- *Corresponding Author:
- J. Caroline Rose
Department of Biotechnology, Arignar Anna Arts and Science College, Krishnagiri, Tamil Nadu 635001, India
E-mail: jcarolinerose@gmail.com
| Date of Received | 21 March 2023 |
| Date of Revision | 07 July 2023 |
| Date of Acceptance | 08 May 2024 |
| Indian J Pharm Sci 2024;86(3):872-881 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Trichophyton rubrum is the major causative organism of dermatophytosis. The methanolic extracts of Pimpinella anisum seed has shown in vitro anti-dermatophytic activity. In the present study, the active compounds present in Pimpinella anisum seed-methanolic extracts was tentatively identified by liquid chromatography/electrospray ionization tandem mass spectrometry. Further molecular docking analysis of these active compounds with Trichophyton rubrum proteins aspartate-beta (β)-semialdehyde dehydrogenase and sialidase was studied. Liquid chromatography/electrospray ionization tandem mass spectrometry analysis revealed the presence of different class of compounds such as polyphenols, flavonoids, tannins, phenolic acids, phenolic acid derivative, fatty acids and lignan. In silico molecular docking analysis of these compounds with Trichophyton rubrum proteins aspartate-β-semialdehyde dehydrogenase revealed the top five compounds with lowest binding energy were phelligridin I (-10.1 kcal/mol), pectolinarin (-9.6 kcal/mol), fortunellin (-9.5 kcal/mol), epigallocatechin gallate (-9.4 kcal/mol) and kaempferol-3-O-glucuronide (-9.2 kcal/mol). In sialidase, the compounds phelligridin I (-10.6 kcal/mol), fortunellin (-9.5 kcal/mol), chicoric acid (-9.2 kcal/mol), epigallocatechin gallate (-9.1 kcal/mol) and kaempferol-3-O-glucuronide (-8.6 kcal/mol) bound with lowest binding energy. All these compounds were found to orient in the active site forming interactions with amino acids involved in catalysis of these proteins. The proteins aspartate-beta-semialdehyde dehydrogenase and sialidase are considered important drug target of Trichophyton rubrum. In silico analysis has shown promising results hence, these compounds identified in the present study might be further studied for its in vitro anti-dermatophytic activity. Also, the results from the present study clearly indicate that the active compounds present in Pimpinella anisum seed-methanolic extracts through its interaction with proteins present in Trichophyton rubrum might have shown in vitro anti-dermatophytic activity.
Keywords
Pimpinella anisum, Trichophyton rubrum, aspartate-β-semialdehyde dehydrogenase, sialidase
The superficial fungal infection called dermatophytosis is limited to the stratum corneum of the epidermis, or to the hair and nails. The infection caused by fungal dermatophytes involves coordinated process such as adhesion, penetration and colonization of keratinized tissues[1]. Dermatophytosis is an important public health problem worldwide particularly in developing countries[2]. Although the disease hardly causes death, it affects the quality of life and it is responsible for high morbidity[3,4]. Onychomycosis, a form of dermatophytosis, is a fungal infection of the fingernails or toenails. Onychomycosis is common in older people and it is related to peripheral vascular disease, immunologic disorders and diabetes mellitus. Onychomycosis is difficult to treat and might lead to cellulites and foot ulcer[5-7]. Trichophyton rubrum (T. rubrum) is the major dermatophyte fungus as well as the most prevalent pathogen that caused human dermatophytoses, which accounts for about 70 % of the total dermatophyte infections[8-10]. This organism may remain viable in the environment for over 6 mo, thus accounting for widespread of infections. Transmission of T. rubrum infection occurs often from person to person through either by shedding of infected skin cells and hair or by direct body contact[11]. Also, the widespread of infections may occur due to viability of T. rubrum in the environment for over 6 mo[12].
At present allylamines, polyenes, echinocandins, azoles and other agents, such as griseofulvin and 5-flucytosine are the antifungals used widely in clinical treatment[8,13,14]. Allyamine (terbinafine) interfere with ergosterol biosynthesis through inhibition of squalene epoxidase. Such lack in ergosterol would affect the fungal cell wall synthesis thereby its growth[15]. Azole (fluconazole) inhibits the cytochrome 450 sterol 14 a-demethylase and affects the conversion of lanosterol into ergosterol. This leads to the disruption of fungal membranes and increase the phospholipids accumulation within the cell[16]. Other than these, spinosins target β-1,3-glucan synthase enzyme of the fungus[17]. Although these antifungal agents are available, the cases of deep infection are increasing in immunosuppressed and immunocompromised patients[18,19]. Also, an increasing number of cases of resistant dermatophyte infections have been reported. In particular, T. rubrum is most persistently described in resistance to standard treatments[20]. Long-term and discontinuing treatments favors resistant acquisition in fungus. The biochemical mechanisms for such resistance to available drugs are point mutations, alteration in drug target sites and increased expression as well as activity of efflux transporter[8]. Also, cross-resistance has also been reported in T. rubrum, that is terbinafine-resistant isolate was found to be resistant to azoles due to overexpression of the multidrug efflux transporter[21]. Hence, identification of novel drugs against dermatophytes are ever increasing. Especially, the search for natural compounds is considered advantageous to overcome multidrug resistant with less side-effects[22].
