, Dono F, Nucera B, Lanzone J, Rinaldi F, Speranza R, et al. Status epilepticus and COVID-19: A systematic review. Epilepsy Behav 2021;118:107887.
[Crossref] [Google Scholar] [PubMed]
- Taquet M, Sillett R, Zhu L, Mendel J, Camplisson I, Dercon Q, et al. Neurological and psychiatric risk trajectories after SARS-CoV-2 infection: an analysis of 2-year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatry 2022;9(10):815-27.
[Crossref] [Google Scholar] [PubMed]
- Misra UK, Tan CT, Kalita J. Viral encephalitis and epilepsy. Epilepsia 2008;49:13-8.
[Crossref] [Google Scholar] [PubMed]
- Kumar H, Gupta R. Neuroinvasion of Severe Acute Respiratory Syndrome Corona Virus-2 (SARS-CoV-2): Future risk of epilepsy. Int J Neurosci 2022;19:1-10.
[Crossref] [Google Scholar] [PubMed]
- Meinhardt J, Radke J, Dittmayer C, Franz J, Thomas C, Mothes R, et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci 2021;24(2):168-75.
[Crossref] [Google Scholar] [PubMed]
- Montalvan V, Lee J, Bueso T, de Toledo J, Rivas K. Neurological manifestations of COVID-19 and other coronavirus infections: A systematic review. Clin Neurol Neurosurg 2020;194:105921.
[Crossref] [Google Scholar] [PubMed]
- Nikbakht F, Mohammadkhanizadeh A, Mohammadi E. How does the COVID-19 cause seizure and epilepsy in patients? The potential mechanisms. Mult Scler Relat Disord 2020;46:102535.
[Crossref] [Google Scholar] [PubMed]
- Remsik J, Wilcox JA, Babady NE, McMillen TA, Vachha BA, Halpern NA, et al. Inflammatory leptomeningeal cytokines mediate COVID-19 neurologic symptoms in cancer patients. Cancer Cell 2021;39(2):276-83.
[Crossref] [Google Scholar] [PubMed]
- Agirman G, Yu KB, Hsiao EY. Signaling inflammation across the gut-brain axis. Science 2021;374(6571):1087-92.
[Crossref] [Google Scholar] [PubMed]
- Fujinami RS. Infiltrating macrophages are key to the development of seizures following virus infection. J Virol 2013;87(3):1849-60.
[Crossref] [Google Scholar] [PubMed]
- Varvel NH, Neher JJ, Bosch A, Wang W, Ransohoff RM, Miller RJ, et al. Infiltrating monocytes promote brain inflammation and exacerbate neuronal damage after status epilepticus. Proceed Natl Acad Sci 2016;113(38):E5665-74.
[Crossref] [Google Scholar] [PubMed]
- Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformat 2008;9(1):1-3.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25(8):1091-3.
[Crossref] [Google Scholar] [PubMed]
- Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59.
[Crossref] [Google Scholar] [PubMed]
- Yang C, Delcher C, Shenkman E, Ranka S. Machine learning approaches for predicting high cost high need patient expenditures in health care. Biomed Eng Online 2018;17(1):131.
[Crossref] [Google Scholar] [PubMed]
- Vejnar CE, Blum M, Zdobnov EM. miRmap web: Comprehensive microRNA target prediction online. Nucl Acids Res 2013;41(1):165-8.
[Crossref] [Google Scholar] [PubMed]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 2006;126(6):1203-17.
[Crossref] [Google Scholar] [PubMed]
- Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucl Acids Res 2015;43(1):146-52.
[Crossref] [Google Scholar] [PubMed]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 2007;13(11):1894-910.
- Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucl Acids Res 2020;48(1):148-54.
[Crossref] [Google Scholar] [PubMed]
- Chen B, Fan W, Liu J, Wu FX. Identifying protein complexes and functional modules-from static PPI networks to dynamic PPI networks. Brief Bioinform 2014;15(2):177-94.
[Crossref] [Google Scholar] [PubMed]
- Somani S, Pati S, Gaston T, Chitlangia A, Agnihotri S. de novo status epilepticus in patients with COVID-19. Ann Clin Transl Neurol 2020;7(7):1240-4.
[Crossref] [Google Scholar] [PubMed]
- Balloy G, Leclair-Visonneau L, Peéreon Y, Magot A, Peyre A, Mahe PJ, et al. Non-lesional status epilepticus in a patient with coronavirus disease-2019. Clin Neurophysiol 2020;131(8):2059-61.
