- *Corresponding Author:
- Song Yunxi
Department of Respiratory and Intensive Care Medicine
People's Liberation Army (PLA) Rocket Force Characteristics Medical Center
Beijing 100088, China
E-mail: songofcloudy@163.com
| Date of Received | 29 June 2021 |
| Date of Revision | 24 April 2022 |
| Date of Acceptance | 12 September 2022 |
| Indian J Pharm Sci 2022;84(5):1269-1278 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Lung cancer is one of the top ten malignant tumors, seriously endangering people’s lives and health, with morbidity and mortality ranking first. Molecular targeted therapy overcomes the shortcomings of traditional chemotherapy. In recent years, the application of tumor therapy has become wider and wider. Gefitinib is the most successful molecular targeted drug for the treatment of lung cancer in recent years. The purpose of this article is to explore the establishment method of gefitinib-resistant lung cancer cell lines and the mechanism of epidermal growth factor receptor signaling pathway change. The method of gradually increasing the concentration of gefitinib was used to successfully establish NC-H1985 gefitinibresistant cells. The strains were also analyzed for epidermal growth factor receptor, messenger ribonucleic acid expression at different times using the anti-tubercular treatment method. The results show that this cell line has a certain difference compared to the parental NC-H1985 cell. With the extension of the culture time, the ribonucleic acid concentration decreased by 51 % and the 18S ribosomal ribonucleic acid, cycle threshold value increased by 39 %. In addition, the higher the relative expression of messenger ribonucleic acid in gefitinib-resistant lung cancer cells, the epidermal growth factor receptor signaling pathway is likely to change. From 24 h to 72 h, the relative expression of messenger ribonucleic acid has increased by 37.5 %. After gefitinib resistance culture of NC-H1985 gefitinib-resistant cells were 44.1 % in resting phase, 36.9 % in growth 1 phase, 11.4 % in synthesis phase, 4.3 % in growth 2 phase and 3.3 % in mitotic phase.
Keywords
Gefitinib, lung cancer cell line, epidermal growth factor, receptor, signaling pathway, chemotherapy, carcinoma
Lung cancer is one of the ten major malignant tumors, seriously harming people’s lives and health, with the highest morbidity and mortality, accounting for about 10 % of total cases and 20 % of total cancer deaths, respectively. Surgical resection is the most effective treatment, but most patients are in the middle or late stage of the disease when they find the tumor and even patients who undergo surgical treatment often require chemotherapy. Therefore, chemotherapy is still the most widely used method in the clinical treatment of lung cancer. However, traditional chemotherapeutic drugs are clinically restricted due to their poor specificity and toxicity, while molecular targeted therapy overcomes the deficiencies of traditional chemotherapeutics and has been widely used in cancer treatment in recent years.
Gefitinib (ZD1839) is an Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor (EGFR-TKI) and is the most successful molecular targeted drug for the treatment of lung cancer. It can actively inhibit the activation of EGFR and its transduction system, prevent the activation of Rat sarcoma virus/Rapidly Accelerated Fibrosarcoma/Mitogen-Activated Protein Kinase (RAS/ RAF/MAPK) transduction system and thus inhibit the activity of EGFR tyrosine kinase[1]. Studies have shown that Non-Small-Cell Lung Carcinoma (NSCLC) that are sensitive to gefitinib usually have EGFR16, 19 and 23 exon mutations and these mutations are mainly found in scan of female and non-smoking lung cancer patients[2]. In contrast, NSCLC for gefitinib usually carries mutations in EGFR exon 19 or k-RAS, B-RAF and certain exons of other genes. It is worth noting that with the delay of gefitinib treatment, some lung cancers initially sensitive to this drug will also develop acquired resistance, which is also the main reason for the failure of the drug treatment. The mechanism of drug action during traditional drug administration includes up regulation of pro drug-related protein expression, abnormal mismatch repair function, enhanced cellular drug efflux and detoxification, and changes in drug targets in cell groups[3]. However, the mechanism of gefitinib acquired resistance may be difficult to explain with the resistance mechanism of traditional chemotherapy. According to reports, the mechanism of acquired resistance of gefitinib in lung cancer mainly includes the following aspects: The secondary mutation of exon EGFR20 causes the conformational change of EGFR, causing gefitinib to lose its target. The exon mutation of the K-RAS and/or B-RAF gene leads to the self-activation or sensitivity of the molecules downstream of the signal transduction pathway and the cancer tissue loses its epithelial tissue characteristics through epithelial-mesenchymal transition, causing the loss of the drug its goal.
This article discusses the establishment of gefitinib resistant lung cancer cell lines and the study of EGFR signaling pathway changes. Among them, Suzuki analyzed the main chemotherapy used in the treatment of lung cancer and pointed out the limitations of molecular targeted therapy. The relationship between gefitinib-resistant micro Ribonucleic Acid (miRNA) expression and kinase signaling was studied. MiR-205 was significantly increased in fentin-resistant cell lines and certain kinase inhibitors were also found to inhibit miR-205 activity in gefitinib-resistant cell lines[4]. Dai established a NSCLC cell line H292 sensitive to gefitinib in the study and screened and analyzed the differentially expressed circular RNA before and after drug resistance and tested the proliferation of cell lines by gefitinib by Cell Counting Kit-8 (CCK-8) analysis inhibition[5]. In the article, Zhang pointed out the key role of EGFR in the treatment of lung cancer and acquired resistance to gefitinib is also very common. A compound called 244-MPT was found through research, which inhibits the growth and colony formation of gefitinib-sensitive or drug-resistant lung cancer cells[6]. Cai clearly pointed out in its research that about 50 % of Asian lung cancer patients will have EGFR mutations, so they are sensitive to EGFR-TKI because patients' EGFR secondary mutations were obtained and resistance to TKI such as gefitinib was obtained[7]. In the study, Lee described that patients with NSCLC with mutations in the EGFR benefited from the treatment of reversible EGFR-TKI such as gefitinib, but may be due to the new type of sexual resistance mechanism cannot significantly improve the overall survival rate of lung cancer patients with EGFR mutation[8].