Currently, there is a lot of activity in the quest for natural products with novel uses, particularly in relation to pest management. Plant extracts with antimicrobial characteristics and a range of secondary metabolites, such as alkaloids, quinones, flavonoids, glycosides, saponins, tannins and terpenoids have gained attention in the field of plant disease prevention. These extracts include aromatic and medicinal plants. Pimpinella anisum is the plant belonging to the Umbelliferae family and its seeds are reported for various biological activities such as antidiabetics, antioxidant, antispasmodic action and antimicrobial effect[22]. Among the many hundreds of soil-borne microorganisms that cause plant diseases, Fusarium oxysporum, Sclerotium rolfsii, and Macrophomina phaseolina are the most prevalent fungal infections.
Previous study has shown the anti-dermatophyte effect of Pimpinella anisum Seed (PAS) extract. Also, the effective treatment for recalcitrant infection of T. rubrum might be combinatorial therapy of available drugs with natural compounds. Accordingly, previous report has shown the synergistic effect of essential oil prepared from PAS along with terbinafine[23]. In addition, our previous study has shown the antifungal activity of PAS-Methanol Extract (PAS-ME) against T. rubrum. However, the active constituents present in PAS-ME and its target protein in T. rubrum has not been determined. Keeping these facts in view, in the present study PAS-ME was prepared and the active compounds present was tentatively identified using Liquid Chromatography/Electrospray Ionization tandem Mass Spectrometry (LC-ESI/MS/MS).
The ability to multiply numerous analytes within a single analytical run at a low incremental cost is a benefit of LC-MS tests. This might make laboratory setup simpler and provide additional valuable information, including metabolite profiles. Further, in silico molecular docking analysis was performed to understand the interaction of identified compounds present in Pimpinella anisum Methanolic Extract (PAME) with T. rubrum proteins, Aspartate-β-Semialdehyde Dehydrogenase (ASADH) and sialidase. Till date, drugs against various other T. rubrum proteins have been identified, however multidrug resistant organisms were developed due to point mutations in these proteins thereby decreasing the affinity of these drugs towards these proteins[8]. Hence, in the present study the potential compounds identified in PAME are studied against these new target proteins of T. rubrum.
Materials and Methods
Materials:
Methanol was purchased from Merck, India.
Preparation of PAME:
Seeds of Pimpinella anisum were collected from Hosur and its associated Ghats region, Krishnagiri District, Tamil Nadu, India. The seeds were shade dried, powdered and stored in dry air-tight containers in dark place. About 10 g of powdered seed sample was extracted using methanol for 8 h in Soxhlet apparatus. The methanol was evaporated using rotary evaporator and PAME obtained was used for compound identification.
LC-ESI/MS/MS:
LC-ESI-MS/MS analysis was performed using the column XSelect CSHT C18 2.5 μm in Xevo TQ-S micro Triple Quadrupole Mass Spectrometry (Waters, USA). The gradient mobile phase consists of 2 solvents, 0.1 % Formic Acid (FA) in water and acetonitrile was used. The injection volume was 2 µl. Spectra was recorded in ESI positive mode between m/z 100 and 1000.
Molecular docking analysis:
The structures of compounds were retrieved from PubChem. The three-dimensional crystal structure of T. rubrum proteins, ASADH (PDB ID:4ZHS) and sialidases (PDB ID: 7P1U) were retrieved from protein data bank[24,25]. The ligand and proteins were prepared using AutoDock tools 1.5.6[26]. Docking study was carried out by AutoDock Vina version 1.1.2[27]. The PyMol software and Discovery Studio were used to analyze the intermolecular interactions between the ligand and protein.
Results and Discussion
The compounds identified in PAS-ME at different retention time using LC-ESI-MS/MS analysis were given in Table 1. The results showed the presence of compounds from various groups such as polyphenols (ligstroside), flavonoids (kaempferol-3-O-neohesperidoside, kaempferol-3-O-galactoside, kaempferol-3-O-glucoside, kaempferol-3-O-glucuronide (KGR), kaempferol-3-O-rhamnoside, isorhamnetin-3-O-arabinose, luteolin-7,3-dimethyl ether, kaempferol, naringenin, isoschaftoside, myricetin, fortunellin, pectolinarin and swertisin), tannins (Epigallocatechin Gallate (EGCG) and 5-galloylquinic acid), phenolic acids (rosmarinic acid, chlorogenic acid and feruloyl glucose), phenolic acid derivative (1,3-O-dicoumaroylglycerol and 1-O-p-coumaroyl-3-O-caffeoylglycerol), fatty acid (1,4-epoxy-1-methoxy-8,13-diacetoxy-10-hydroxygermacra-5(E),7(11)-dien-6,12-olide and linoleic acid), lignan (chrysoeriol-7-O-neohesperidoside and secoisolariciresinol) and other compounds (esculin and phelligridin I).