- Vollono C, Rollo E, Romozzi M, Frisullo G, Servidei S, Borghetti A, et al. Focal status epilepticus as unique clinical feature of COVID-19: A case report. Seizure 2020;78:109-12.
[Crossref] [Google Scholar] [PubMed]
- Carroll E, Neumann H, Aguero-Rosenfeld ME, Lighter J, Czeisler BM, Melmed K, et al. Post-COVID-19 inflammatory syndrome manifesting as refractory status epilepticus. Epilepsia 2020;61(10):e135-9.
[Crossref] [Google Scholar] [PubMed]
- Laxer KD, Trinka E, Hirsch LJ, Cendes F, Langfitt J, Delanty N, et al. The consequences of refractory epilepsy and its treatment. Epilepsy Behav 2014;37:59-70.
[Crossref] [Google Scholar] [PubMed]
- Desforges M, Le CA, Dubeau P, Bourgouin A, Lajoie L, Dube M, et al. Human coronaviruses and other respiratory viruses: Underestimated opportunistic pathogens of the central nervous system? Viruses 2019;12(1):14.
[Crossref] [Google Scholar] [PubMed]
- Steardo L, Steardo Jr L, Zorec R, Verkhratsky A. Neuroinfection may contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol (Oxford, England). 2020;229(3):e13473.
[Crossref] [Google Scholar] [PubMed]
- van Vliet EA, da Costa Araujo S, Redeker S, van Schaik R, Aronica E, Gorter JA. Blood-brain barrier leakage may lead to progression of temporal lobe epilepsy. Brain 2007;130(2):521-34.
[Crossref] [Google Scholar] [PubMed]
- Rana A, Musto AE. The role of inflammation in the development of epilepsy. J Neuroinflammat 2018;15:1-2.
- Vezzani A, Fujinami RS, White HS, Preux PM, Blumcke I, Sander JW, et al. Infections, inflammation and epilepsy. Acta Neuropathol 2016;131:211-34.
[Crossref] [Google Scholar] [PubMed]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497-506.
[Crossref] [Google Scholar] [PubMed]
- Varatharaj A, Galea I. The blood-brain barrier in systemic inflammation. Brain Behav Immun 2017;60:1-2.
[Crossref] [Google Scholar] [PubMed]
- Basile MS, Cavalli E, McCubrey J, Hernández-Bello J, Muñoz-Valle JF, Fagone P, et al. The PI3K/Akt/mTOR pathway: A potential pharmacological target in COVID-19. Drug Discov Today 2022;27(3):848-56.
[Crossref] [Google Scholar] [PubMed]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010;11(5):329-41.
[Crossref] [Google Scholar] [PubMed]
- Ostendorf AP, Wong M. mTOR inhibition in epilepsy: Rationale and clinical perspectives. CNS Drugs 2015;29:91-9.
[Crossref] [Google Scholar] [PubMed]
- Cardamone M, Flanagan D, Mowat D, Kennedy SE, Chopra M, Lawson JA. Mammalian target of rapamycin inhibitors for intractable epilepsy and subependymal giant cell astrocytomas in tuberous sclerosis complex. J Pediatr 2014;164(5):1195-200.
[Crossref] [Google Scholar] [PubMed]
- Wang YF, Kelly CG, Singh M, McGowan EG, Carrara AS, Bergmeier LA, et al. Stimulation of Th1-polarizing maturation of dendritic cells, cytokines, CC chemokines, and adjuvant function by the peptide binding fragment of heat shock protein 70. J Immunol 2002;169(5):2422-9.
[Crossref] [Google Scholar] [PubMed]
- Dodge GR, Kovalszky I, McBride OW, Yi HF, Chu ML, Saitta B, et al. Human Clathrin heavy Chain (CLTC): Partial molecular cloning, expression, and mapping of the gene to human chromosome 17q11-qter. Genomics 1991;11(1):174-8.
[Crossref] [Google Scholar] [PubMed]
- Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 2018;19(5):313-26.
- Nabais Sá MJ, Venselaar H, Wiel L, Trimouille A, Lasseaux E, Naudion S, et al. de novo CLTC variants are associated with a variable phenotype from mild to severe intellectual disability, microcephaly, hypoplasia of the corpus callosum, and epilepsy. Genet Med 2020;22(4):797-802.