The main research content of this article is the establishment of gefitinib-resistant lung cancer cell lines and the study of EGFR signaling pathway changes. Based on the research of previous scholars, this paper has made some innovations and the innovations are roughly as follows; the first point is that this article has gradually established the method by gradually increasing the concentration of gefitinib and successfully established the NC-H1985 gefitinib resistant cell line. The second point is to use the Anti- Tubercular Treatment (ATT) method to analyze the expression of EGFR, messenger RNA (mRNA) at different times, strictly ensuring the authenticity and reliability of the experimental data. The third point is the use of a new type of statistical-related software. The experimental data is analyzed using Statistical Package for the Social Sciences (SPSS) 13.0 statistical software. The data of the detection indicators are described by the standard deviation (x±s). NC-H1985 cells are in each gene. The difference in mRNA level expression was determined by t test between two independent samples.
Theoretical Study on Gefitinib-Resistant Lung Cancer Cell Lines and EGFR Signaling Pathway
Advantages and limitations of gefitinib in the treatment of lung cancer:
To establish a gefitinib-resistant lung cancer cell line, it is first necessary to understand the advantages of gefitinib over other lung cancer treatments and the limitations of gefitinib. Gefitinib can shrink the tumors of patients with EGFR-positive NSCLC[9]. Compared with docetaxel, gefitinib achieved a higher remission rate and significantly improved the quality of life of patients[10]. Due to the high rate of EGFR mutations in female lung cancer patients with no history of smoking, gefitinib can achieve a remission rate of up to 70 % in the treatment of NSCLC, EGFR-TKI that has failed previous chemotherapy. Sexual patients have better efficacy and safety. Therefore, it is clinically recommended to use EGFR-TKI drugs, such as gefitinib, until the disease progression or toxicity intolerance of lung cancer patients with advanced EGFR mutation[11].
Although EGFR-TKI (such as gefitinib) shows good therapeutic effects on lung cancer patients, they are only effective for a small number of patients with EGFR gene mutations or increased copy number and there are also problems of secondary drug resistance[12]. Secondary drug resistance is the cause of the failure of EGFR-TKI targeted therapy in NSCLC patients. At present, the research on secondary drug resistance is generally regarded as the theory of secondary mutations of the EGFR gene[13]. Therefore, although gefitinib has achieved certain therapeutic effects in some lung cancer patients, due to the problem of drug resistance, this type of treatment based on tyrosine kinase inhibition cannot completely cure the tumor. From clinical experience and mechanism research data, drug resistance that is unavoidable after continuous application is the bottleneck restricting the progress of gefitinib application.
Mechanism of resistance of lung cancer cells to gefitinib:
According to existing assumptions, the resistance mechanism of lung cancer cells to gefitinib may have the following aspects. First, changes in drug targeting sites, such as EGFR mutation (T790M), prevent gefitinib from binding to the Adenosine Triphosphate (ATP) binding site and activate downstream signaling pathways[14]. Second, the activation of EGFR bypass (such as Insulin Growth Factor 1 Receptor (IGF1R)) directly activates the downstream signaling pathway of EGFR. The third is the inactivation of functional Phosphatase and Tensin Homolog (PTEN), which leads to excessive activation of Phosphatidylinositol 3-Kinase (PI3K)/Protein Kinase B (AKT) pathway and loss of coupling with EGFR. Despite many assumptions, the mechanism of EGFR-TKI resistance in lung cancer cells has not been fully elucidated. However, scientists know that the Forkhead Box O (FOXO) transcription factor is an important substrate for the survival kinase AKT. FOXO3a is the main member of the permanent family of FOXO and belongs to subtype 0 of the forehead transcription factor[15]. FOXO transcription factors regulate the proliferation, apoptosis and cell cycle of tumor cells by inhibiting or activating the transcription of many tumor-related genes (such as p27kip1 and cyclin D). In addition, the FOXO transcription factor family is also involved in the formation of tumor resistance and resistance mechanisms. Members of the FOXO family play different roles in various tumor cells. FOXO3a regulates the sensitivity of tumor cells to platinum drugs. The abnormal expression of FOXO3a may be closely related to the occurrence of tumors and FOXO3a is related to the drug resistance mechanism of breast cancer. For example, the activation of FOXO3a can reverse the resistance of chronic myelogenous leukemia to tyrosine kinase inhibition. In general, FOXO3a transcription factor plays an important and complex regulatory role in various physiological and biochemical processes of tumor cells[16]. Therefore, it is speculated that it may also play a role in the resistance of lung cancer cells to, for example, EGFR-TKI targeted drugs.
Experiments on the Establishment of Gefitinib-Resistant Lung Cancer Cell Line and Its EGFR Signaling Pathway
Experimental materials:
Cell line: Human lung cancer cell line NC-H1985, purchased from the Institute of Basic Research of the Chinese Academy of Medical Sciences and kept by a molecular biology laboratory of a hospital.