| Retention time | Predicted compound | Molecular weight | m/z [M]/[M+H] |
|---|---|---|---|
| 5.2 | Ligstroside | 524 | 523.92 |
| 5.529 | Kaempferol-3-O-neohesperidoside | 594 | 595.1 |
| 6.009 | Rosmarinic acid | 360 | 361.22 |
| 6.303 | Phelligridin I | 624 | 625.13 |
| 6.54 | Chlorogenic acid | 354 | 355.5 |
| 6.798 | Chrysoeriol-7-O-neohesperidoside | 608 | 609.1 |
| 6.927 | Kaempferol-3-O-galactoside | 448 | 449.23 |
| 7.049 | Kaempferol-3-O-glucoside | 448 | 449 |
| 7.443 | KGR | 462 | 463.44 |
| 7.6 | Kaempferol-3-O-rhamnoside | 432 | 434.17 |
| 8.21 | Isorhamnetin -3-O-arabinose | 448 | 449.02 |
| 8.647 | Luteolin-7,3'-dimethyl ether | 314 | 315.21 |
| 9.12 | Chicoric acid | 474 | 475.55 |
| 9.313 | Kaempferol | 286 | 288.25 |
| 10.395 | Naringenin | 272 | 272.21 |
| 11.686 | Isoschaftoside | 564 | 565.29 |
| 11.886 | Myricetin | 318 | 319.14 |
| 12.481 | Naringenin | 272 | 274.24 |
| 12.811 | Kaempferol | 286 | 288.25 |
| 12.976 | Fortunellin | 592 | 593.21 |
| 13.291 | Fortunellin | 592 | 593 |
| 14.309 | Pectolinarin | 622 | 623.31 |
| 14.588 | Pectolinarin | 622 | 623.31 |
| 15.176 | Swertisin | 446 | 447.34 |
| 15.706 | Esculin | 340 | 341.19 |
| 16.695 | 5-Galloylquinic acid | 344 | 344.28 |
| 16.881 | Chlorogenic acid | 354 | 355.27 |
| 18.078 | Linoleic acid | 281 | 282.23 |
| 18.286 | 1,3-O-Dicoumaroylglycerol | 384 | 385.38 |
| 18.566 | 1-O-p-Coumaroyl-3-O-caffeoylglycerol | 400 | 401.34 |
| 19.168 | EGCG | 458 | 458.05 |
| 20.701 | Feruloyl glucose | 356 | 357.16 |
| 21.905 | 1,4-epoxy-1-methoxy-8,13-diacetoxy-10-hydroxygermacra-5(E),7(11)-dien-6,12-olide | 410 | 411.49 |
| 22.579 | Secoisolariciresinol | 362 | 363.39 |
Table 1: Tentative Identification of Compounds Presents in PAS-ME by LC-MS/MS analysis
The binding energy of compounds present in PAS-ME with T. rubrum ASADH is given in Table 2. The top five compounds with lowest binding energy were found to be phelligridin I (-10.1 kcal/ mol), pectolinarin (-9.6 kcal/mol), fortunellin (-9.5 kcal/mol), EGCG (9.4 kcal/mol) and KGR (-9.2 kcal/mol).
| Name of the ligands | Dock scores (kcal/mol) | |
|---|---|---|
| ASADH (PDB ID: 4ZHS) | Sialidase (PDB ID: 7P1U) | |
| Ligstroside | -8.2 | -8 |
| Kaempferol-3-O-neohesperidoside | -7.4 | -8.5 |
| Rosmarinic acid | -7.9 | -7.9 |
| Phelligridin I | -10.1 | -10.6 |
| Chlorogenic acid | -7.9 | -8.5 |
| Chrysoeriol-7-O-neohesperidoside | -9 | -8.4 |
| Kaempferol-3-O-galactoside | -7.4 | -7.1 |
| Kaempferol-3-O-glucoside | -8.3 | -7.5 |
| KGR | -9.2 | -8.6 |
| Kaempferol-3-O-rhamnoside | -7.8 | -7.3 |
| Isorhamantin-3-O-arabinoside | -7.6 | -8 |
| Luteolin-7,3'-dimethyl ether | -7.9 | -7.4 |
| Chicoric acid | -8.4 | -9.2 |
| Kaempferol | -7.9 | -7.5 |
| Naringenin | -7.8 | -7.4 |
| Isoschaftoside | -7.8 | -8.3 |
| Myricetin | -8 | -8.1 |
| Fortunellin | -9.5 | -9.5 |
| Pectolinarin | -9.6 | -8.5 |
| Swertisin | -8.8 | -7.4 |
| Esculin | -8.2 | -7.8 |
| 5-Galloylquinic acid | -8.1 | -6.8 |
| Linoleic acid | -5.4 | -5 |
| 1-O-p-Coumaroyl-3-O-caffeoylglycerol | -8.2 | -7.8 |
| 1,3-O-dicoumaroylglycerol | -8.1 | -8.1 |
| EGCG | -9.4 | -9.1 |
| Feruloyl glucose | -8 | -7.3 |
| 1,4-epoxy-1-methoxy-8,13-diacetoxy-10-hydroxygermacra-5(E),7(11)-dien-6,12-olide | -7.1 | -6.8 |
| Secoisolariciresinol | -7.2 | -7.2 |
Table 2: Dock Score Obtained for Various Compounds Identified in PAS-ME by Molecular docking. The Dock Score of First Five Compounds with Lowest Binding Energy is Given in Bold
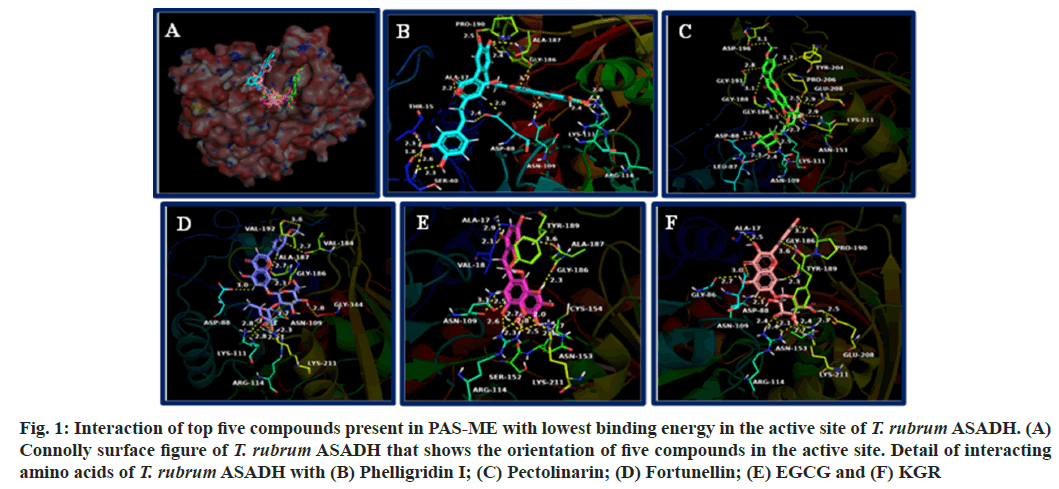
The connolly surface figure of T. rubrum ASADH that shows the orientation of top five compounds with lowest binding energy is shown in fig. 1A. The interaction of the compounds with amino acids of T. rubrum ASADH is shown in fig. 1B-fig. 1F. Detail of amino acids that forms hydrogen bonding, hydrophobic interaction and electrostatic interactions with the compounds are given in Table 3.