[Crossref] [Google Scholar] [PubMed]
- Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med 2010;12(7):385-95.
[Crossref] [Google Scholar] [PubMed]
- Sun AX, Yuan Q, Fukuda M, Yu W, Yan H, Lim GG, et al. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science 2019;366(6472):1486-92.
[Crossref] [Google Scholar] [PubMed]
- Ozkara C, Altintas A, Yilmaz E, Eskazan E, Erkol G, Ozyurt E, et al. An association between mesial temporal lobe epilepsy with hippocampal sclerosis and human leukocyte antigens. Epilepsia 2002;43(3):236-9.
[Crossref] [Google Scholar] [PubMed]
- Augusto DG, Hollenbach JA. HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection. Curr Opin Immunol 2022;76:102178.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Guan F, Miller H, Byazrova MG, Candotti F, Benlagha K, et al. The role of dendritic cells in COVID-19 infection. Emerg Microbes Infect 2023;12(1):2195019.
- Laing AG, Lorenc A, Del Molino, Del Barrio I, Das A, Fish M, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med 2020;26(10):1623-35.
[Crossref] [Google Scholar] [PubMed]
- Reis e Sousa C. Dendritic cells in a mature age. Nat Rev Immunol 2006;6(6):476-83.
[Crossref] [Google Scholar] [PubMed]
- Colton CA. Immune heterogeneity in neuroinflammation: Dendritic cells in the brain. J Neuroimmun Pharmacol 2013;8:145-62.
- Liu JQ, Nishimura H. Estimating the dissipative factors of synaptic exocytosis in Drosophila using a filter based reverse engineering method. Nano Commun Networks 2017;11:1-10.
- Khodaverdian S, Dashtban-Moghadam E, Dabirmanesh B, Mirnajafi-Zadeh J, Taleb M, Khajeh K, et al. CD38 and MGluR1 as possible signaling molecules involved in epileptogenesis: A potential role for NAD+homeostasis. Brain Res 2021;1765:147509.
[Crossref] [Google Scholar] [PubMed]
- Horenstein AL, Faini AC, Malavasi F. CD38 in the age of COVID-19: A medical perspective. Physiol Rev 2021;101(4):1457-86.
[Crossref] [Google Scholar] [PubMed]
- Qin X, Huang C, Wu K, Li Y, Liang X, Su M, et al. Anti-Coronavirus Disease-2019 (COVID-19) targets and mechanisms of puerarin. J Cell Mol Med 2021;25(2):677-85.
[Crossref] [Google Scholar] [PubMed]
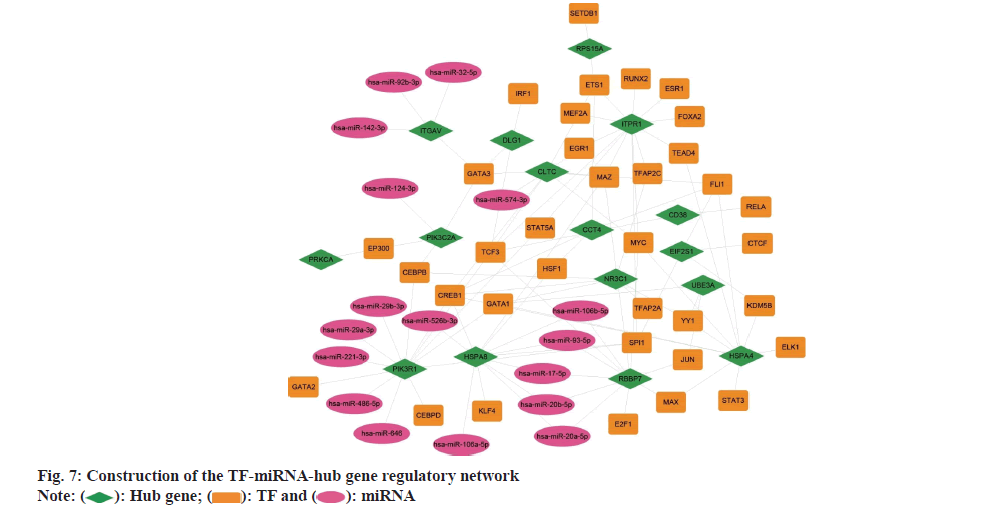
 ): Hub gene; (
): Hub gene; ( ): TF and (
): TF and ( ): miRNA
): miRNA