Experimental reagents: Roswell Park Memorial Institute (RPMI)-1640 medium (Beijing fly, biochemical products, China) and Fetal Bovine Serum (FBS) standard (Tianjin Hao Ocean Biotechnology Co., Ltd., China), trypsin (Shanghai SsangYong Pharmaceutical Co., Ltd., China), Sajida Oxathiazole blue (measured by 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide (MTT)) (Sigma, United States of America (USA)), Dimethyl Sulfoxide (DMSO) (China Tianjin Fine Chemical Industry Research Institute), mouse anti-human EGFR monoclonal antibody (clone number 1116, Netmakers, USA), mouse antihuman P-EGFR monoclonal antibody (Cell Signaling Technology, USA), mouse anti-human; human MutS Homolog 2 (hMSH2) monoclonal antibody (clone number FE11, Invitrogen, USA), anti-human K-RAS polyclonal antibody (China Sino Bio Technology Co., Ltd., China), Acrylamide (Sigma, USA), sodium dihydrogen phosphate chemicals (Beijing Chemical Factory), disodium hydrogen phosphate (Beijing Chemical Factory), Glycerin (Beijing Chemical Factory), Sodium Dodecyl Sulfate (SDS) (Leboa Technology Co., Ltd.), Glycine (Leboa Technology Co., Ltd.), Tris (Leboa Technology Co., Ltd.), Skimmed Milk Powder (Yilin Dairy) and Ammonium Persulfate (APS) (Beijing Bosin Bio Technology Co., Ltd).
Experimental equipment: Ultra-clean workbench (Suzhou Purification Equipment Factory, China), inverted microscope (Chongqing XCZ-D2), sharp general optical microscope (Olympus, Japan), micro plate reader (Thermo Lab system, Finland), MMZ trace oscillation instrument (Jiangnan Jinan Medical Instrument, China), nucleic acid quantifier (Eppendorf, LTD, Germany), light scanner HR196 (Idaho Technology Company), Polymerase Chain Reaction (PCR) amplifier (Eppendorf, Germany), other experimental instruments used were shown in the Table 1.
| Name | Type of machine | Producer |
|---|---|---|
| Hemocytometer | 1/400 mm2 | Yuquan County Hardware Optical Instrument Factory |
| Hand-held steam sterilizer | YXQG02 | Shandong Dongle Medical Technology Co., Ltd. |
| Dry box | DHG-9146A | Shanghai Junhong Test Equipment Co., Ltd. |
| Cell incubator | HERA Cell | German Heraeus Company |
Table 1: Other Instruments Used in the Experiment
Preparation of main reagents: The molecular weight of gefitinib is 446.90. 10 mg, pure gefitinib powder is dissolved in 559.40 μl DMSO solution to make a storage solution of 40 mmol/l. Separate and store at 20° for use[17]. Complete medium RPMI-1640 contains 10 % FBS and 1 % streptomycin. Cell cryopreservation solution RPMI-1640 contains 20 % FBS and 10 % DMSO. Gefitinib’s medium contains 10 % FBS, 1 % penicillin-streptomycin and the corresponding concentration of gefitinib. The order of administration in RPMI-1640 was, slowly add gefitinib to RPMI-1640, that is, after adding 4 ml of complete medium to a 25 cm2 medium, add a storage medium with a calculated dose of 40 mmol/l[18]. Preparation of different concentrations of gefitinib working solution (DMSO concentration within 0.3 %), take sterile 15 ml centrifuge tubes and mark them as tubes 1-10, respectively. The corresponding concentrations are marked as 0, 2.5, 5, 10, 20, 30, 40, 60, 80, 120 μmol/l. Add gefitinib while shaking gently to prevent local precipitation of excessive precipitation crystallization.
Experimental method:
Cell culture and medium exchange: After purchasing NC-H1985 cells, cells were cultured in an incubator at 37° and 5 % Carbon dioxide (CO2) using RPMI 1640 (Sigma) medium containing 10 % FBS (Sigma). If the degree of cell fusion is low or the growth is slow and the color of the medium does not change significantly, the fluid can be changed every 2 d or once a day[19].
Cell digestion and passage: When the cells grow to 70 %-90 % saturation in a 25 cm2 culture flask, they are washed twice with Phosphate Buffered Saline (PBS). When the cells become round and pushed, immediately add about 4 ml of complete culture medium to stop the digestion and gently blow the wall of the bottle with a disposable pipette (to avoid air bubbles). Transfer the liquid in the bottle to a 15 ml centrifuge tube at a speed of 1200 r/min and centrifuge for 5 min. The original flask was washed twice with PBS solution. Discard the supernatant and add complete medium. The medium was added to 5 ml and the medium was placed in a constant temperature incubator at 37°, 5 % CO2 and 85 % humidity for further cultivation.
Cryopreservation of cells: The number of cryopreserved cells should be at least 3 times that of the plated cells and the number of cells in the cryopreservation tube should be determined based on the cell count results. Then, the cells in good condition in the exponential growth phase were centrifuged at 1200 r/min for 5 min and the supernatant was discarded. Next, suspend the cells with the prepared cell cryopreservation solution and transfer the cell suspension to a 2 ml cryopreservation tube, each of which has a volume of approximately 1.5 ml. The cells were placed sequentially at 4° for 30 min, 20° for 2 h and 80° overnight.