| Ligand | Interacting amino acid residues (Sialidase; PDB ID: 7P1U) | ||
|---|---|---|---|
| Hydrogen bond | Hydrophobic interaction | Electrostatic interaction | |
| Phelligridin | Arg62, Gly79, Asn85, Arg86, Asn127, Gln150, Arg173, Trp204, Arg267, Arg390 | Trp204, Ala206 | Asp87, Glu251, Arg267 |
| Fortunellin | Arg62, Asp87, Asn127, Asn154, Trp204, Arg267, Pro268, Gly269 | Arg86 | Arg86, Asp87 |
| Chicoric acid | Arg62, Asp87, Asn127, Arg324, Arg390 | - | Asp87, Glu251 |
| EG | Arg62, Asp87, Asn127, Gln150, Tyr360 | Trp204, Ala206, Arg267 | Asp87, Glu251 |
| KGR | Arg62, Asp87, Trp204, Arg267, Arg324, Arg390 | - | Asp87, Glu251, Arg267 |
Table 3: Details of Interaction between Compounds and Amino Acids Present in T. rubrum ASADH
Fig. 1: Interaction of top five compounds present in PAS-ME with lowest binding energy in the active site of T. rubrum ASADH. (A) Connolly surface figure of T. rubrum ASADH that shows the orientation of five compounds in the active site. Detail of interacting amino acids of T. rubrum ASADH with (B) Phelligridin I; (C) Pectolinarin; (D) Fortunellin; (E) EGCG and (F) KGR
The compounds formed hydrogen bonding with amino acids such as Thr15, Ala17, Ser40, Asp88, Asn109, Lys111, Arg114, Asn153, Gly186, Ala187, Gly188, Pro190, Gly191, Asp196, Tyr204, Pro206, Lys211 and Gly334. Hydrophobic interactions were formed with the amino acids Ala17, Val18, Lys55, Cys154, Val184, Gly188, Tyr189, Val192, Pro206 and Ala345. The compounds formed electrostatic interaction with amino acids Lys55, Asp88 and Lys211.
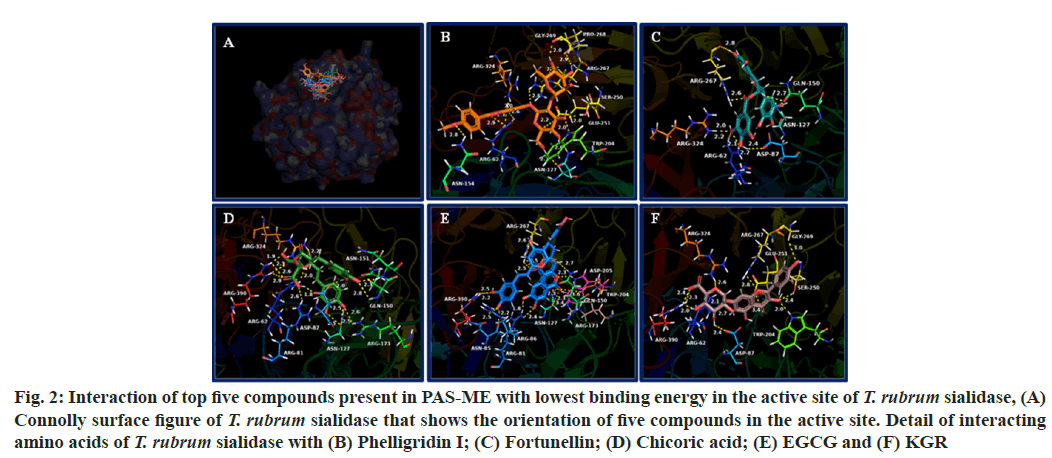
The binding energy of compounds present in PAS-ME with T. rubrum sialidase is given in Table 2. The top 5 compounds with lowest binding energy were found to be phelligridin I (-10.6 kcal/mol), fortunellin (-9.5 kcal/mol), chicoric acid (-9.2 kcal/mol), EGCG (-9.1 kcal/mol) and KGR (-8.6 kcal/mol). The orientation of top 5 compounds with lowest binding energy in the T. rubrum sialidase is shown in fig. 2A. The detail of interaction between the compounds and amino acids of T. rubrum sialidase is shown in fig. 2B-fig. 2F. Detail of interacting amino acids of T. rubrum sialidase through hydrogen bonding, hydrophobic interaction and electrostatic interactions are given in Table 4.