Frozen recovery of cells: First, quickly remove the NC-H1985 cells in liquid nitrogen and immediately transfer the NC-H1985 cells to a 37° water bath, shaking in a water bath to quickly melt. The frozen storage tube was then sterilized with 75 % ethanol and then transferred to an ultra-clean workbench. Transfer the thawed cell solution to a 15 ml centrifuge tube and add approximately 5 ml of fresh culture fluid. Then the solution was centrifuged at 1200 r/min for 5 min. Finally, the cells were transferred to a sterile culture bottle; the medium was added to 5 ml and stored in a constant temperature incubator at 37°, 5 % CO2 and 85 % humidity.
Drug-resistant culture of cells: In this study, drug resistant cell lines were established by gradually increasing the concentration of gefitinib[20]. Take cells in logarithmic growth phase, wash twice with PBS and add gefitinib at a concentration of 5 μmol/l. After culturing in the incubator for 48 h, the drug solution was discarded and complete medium without gefitinib was added to the solution for further cultivation. When the cell growth and fusion reach 70 %-80 % saturation, the above process is repeated until NC-H1985 cells grow stably in this concentration of drug solution. The remaining cells were further cultured with 10 μmol/l gefitinib solution. Increase the concentration of the solution gradually during the treatment. The concentrations are 15 μmol/l, 20 μmol/l, 25 μmol/l, 30 μmol/l and the drug addiction process is repeated continuously. After the concentration of the cells has increased, the concentration of the cells can be stabilized. One was that, until NC-H1985 cells grew steadily in a solution with a concentration of 30 μmol/l, the whole process lasted about 10 mo before the resistance of NC-H1985 cells could be established. Finally, NC-H1985 cells were cultured in a non-toxic medium for 2 w and the logarithmic phase cells were taken for subsequent experiments to eliminate the effects of gefitinib[20].
Half-maximal Inhibitory Concentration (IC50) value and NC-H1985 resistance to gefitinib: Cell resistance were measured by the CCK-8 method. NC-H1985 cells in the logarithmic growth phase were seeded into 96- well cell culture plates (2×103/well). Each well contains 100 μl cell suspension (100 μl PBS only) and add the solution to the blank group, then incubate for 24 h in an incubator at 37°, 5 % CO2 and 85 % humidity. Absorb liquid from each well and add 10 μl of CCK-8 liquid and 90 μl medium (after mixing) to each well and continue culturing for 3 h in a constant temperature incubator. To determine the IC50 value, first select a wavelength of 450 nm and measure the absorption value (Optical Density (OD50)) of each well on the Enzyme-Linked Immunosorbent Assay (ELISA). Then use the drug concentration as the horizontal axis and the cell survival rate as the vertical axis to draw the cell resistance curve[21]. Throughout the experiment, SPSS 13.0 software was used to calculate the IC50 of each drug through the linear regression of the inhibition concentration value and Vascular Resistance (VR) and the resistance index IC50 was calculated.
Cell growth measurement and calculation of doubling time: First, trypsinogen and centrifuge NC-H1985 cells into a logarithmic phase after cell suspension, inoculate 96 cell culture plates according to 5×102 wells/vaccine, using RPMI 1640 and 10 % FBS. The cultivation was carried out at 37° and the cultivation was carried out in the 5 % CO2 incubator 1, 2, 3, 4, 5, 6 and 7 d after sampling (repeat 12 times per well), giving up original culture, add 100 μl CCK-8 mixture (CCK 8-10 μl, 90 μl) to each well, continue to incubate at 37° for 3 h, 450 m wavelength, measure each well on ELISA. The absorbance (OD450) value (the method is same as the above experiment). Finally, draw a cell growth curve (the number of cells is proportional to the OD value) and calculate the cell population doubling time (cell population ion doubling time T).
Observation of cell morphology: This study used cell slide Hematoxylin and Eosin (HE) staining method to observe the cell morphology. The specific operation method is; before sample preparation, select the logarithmic phase NC-H1985 cells and use them in a medium containing 10 % FBS digest with 0.25 % trypsin, then tap the cells gently with a disposable pipette, transfer the cell suspension to a sterile centrifuge tube, centrifuge at 1000 RPM for 10 min, pour off the clear solution and suspend the cells[22]. After counting the cells by staining, the cell concentration was adjusted to about 1×105 ml and the cells were seeded into a 24- well plate with a cover glass placed until the cover glass was completely submerged. The protective glass has been treated with conventional foam acid and treated with an autoclave at high temperature. The inoculated cells were cultured at 37° in 5 % CO2. When the cells were covered with a cover glass of about 70 %-80 %, the original medium can be aspirated and washed 3 times with 5 ml PBS and then the cell color separation starts. First observe the condition of the cells under a microscope. If the nuclei are too dark, stain them with a 1 % hydrochloric acid alcohol solution for a few seconds and then wash them in tap water. Cytoplasmic staining method was done by immersing the coverslip in eosin dye for 1 min and then washes with tap water. After the cover slip is purged or naturally dried, it is sealed with neutral glue. Finally, observe the morphology of the cells under a microscope and take pictures for comparison.