| Ligand | Interacting amino acid residues (ASADH; PDB ID: 4ZHS) | ||
|---|---|---|---|
| Hydrogen bond | Hydrophobic interaction | Electrostatic interaction | |
| Phelligridin | Thr15, Ala17, Ser40, Asp88, Asn109, Lys111, Arg114, Gly186, Ala187, Pro190 | Ala17, Val18, Lys55, Cys154, Val184, Ala345 | Lys55, Asp88 |
| Pectolinarin | Leu87, Asp88, Asn109, Lys111, Asn153, Gly186, Gly188, Gly191, Asp196, Tyr204, Pro206, Lys211 | Ala17, Val18, Tyr189, Val192, Pro206 | - |
| Fortunellin | Asp88, Asn109, Lys111, Arg114, Val184, Ala187, Gly186, Val192, Lys211, Gly334 | Ala17, Val18, Pro89, Val184, Tyr189 | - |
| EG | Ala17, Val18, Asn109, Lys111, Arg114, Ser152, Asn153, Cys154, Gly186, Ala187, Tyr189 | Pro89, Lys111, Cys154 | Lys211 |
| KGR | Ala17, Gly86, Asp88, Asn109, Arg114, Asn153, Gly186, Thr189, Pro190, Glu208, Lys211 | Ala17, Gly188 | Asp88 |
Table 4: Details of Interaction between Compounds and Amino Acids Present in T. rubrum Sialidase
Fig. 2: Interaction of top five compounds present in PAS-ME with lowest binding energy in the active site of T. rubrum sialidase, (A) Connolly surface figure of T. rubrum sialidase that shows the orientation of five compounds in the active site. Detail of interacting amino acids of T. rubrum sialidase with (B) Phelligridin I; (C) Fortunellin; (D) Chicoric acid; (E) EGCG and (F) KGR
The compounds formed hydrogen bonding with amino acids such as Arg62, Gly79, Asn85, Arg86, Asn127, Gln150, Arg173, Trp204, Arg267, Pro268, Gly269, Arg324, Tyr360 and Arg390. Hydrophobic interactions were formed with the amino acids Arg86, Trp204, Ala206 and Arg267. The compounds formed electrostatic interaction with amino acids Arg86, Asp87, Glu251 and Arg267.
To understand the molecular mechanism of antifungal activity of PAS-ME against T. rubrum, the active molecules in extract were identified and interaction of these identified compounds with proteins of T. rubrum was studied. Till date, the proteins such as squalene epoxidase, cytochrome 450 sterol 14 a-demethylase and β-1,3-glucan synthase of fungus are targeted by the antifungal drugs[8,13-17]. However, point mutations in these proteins as well as increased expression of drug efflux proteins has increased the incidence of fungal infection[8]. Also, the side-effects of available drugs are making it difficult to treat the dermatophyte and leads to deep infection[28]. In addition, fungi are eukaryotic organism like mammals, hence it is necessary to target the structure unique to fungi[29]. Hence, two new protein targets such as ASADH and sialidase have been targeted in the present study. Also, till date, the crystal structure of these proteins of T. rubrum are only available in protein data bank. Hence interaction of the compounds in PAS-ME with these proteins might give a definitive understanding of antifungal activity of these compounds against T. rubrum.
The fungal aspartate pathway is required for the biosynthesis of amino acids such as threonine, isoleucine and methionine. These are essential for fungal viability and is found to be indispensable[24]. Also, the aspartate pathway is involved in cell-wall biosynthesis, the protective dormancy process and virulence factor production by providing the source for these processes[24,30-32]. ASADH catalyze, the second reaction in aspartate pathway and does not have homologous in mammalian cells[29,33]. It has been found that ASADH expression is increased upon exposure of T. rubrum to human skin, suggesting its role in virulence of the fungus[34]. Inhibitors are targeting ASADH could act as potent antifungal agent against T. rubrum[24]. The amino acids Cys154 and His251 is considered catalytically important amino acids in T. rubrum ASADH. Further, the open loop structure is required for the proper binding of the substrate and catalytic activity of T. rubrum ASADH. The amino acids such as Gly188, Tyr189, Pro190 and Gly191 are important residues present in loop structure. The first five compounds with lowest binding energy in docking such as phelligridin-I, pectolinarin, fortunellin, EGCG and KGR oriented spanning the entire loop structure into the active site. Among these compounds, EGCG showed both hydrogen bonding and hydrophobic interaction with catalytic amino acid Cys154. As well as phelligridin formed hydrophobic interaction with Cys154. All the compounds interacted with amino acids present in loop structure either through hydrogen bonding and hydrophobic interaction or through both interactions. Such interaction with these compounds would hinder the binding the positioning of substrate for the catalytic function of the T. rubrum ASADH. In addition, it might also disturb the binding of coenzyme, Nicotinamide Adenine Dinucleotide Phosphate (NADP) to the enzyme thereby affecting its function.