Judgment criteria for protein expression and mRNA expression: First, the expression of EGFR protein, take the yellow particles in the cell membrane or cytoplasm as positive expression, record the cell expression site separately and compare the expression intensity with the corresponding expression site[22]. The expression standard of P-EGFR protein is as follows; first, the positive expression is recorded in the cell membrane or cytoplasm with yellow particles and the expression intensity is compared with the parental cell line. The standard of k-RAS protein expression; the yellow particles in the cytoplasm are used as positive expression and the expression intensity is compared with the expression intensity of the parent cell[23]. The standard of b-RAF protein expression; the yellow particles in the cytoplasm are used as positive expressions and the expression intensity is compared with the parent cells. The standard expression of E-cadherin protein is as follows; the expression of E-cadherin protein is positive for yellow particles in the cell membrane or cytoplasm and the expression site is recorded separately and compared with the corresponding expression intensity. Finally, the expression standard of the mismatch repair protein hMSH2 is as follows; the yellow particles are positive in the nucleus when compared with the parent cells.
Statistical analysis:
Suspend the cells obtained after digestion with 2 ml of culture solution, take a clean cell counting plate, cover with a coverslip, shake the cells and suck up 10 μl single cell suspensions and 10 μl trypan blue staining. After mixing the liquid, suck 10 μl and slowly inject along the edge of the cover glass to avoid the generation of air bubbles. Place it under an inverted microscope for observation, look for a large square under a low-power microscope, count the number of cells in four large squares under a high-power microscope and count it with an electronic cell counter.
SPSS 13.0 statistical software was used for analysis. The data of the detection indicators in this study were described by the standard deviation (x±s). The difference in the mRNA expression of each gene of NC-H1985 cells was measured by t-test between two independent samples.
Results and Discussion
Analysis of drug resistance characteristics of cells after culturing NC-H1985 cells with different concentrations of gefitinib for 24 h, 48 h and 72 h, the cell survival rate can be measured and recorded to make a table. It can be seen from the research results that with the increase of gefitinib concentration and culture time, the survival rate of NC-H1985 cells gradually decreased and the specific data are shown in Table 2.
| Time | 2.5 µmol/l | 5 µmol/l | 10 µmol/l | 20 µmol/l | 30 µmol/l | 40 µmol/l | 60 µmol/l | 80 µmol/l | 120 µmol/l |
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 0.99 | 0.974 | 0.955 | 0.93 | 0.912 | 0.896 | 0.758 | 0.714 | 0.655 |
| 48 h | 0.972 | 0.943 | 0.916 | 0.886 | 0.841 | 0.779 | 0.654 | 0.606 | 0.552 |
| 72 h | 0.955 | 0.932 | 0.894 | 0.812 | 0.773 | 0.607 | 0.425 | 0.295 | 0.12 |
Table 2: The Survival Rate of NC-H1985 Cells after Different Concentrations of Gefitinib After 24 H, 48 H and 72 H
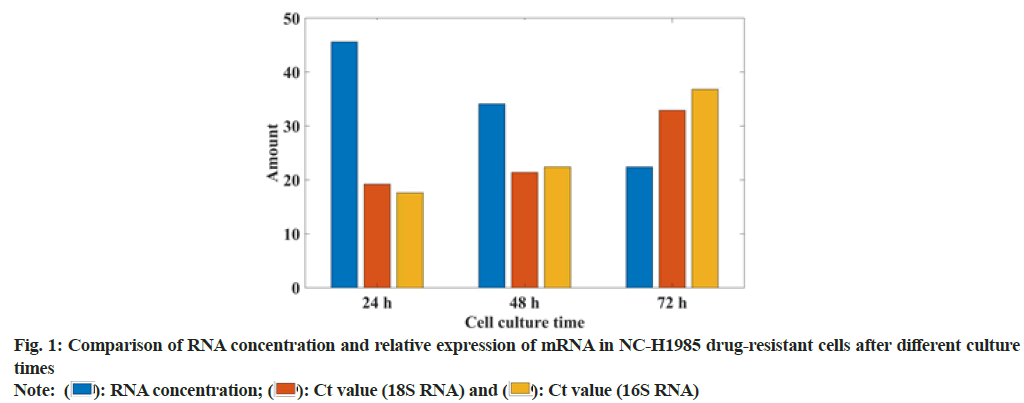
In this study, the gefitinib resistant strain NC-H1985 was established by gradually increasing the concentration of gefitinib. The sensitivity of gefitinib to the parental cell NC-H1985 and the drug-resistant cell NC-H1985 was compared. It was found that the IC50 of drug-resistant cells was significantly higher than that of parental cells and there was no significant change in the expression levels of Extracellular Signal-Regulated Protein Kinase (ERK1/2) and phosphorylated Signal Transducer and Activator of Transcription 3 (p-STAT3) in NC-H1985 cells, but the expression of p-AKT was significantly reduced and it was treated in NC-H1985 drug-resistant strains. It was also found that the treatment after the expression of p-AKT and p-STAT3 was suppressed and almost no expression of pERK1/2 was moderately suppressed. It can be seen that NC-H1985 parental cells and gefitinib-resistant strain NC-H1985 have significant differences in drug resistance signaling mechanism. In this study, Western blot was used to determine the RNA concentration of NC-H1985 drug-resistant cells and the mRNA expression was analyzed. The specific data was shown in fig. 1.
From the data in fig. 1, it can be seen that the NC-H1985 gefitinib cell line has a certain difference compared to the parental NC-H1985 cell. With the extension of the culture time, the RNA concentration decreased by 51 % and the 18 transfer RNA (tRNA), Cycle threshold (Ct) value increased 39 %, the Ct value of IGF1R increased by 44 %, which shows that the NC-H1985 gefitinib cell line has a certain resistance. It proves that this study successfully established the NC-H1985 drug-resistant cell line.
Most NSCLC patients with EGFR mutation activators (such as exon 19 deletion (Del E746-A750) and exon 21 mutation (L858R) are initially sensitive to EGFR-TKI. Gefitinib can suppress cancer throughout Resting (G0) stage. It plays a role in the rapid development of cells, thereby reducing the ability of cancer cells to invade, metastasize and infiltrate. However, most of them will eventually become resistant to drugs. At present, the mechanism of resistance to gefitinib in NSCLC patients is mainly divided. It is a secondary mutation of EGFR (the most common mutation is T790M), abnormal activation of STAT3 signaling pathway and continuous activation of PI3K/AKT signaling pathway. The growth of NC-H1985 drug-resistant cell lines at different concentrations was shown in fig. 2.
From the data in fig. 2, it can be seen that the growth rate of the NC-H1985 drug-resistant cell line successfully established in this study is related to time and the concentration of gefitinib solution. With the extension of time, NC-H1985 drug-resistant cells at 10 μmol/l, 40 μmol/l and 120 μmol/l gefitinib solution increased 6 times, 5 times and 4.5 times respectively, it can be seen that the higher the concentration of gefitinib solution, the slower the growth of NC-H1985 drug-resistant cells. The longer the time, the slower the growth of NCH1985 drug-resistant cells.
EGFR is a protein encoding the proto-oncogene Human Epidermal growth factor Receptor 1 (HER1). It is synthesized in the cytoplasm and then moved to the cell membrane to perform its function. It is the starting point of multiple signal transduction pathways in the cell. The human EGFR gene is located in the p13-22 region of chromosome 7 and is composed of 28 exons, which belong to HER2 (ERBB2/NEU), ERBB-3, (HER3) and ERBB-4 (HER4). However, when EGFR protein is overexpressed and/or mutated, ligand-independent tyrosine kinase activation occurs. Activated EGFR activates a series of related genes, leading to cell proliferation and inhibiting apoptosis. It is known that EGFR protein overexpression occurs in 40 %-80 % of NSCLC patients. The mechanism of EGFR overexpression is gene amplification (95 %), increased transcription (5 %), or gene mutation. In this study, the ATT method was used to analyze the expression of EGFR, mRNA at different times, as shown in fig. 3.
From fig. 3, it can be seen that the change of EGFR signaling pathway of gefitinib-resistant lung cancer cells is related to the relative expression of mRNA. The higher the relative mRNA expression, the easier the EGFR signaling pathway changes. From 24-72 h, mRNA relative expression increased by 37.5 %.
There are 486 mutations in the ATP-binding domain of EGFR tyrosine kinase and the main mutations are located in exons 18, 19, 20 and 21 of the EGFR gene. In this study, immunocytochemistry was used to test and compare the differences in the expression of EGFR and P-EGFR proteins in the NC-H1985 cell line. It was found that although these two cell lines have the same strength, they express both in the cell membrane and cytoplasm protein cell membrane expression, but the cytoplasmic expression of both in NC-H1985 resistant strain is stronger than the parental strain NC-H1985, that is EGFR and P-EGFR protein in gefitinib obtained cytoplasmic expression of drug-resistant cell lines enhanced. The reason may be that the growth rate of drug-resistant cells slows down and cells continuously synthesize EGFR to promote cell growth. However, EGFR-TKI weakens the activation of EGFR, which leads to the failure of signal activation of downstream signal transduction pathways. As a result, the EGFR protein synthesized in the cytoplasm cannot be efficiently transported to the cell membrane. In this study, the phase distribution of NC-H1985 gefitinib-resistant cells was observed through an inverted phase contrast microscope. The specific data was shown in As shown in fig. 4, 44.1 % of NC-H1985 gefitinibresistant cells cultured after gefitinib resistance were in G0 phase, 36.9 % were in Growth 1 (G1) phase, 11.4 % were in Synthesis (S) phase and 4.3 % were in Growth 2 (G2) phase and 3.3 % in the Mitosis (M) period.
This article analyzes the drug resistance problems in the current common methods of treating lung cancer and discusses the solutions to these problems and establishes the corresponding drug-resistant cells. It also briefly introduces the development and prospects of molecular targeted therapy, analyzes the advantages and disadvantages of the commonly used drug gefitinib and provides solutions for solving the problems in application.
This paper analyzes the establishment of gefitinibresistant lung cancer cell lines and the changes in EGFR signaling pathway, explains the benefits and limitations of gefitinib in the treatment of lung cancer and successfully established NC-H1985 gefitinib-resistant cell line. It was found to have a certain difference compared to the parental NC-H1985 cell line. With the extension of the culture time, the RNA concentration decreased by 51 % and the 18S ribosomal RNA (18 sRNA) Ct value increased by 39 %. In addition, the higher the concentration of gefitinib solution, the slower the growth of NC-H1985 drug-resistant cells. At the same time, the growth rate of gefitinib solution in 10 μmol/l, 40 μmol/l and 120 μmol/l, respectively increased by 6 times, 5 times and 4.5 times.
This study explored the establishment of gefitinib-resistant lung cancer cell lines and the mechanism of EGFR signaling pathway changes. It has been verified by research that a gefitinib resistant cell line can be successfully established by gradually increasing the concentration of gefitinib. It was also found that the higher the relative expression of mRNA in gefitinib-resistant lung cancer cells, the EGFR signaling pathway is likely to change. From 24 h to 72 h, the relative expression of mRNA has increased by 37.5 %. After gefitinib-resistant culture out of NC-H1985 gefitinib-resistant cells, 44.1 % were in G0 phase, 36.9 % were in G1 phase, 11.4 % were in S phase, 4.3 % were in G2 phase and 3.3 % were in M phase.