The sialidase from T. rubrum prefer the sialic acid substrate, 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid (KDN); hence it is a sialidase (Kdnase)[25]. Although sialic acid is absent in T. rubrum, the C8 monosaccharide 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) was found in T. rubrum samples in a recent study[25]. Although the role of Kdo in T. rubrum virulence has yet to be determined, Kdo is a component of endotoxin lipopolysaccharide in bacteria[35]. In addition, it is crucial for growth and survival of microorganism. Other than T. rubrum, sialidase from Aspergillus fumigatus is also a Kdnase and its activity was not inhibited by the classical sialidase inhibitor, 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. Also, Kdnase from A. fumigatus was found to be important for fungal cell wall integrity and virulence[36]. In addition, Kdnase is not present in the host hence it may be potential target for the development of novel antifungal agents. In accordance, identifying compounds that could target sialidase from T. rubrum would inhibit its survival. The top five compounds with lowest binding energy with T. rubrum sialidase such as phelligridin I, fortunellin, chicoric acid, EGCG and KGR bound in the substrate, KDN binding site. In general, the fungal sialidase consists of key active site residues an arginine triad that interacts with the carboxylic acid group of KDN, a nucleophilic tyrosine, its associated general acid (Glu), and an acid/base (Asp). The compounds from PAS-ME formed hydrogen bonding, hydrophobic interaction or electrostatic interactions with amino acids such as Arg62, Asp87, Gln150, Glu250 and Arg267, which play significant role in the orientation of substrate KDN in the active site as well as in its catalysis[25,37]. Hence, the interaction of these compounds would inhibit the catalytic activity of T. rubrum Kdnase. Such inhibition would disturb the cell wall thereby the virulence of T. rubrum. The top scored compound phelligridin that interacted with both ASADH and sialidase has previously been found to inhibit neuraminidase (also known as sialidase) present in influenza viruses[38]. In addition, phelligridin were reported for anticancer, inhibition of oxidative stress, and anti-inflammatory activity (through inhibiting amyloid beta aggregation)[39-41]. Pectolinarin that interacted with T. rubrum ASADH was reported for beneficial effects such as antidepressant antidiabetic, antitumor, antiviral, anti-rheumatoid arthritis, analgesic, anti-inflammatory, hepatoprotective and neuroprotective activity[42]. Fortunellin, EGCG and KGR interacted with both ASADH and sialidase with lowest binding score was found to have various other biological activities such as anticancer, antidiabetic, antioxidant, antiviral, hepatoprotection and antiinflammation[43-46]. Chicoric acid that interacted with T. rubrum Kdnase is known for hepatoprotective, nephroprotective, neuroprotective, antioxidative and anti-inflammatory activity[47-49]. Other than these compounds, most of the compounds identified in PAS-ME interacted in the active site of both T. rubrum ASADH and Kdnase with low binding score. Hence, synergistically, these compounds might have shown toxicity against T. rubrum and may be further studied for potential anti-dermatophyte agents.
In conclusion, the active compounds present in PAS-ME were identified by LC-ESI-MS/MS analysis. The compounds identified belonged to various groups such as polyphenols, flavonoids, tannins, phenolic acids, phenolic acid derivatives, fatty acids and lignan. In silico molecular docking analysis of these compounds with T. rubrum proteins ASADH and KDNase revealed the compounds phelligridin I, pectolinarin, fortunellin, chicoric acid, EGCG and KGR to interact with these proteins with low binding energy. These compounds oriented in the active site forming interactions with amino acids involved in catalysis of these proteins. Since, the proteins ASADH and Kdnase is an important drug target to identify drug against T. rubrum, the compounds identified in the present study interacting with these proteins might be further studied for its in vitro anti-dermatophytic activity. Further, the molecular mechanism of PAS-ME for its anti-dermatophyte activity might be due the interaction of these active compounds present in the extract with these proteins.
Conflict of interests:
The authors declared no conflict of interests.
References
- Bitencourt TA, Neves-da-Rocha J, Martins MP, Sanches PR, Lang EA, Bortolossi JC, et al. StuA-regulated processes in the dermatophyte Trichophyton rubrum: Transcription profile, cell-cell adhesion, and immunomodulation. Front Cell Infect Microbiol 2021;11:643659.
[Crossref] [Google Scholar] [PubMed]
- Bitew A. Dermatophytosis: Prevalence of dermatophytes and non-dermatophyte fungi from patients attending Arsho advanced medical laboratory, Addis Ababa, Ethiopia. Dermatol Res Pract 2018;12(9):119-21.
[Crossref] [Google Scholar] [PubMed]
- Anuthara R, Midhun SJ, Mathew J. An in vitro and in silico study of anti-dermatophytic activity of gossypol from fruits of Thespesia populnea (L.) Sol. ex Correa. Asian Pac J Trop Biomed 2021;11(12):543-52.
- Sharma R, Adhikari L, Sharma RL. Recurrent dermatophytosis: A rising problem in Sikkim, a Himalayan state of India. Indian J Pathol Microbiol 2017;60(4):541-5.
[Crossref] [Google Scholar] [PubMed]
- Boyko EJ, Ahroni JH, Cohen V, Nelson KM, Heagerty PJ. Prediction of diabetic foot ulcer occurrence using commonly available clinical information: The seattle diabetic foot study. Diabetes care 2006;29(6):1202-7.
[Crossref] [Google Scholar] [PubMed]
- Gaziano R, Campione E, Iacovelli F, Marino D, Pica F, di Francesco P, et al. Antifungal activity of Cardiospermum halicacabum L.(Sapindaceae) against Trichophyton rubrum occurs through molecular interaction with fungal Hsp90. Drug Des Devel Ther 2018:2185-93.
[Crossref] [Google Scholar] [PubMed]
- Roujeau JC, Sigurgeirsson B, Korting HC, Kerl H, Paul C. Chronic dermatomycoses of the foot as risk factors for acute bacterial cellulitis of the leg: A case-control study. Dermatology 2004;209(4):301-7.