Conflict of interests:
The authors declared no conflict of interests.
References
- Huh YH, Oh S, Yeo YR, Chae IH, Kim SH, Lee JS, et al. Swiprosin-1 stimulates cancer invasion and metastasis by increasing the Rho family of GTPase signaling. Oncotarget 2015;6(15):13060-71.
[Crossref] [Google Scholar] [PubMed]
- Castillo G, Jiang X. Association of the hedgehog signal pathway to gefitinib sensitivity in a 3D cancer cell lines screening. Cancer Res 2015;75(15):5447.
- Jeong CH. Inhibition of ubiquitin-specific peptidase 8 suppresses growth of gefitinib-resistant non-small cell lung cancer cells by inducing apoptosis. J Cancer Prev 2015;20(1):57-63.
[Crossref] [Google Scholar] [PubMed]
- Suzuki T, Nagasawa I, Yamaoka T, Ohmori T, Nishio K, Koyama K, et al. The effect of kinase signaling for miR-205 regulation in gefitinib-resistant lung cancer cell lines. Cancer Res 2016;76(14):1893.
- Dai Y. Differential expression of circular RNAs in gefitinib-acquired resistant non-small cell lung cancer cells. Tumor 2017;37(11):1128-35.
- Zhang Y, Yao K, Shi C, Jiang Y, Liu K, Zhao S, et al. 244-MPT overcomes gefitinib resistance in non-small cell lung cancer cells. Oncotarget 2015;6(42):44274-88.
[Crossref] [Google Scholar] [PubMed]
- XueTing CA, Jie YA, ChunPing HU, Peng CA. Dihydroartemisinin enhances the sensitivity of gefitinib in non-small cell lung cancer cells by inhibiting STAT3. Chin Sci Bull 2017;62(18):2013-9.
- Lee Y, Wang Y, James M, Jeong JH, You M. Inhibition of IGF1R signaling abrogates resistance to afatinib (BIBW2992) in EGFR T790M mutant lung cancer cells. Mol Carcinog 2016;55(5):991-1001.
[Crossref] [Google Scholar] [PubMed]
- Kang AR, An HT, Ko J, Choi EJ, Kang S. Ataxin-1 is involved in tumorigenesis of cervical cancer cells via the EGFR–RAS–MAPK signaling pathway. Oncotarget 2017;8(55):94606.
[Crossref] [Google Scholar] [PubMed]
- Fernando RI, Hamilton DH, Dominguez C, David JM, McCampbell KK, Palena C. IL-8 signaling is involved in resistance of lung carcinoma cells to erlotinib. Oncotarget 2016;7(27):42031-44.
[Crossref] [Google Scholar] [PubMed]
- Jia FE, Xueyan WE, Chuang LI, Mingxiong GU, Min PE, Qibin SO, et al. Mechanism of EGFR over-expression and mutations leading to biological characteristics changes of human lung adenocarcinoma cells through CXCR4/CXCL12 signaling pathway. Zhongguo Fei Ai Za Zhi 2018;21(7):503-12.
[Crossref] [Google Scholar] [PubMed]
- Lopes GL, Vattimo EF, Castro Junior GD. Identifying activating mutations in the EGFR gene: Prognostic and therapeutic implications in non-small cell lung cancer. J Bras Pneumol 2015;41(4):365-75.
[Crossref] [Google Scholar] [PubMed]
- Hu Y, Dong XZ, Liu X, Liu P, Chen YB. Enhanced antitumor activity of cetuximab in combination with the Jak inhibitor CYT387 against non-small-cell lung cancer with various genotypes. Mol Pharm 2016;13(2):689-97.
[Crossref] [Google Scholar] [PubMed]
- Lin YM, Kuo WW, Velmurugan BK, Hsien HH, Hsieh YL, Hsu HH, et al. Helioxanthin suppresses the cross talk of COX-2/PGE2 and EGFR/ERK pathway to inhibit arecoline-induced oral cancer cell (T28) proliferation and blocks tumor growth in xenografted nude mice. Environ Toxicol 2016;31(12):2045-56.
[Crossref] [Google Scholar] [PubMed]
- Zhong D, Ru Y, Wang Q, Zhang J, Zhang J, Wei J, et al. Chimeric ubiquitin ligases inhibit non-small cell lung cancer via negative modulation of EGFR signaling. Cancer Lett 2015;359(1):57-64.
[Crossref] [Google Scholar] [PubMed]
- Xiao X, He Z, Cao W, Cai F, Zhang L, Huang Q, et al. Oridonin inhibits gefitinib-resistant lung cancer cells by suppressing EGFR/ERK/MMP-12 and CIP2A/Akt signaling pathways. Int J Oncol 2016;48(6):2608-18.
[Crossref] [Google Scholar] [PubMed]
- Kobayashi I, Takahashi F, Nurwidya F, Nara T, Hashimoto M, Murakami A, et al. Oct4 plays a crucial role in the maintenance of gefitinib-resistant lung cancer stem cells. Biochem Biophys Res Commun 2016;473(1):125-32.
[Crossref] [Google Scholar] [PubMed]
- Hao GO, Yin YU, Yongwen LI, Zhang H, Ying LI, Weiting LI, et al. Role of EZH2 inhibitor combined with gefitinib in EGFR-TKIs resistant lung cancer cells. Zhongguo Fei Ai Za Zhi 2019;22(5):255-63.
[Crossref] [Google Scholar] [PubMed]
- Lee TB, Seo EJ, Lee JY, Jun JH. Synergistic anticancer effects of curcumin and hinokitiol on gefitinib resistant non-small cell lung cancer cells. Nat Product Commun 2018;13(12):1667-71.
- Dhillon S. Gefitinib: A review of its use in adults with advanced non-small cell lung cancer. Targeted Oncol 2015;10(1):153-70.
[Crossref] [Google Scholar] [PubMed]
- Soria JC, Wu YL, Nakagawa K, Kim SW, Yang JJ, Ahn MJ, et al. Gefitinib plus chemotherapy vs. placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): A phase 3 randomised trial. Lancet Oncol 2015;16(8):990-8.
[Crossref] [Google Scholar] [PubMed]
- Fukuhara T, Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, et al. Factors associated with a poor response to gefitinib in the NEJ002 study: Smoking and the L858R mutation. Lung Cancer 2015;88(2):181-6.
[Crossref] [Google Scholar] [PubMed]
- Kaneko T, Shimizu A, Aoki M, Tsuruoka S. A case of gefitinib-associated membranous nephropathy in treatment for pulmonary adenocarcinoma. CEN Case Rep 2015;4(1):31-7.
[Crossref] [Google Scholar] [PubMed]
 Ctvalue (16S RNA)
Ctvalue (16S RNA)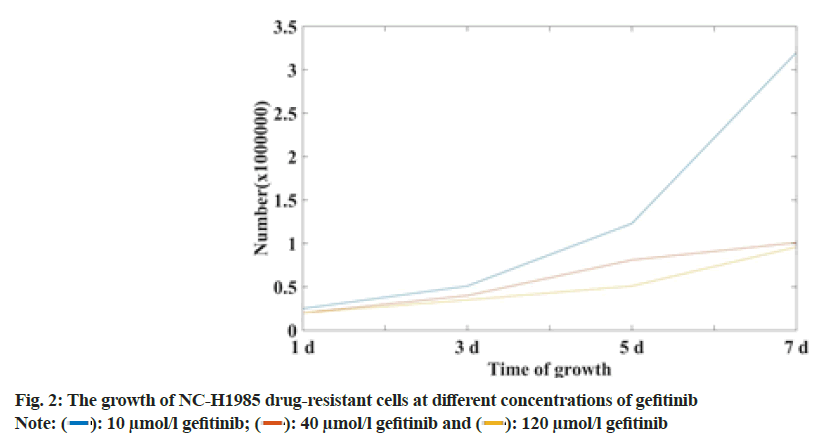
 10 µmol/l gefitinib;
10 µmol/l gefitinib;  120 µmol/l gefitinib
120 µmol/l gefitinib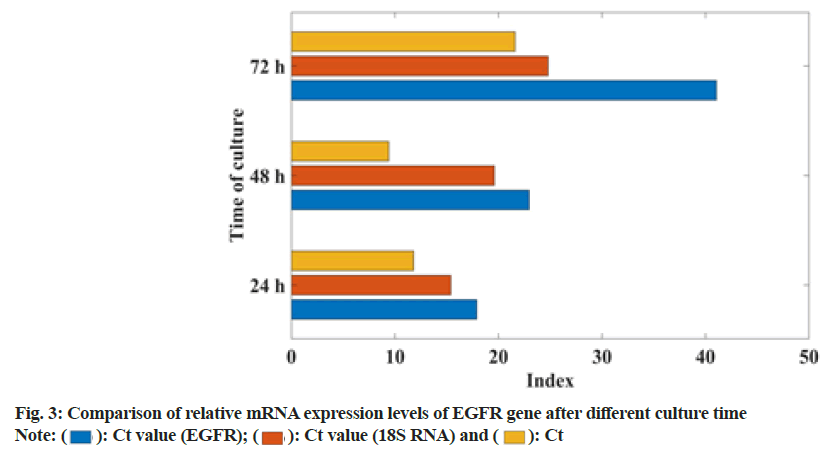
 Ct
Ct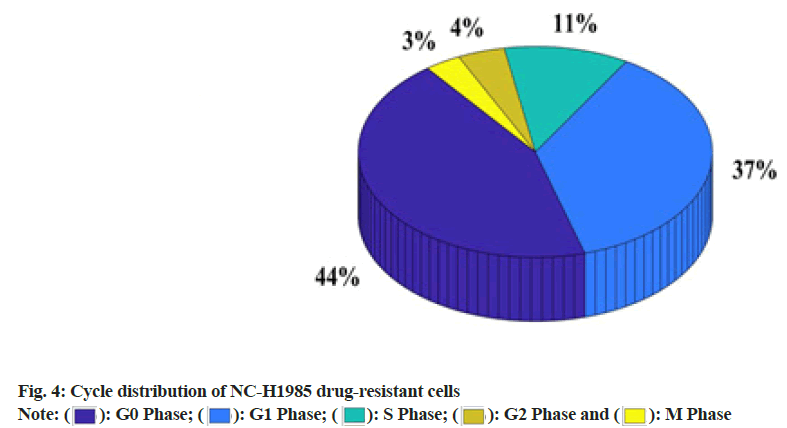
 M Phase
M Phase