[Crossref] [Google Scholar] [PubMed]
- Martinez-Rossi NM, Peres NT, Bitencourt TA, Martins MP, Rossi A. State-of-the-art dermatophyte infections: Epidemiology aspects, pathophysiology, and resistance mechanisms. J Fungi 2021;7(8):629.
[Google Scholar] [PubMed]
- Verma SB, Panda S, Nenoff P, Singal A, Rudramurthy SM, Uhrlass S, et al. The unprecedented epidemic-like scenario of dermatophytosis in India: I. Epidemiology, risk factors and clinical features. Indian J Dermatol Venereol Leprol 2021;87(2):154-75.
[Crossref] [Google Scholar] [PubMed]
- Wang R, Huang C, Zhang Y, Li R. Invasive dermatophyte infection: A systematic review. Mycoses 2021;64(4):340-8.
[Crossref] [Google Scholar] [PubMed]
- Leung AK, Lam JM, Leong KF, Hon KL. Tinea corporis: An updated review. Drugs Context 2020;9.(12):819-68.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Ma L, Leng W, Liu T, Yu L, Yang J, et al. Analysis of the dermatophyte Trichophyton rubrum expressed sequence tags. BMC Genomics 2006;7:1-3.
[Crossref] [Google Scholar] [PubMed]
- K Mazu T, A Bricker B, Flores-Rozas H, Y Ablordeppey S. The mechanistic targets of antifungal agents: An overview. Mini Rev Med Chem 2016;16(7):555-78.
[Crossref] [Google Scholar] [PubMed]
- Xie H, Yang X, Lyu C, Ke C. Antifungal drugs and their mechanisms of action. Zhongguo Weishengtaixue Zazhi Chin J Microecol 2015;27(12):1477-88.
- Kathiravan MK, Salake AB, Chothe AS, Dudhe PB, Watode RP, Mukta MS, et al. The biology and chemistry of antifungal agents: A review. Bioorg Med Chem 2012;20(19):5678-98.
[Crossref] [Google Scholar] [PubMed]
- Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clin Microbiol Rev 1999;12(1):40-79.
[Crossref] [Google Scholar] [PubMed]
- Huang KX, Xia L, Zhang Y, Ding X, Zahn JA. Recent advances in the biochemistry of spinosyns. Appl Microbiol Biotechnol 2009;82:13-23.
[Crossref] [Google Scholar] [PubMed]
- Costa JE, Neves RP, Delgado MM, Lima-Neto RG, Morais VM, Coêlho MR. Dermatophytosis in patients with human immunodeficiency virus infection: Clinical aspects and etiologic agents. Acta Trop 2015;150:111-5.
[Crossref] [Google Scholar] [PubMed]
- Kershenovich R, Sherman S, Reiter O, Huss SR, Didkovsky E, Mimouni D, et al. A unique clinicopathological manifestation of fungal infection: A case series of deep dermatophytosis in immunosuppressed patients. Am J Clin Dermatol 2017;18:697-704.
[Crossref] [Google Scholar] [PubMed]
- Bontems O, Fratti M, Salamin K, Guenova E, Monod M. Epidemiology of dermatophytoses in Switzerland according to a survey of dermatophytes isolated in Lausanne between 2001 and 2018. J Fungi 2020;6(2):95.
[Crossref] [Google Scholar] [PubMed]
- Monod M, Feuermann M, Salamin K, Fratti M, Makino M, Alshahni MM, et al. Trichophyton rubrum azole resistance mediated by a new ABC transporter, TruMDR3. Antimicrob Agents Chemother 2019;63(11):10-128.
[Crossref] [Google Scholar] [PubMed]
- Sun W, Shahrajabian MH, Cheng Q. Anise (Pimpinella anisum L.), a dominant spice and traditional medicinal herb for both food and medicinal purposes. Cogent Biol 2019;5(1):1673688.
- Trifan A, Luca SV, Bostanaru AC, Brebu M, Jitareanu A, Cristina RT, et al. Apiaceae essential oils: Boosters of terbinafine activity against dermatophytes and potent anti-inflammatory effectors. Plants 2021;10(11):2378.
[Crossref] [Google Scholar] [PubMed]
- Li Q, Mu Z, Zhao R, Dahal G, Viola RE, Liu T, et al. Structural insights into the tetrameric state of aspartate-β-semialdehyde dehydrogenases from fungal species. Sci Rep 2016;6(1):21067.
[Crossref] [Google Scholar] [PubMed]
- Nejatie A, Steves E, Gauthier N, Baker J, Nesbitt J, McMahon SA, et al. Kinetic and structural characterization of sialidases (Kdnases) from ascomycete fungal pathogens. ACS Chem Biol 2021;16(11):2632-40.
[Crossref] [Google Scholar] [PubMed]
- Sanner MF. Python: A programming language for software integration and development. J Mol Graph Model 1999;17(1):57-61.
[Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Benitez LL, Carver PL. Adverse effects associated with long-term administration of azole antifungal agents. Drugs 2019;79(8):833-53.
[Crossref] [Google Scholar] [PubMed]
- Su H, Han L, Huang X. Potential targets for the development of new antifungal drugs. J Antibiot 2018;71(12):978-91.
[Crossref] [Google Scholar] [PubMed]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 2004;25(9):1389-403.
[Crossref] [Google Scholar] [PubMed]
- Ragkousi K, Eichenberger P, Van Ooij C, Setlow P. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J Bacteriol 2003;185(7):2315-29.
[Crossref] [Google Scholar] [PubMed]
- van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep 2001;18(5):503-19.
[Crossref] [Google Scholar] [PubMed]
- Bareich DC, Nazi I, Wright GD. Simultaneous in vitro assay of the first four enzymes in the fungal aspartate pathway identifies a new class of aspartate kinase inhibitor. Chem Biol 2003;10(10):967-73.
[Crossref] [Google Scholar] [PubMed]
- Liu T, Xu X, Leng W, Xue Y, Dong J, Jin Q. Analysis of gene expression changes in Trichophyton rubrum after skin interaction. J Med Microbiol 2014;63(5):642-8.
[Crossref] [Google Scholar] [PubMed]
- Lodowska J, Wolny D, Węglarz L. The sugar 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) as a characteristic component of bacterial endotoxin-a review of its biosynthesis, function, and placement in the lipopolysaccharide core. Can J Microbiol 2013;59(10):645-55.
[Crossref] [Google Scholar] [PubMed]
- Nesbitt JR, Steves EY, Schonhofer CR, Cait A, Manku SS, Yeung JH, et al. The Aspergillus fumigatus sialidase (Kdnase) contributes to cell wall integrity and virulence in amphotericin B-treated mice. Front Microbiol 2018;8:327451.
[Crossref] [Google Scholar] [PubMed]
- Warwas ML, Yeung JH, Indurugalla D, Mooers AO, Bennet AJ, Moore MM. Cloning and characterization of a sialidase from the filamentous fungus, Aspergillus fumigatus. Glycoconj J 2010;27:533-48.
[Crossref] [Google Scholar] [PubMed]
- Kim JY, Kim DW, Hwang BS, Woo EE, Lee YJ, Jeong KW, et al. Neuraminidase inhibitors from the fruiting body of Phellinus igniarius. Mycobiology 2016;44(2):117-20.
[Crossref] [Google Scholar] [PubMed]
- Kim JE, Takanche JS, Yun BS, Yi HK. Anti‐inflammatory character of Phelligridin D modulates periodontal regeneration in l ipopolysaccharide‐induced human periodontal ligament cells. J Periodontal Res 2018;53(5):816-24.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Zhou Y, Wu J, Li J, Yao H. Phelligridin D from inonotus obliquus attenuates oxidative stress and accumulation of ECM in mesangial cells under high glucose via activating Nrf2. J Nat Med 2021;75(4):1021-9.
[Crossref] [Google Scholar] [PubMed]
- Mo S, Wang S, Zhou G, Yang Y, Li Y, Chen X, et al. Phelligridins C-F: cytotoxic pyrano [4, 3-c][2] benzopyran-1, 6-dione and furo [3, 2-c] pyran-4-one derivatives from the fungus Phellinus igniarius. J Nat Prod 2004;67(5):823-8.
- Patel DK. Biological importance, therapeutic benefit and analytical aspects of bioactive flavonoid pectolinarin in the nature. Drug Metab Lett 2021;14(2):117-25.
[Crossref] [Google Scholar] [PubMed]
- Alam M, Ali S, Ashraf GM, Bilgrami AL, Yadav DK, Hassan MI. Epigallocatechin 3-gallate: From green tea to cancer therapeutics. Food Chem 2022;379:132135.
[Crossref] [Google Scholar] [PubMed]
- Deng Y, Ma J, Weng X, Wang Y, Li M, Yang T, et al. Kaempferol-3-O-glucuronide ameliorates non-alcoholic steatohepatitis in high-cholesterol-diet-induced larval zebrafish and HepG2 cell models via regulating oxidation stress. Life 2021;11(5):445.
[Crossref] [Google Scholar] [PubMed]
- Xiong Y, Qiu J, Li C, Qiu Y, Guo L, Liu Y, et al. Fortunellin-induced modulation of phosphatase and tensin homolog by MicroRNA-374a decreases inflammation and maintains intestinal barrier function in colitis. Front Immunol2018;9:83.
[Crossref] [Google Scholar] [PubMed]
- Zhao C, Zhang Y, Liu H, Li P, Zhang H, Cheng G. Fortunellin protects against high fructose-induced diabetic heart injury in mice by suppressing inflammation and oxidative stress via AMPK/Nrf-2 pathway regulation. Biochem Biophys Res Commun 2017;490(2):552-9.
[Crossref] [Google Scholar] [PubMed]
- Ding X, Jian T, Li J, Lv H, Tong B, Li J, et al. Chicoric acid ameliorates nonalcoholic fatty liver disease via the AMPK/Nrf2/NFκB signaling pathway and restores gut microbiota in high-fat-diet-fed mice. Oxid Med Cell Longev 2020;63(2):56-9.
[Crossref] [Google Scholar] [PubMed]
- Li Z, Feng H, Han L, Ding L, Shen B, Tian Y, et al. Chicoric acid ameliorate inflammation and oxidative stress in lipopolysaccharide and d‐galactosamine induced acute liver injury. J Cell Mol Med 2020;24(5):3022-33.
[Crossref] [Google Scholar] [PubMed]
- Wang N, Li R, Feng B, Cheng Y, Guo Y, Qian H. Chicoric acid prevents neuroinflammation and neurodegeneration in a mouse Parkinson’s disease model: Immune response and transcriptome profile of the spleen and colon. Int J Mol Sci 2022;23(4):2031.
[Crossref] [Google Scholar] [PubMed]