- *Corresponding Author:
- Lixin Wang
Department of Stomatology, Beijing Rehabilitation Hospital, Capital Medical University, Shijingshan, Beijing 100144, China
E-mail: wanglixinlilly@126.com
| Date of Received | 22 February 2023 |
| Date of Revision | 27 November 2023 |
| Date of Acceptance | 06 May 2024 |
| Indian J Pharm Sci 2024;86(3):882-889 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Numerous papers stated that Marsdenia tenacissima extract possessed tumor-suppression properties. Herein, we aimed to investigate the influence and underlying mechanism of Marsdenia tenacissima extract on oral squamous cell carcinoma. After being cultured, CAL-27 cells were treated with various doses of Marsdenia tenacissima extract. Proliferation, apoptosis, and migration were assessed using cell counting kit-8, clone formation, scratch, and flow cytometry assays. Western blot detected protein levels. Quantitative reverse transcription polymerase chain reaction method analyzed hsa_circular_0003645 and microRNA-335 expression. Their interaction was validated using dual-luciferase reporter gene experiment. 1.0 and 1.5 mg/ml Marsdenia tenacissima extract or si-hsa_circular_0003645 could suppress CAL-27 cell proliferation, migration, N-cadherin, and hsa_circ_0003645, whereas increase apoptosis, E-cadherin, and microRNA-335. Furthermore, the upregulation of hsa_circular_0003645 might abolish Marsdenia tenacissima extract exposure-mediated oral squamous cell carcinoma cell proliferation and migration inhibition and apoptosis promotion. Besides, hsa_circular_0003645 could target microRNA-335. Marsdenia tenacissima extract treatment might hinder oral squamous cell carcinoma progression via regulating hsa_circular_0003645 and microRNA-335.
Keywords
Marsdenia tenacissima extract, oral squamous cell carcinoma, hsa_circular_0003645, microRNA-335, cell proliferation, apoptosis, migration
As a prevalent head and neck malignant disease worldwide, Oral Squamous Cell Carcinoma (OSCC) is characterized by local invasiveness, high recurrence, and ease of metastasis, with approximately 58 450 newly diagnosed cases and 12 230 deaths in the United States[1]. Nevertheless, the pathogenesis of OSCC has not yet been clarified. Although significant advances in surgery, radiotherapy, and chemotherapy have recently acquired some benefits, most sufferers with advanced or metastatic OSCC are responsible for the poor prognosis[2,3]. Accordingly, exploring the mechanisms underlying the OSCC process is worthy of the development of new therapeutic targets. A Traditional Chinese Medicine (TCM) and Dai herbal medicine, Marsdenia tenacissima (M. tenacissima) contains complex ingredients, such as alkaloids, steroidal ester glycosides, and resins, which have been widely used in the treatment of asthma, bronchitis, and other diseases[4]. Beneficially, M. tenacissima Extract (MTE) has presented strong anti-tumor properties in different human cancers through multiple pathways in vitro[5,6]. Yet, its function in OSCC is still unknown. Of interest, recent studies have shown that MTE might prevent the malignant behaviors of glioma via modulating non-coding Ribonuclic Acid (RNA), such as Long noncoding RNA (LncRNA), Maternally Expressed Gene 3 (MEG3) and microRNA (miR)-542-3p[7]. Different from other non-coding RNAs, circular RNAs (circRNAs) were generated and formed by alternative splicing of pre-messenger RNA (mRNA), with a covalently closed-loop structure[8]. It has been reported that dysregulated circRNAs were widely involved in the pathogenesis of diverse tumors[9]. Previous laboratory work has demonstrated that hsa_circ_0003645 functions as a well-known carcinogenic factor in various tumors[10,11], but its expression profiles and functions in OSCC remain largely unclear. In terms of molecular mechanisms, circRNAs have been pointed out to exert key roles by regulating the downstream-target miRNA[12]. Herein, circRNA interactive bioinformatics software found miR-335 as a probable target of hsa_circ_0003645. In fact, some reports have indicated that miR-335 might restrain the aggressive phenotypes of breast cancer and ovarian cancer[13,14]. As a metastasis suppressor miRNA, miR-335 has confirmed to repress tongue squamous carcinoma cell proliferative ability and induce cell cycle arrest[15]. Herein, this project focused on whether MTE might control OSCC progression via modulating hsa_circ_0003645/ miR-335.
Materials and Methods
Cell culture and reagents:
MTE was provided by Sciphar Limited Company (Shaanxi, China). OSCC cell line (CAL-27, Chinese Academy of Sciences, Shanghai, China) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Solarbio, Beijing, China) and 10 % Fetal Bovine Serum (FBS) (Invitrogen, Paisley Scotland, United Kingdom (UK)). LipofectamineTM 2000 reagent, Cell Counting Kit-8 (CCK-8), and Bicinchoninic Acid (BCA) Kit were offered by Solarbio. Takara (Liaoning, Dalian) provided reverse transcription and Polymerase Chain Reaction (PCR) kits. Invitrogen offered Trizol reagent. Besides, Genepharma (Shanghai, China) offered PCR primers, si- hsa_circ_0003645, si-Negative Control (NC), plasmid cloning Deoxyribonucleic Acid (pcDNA)- hsa_circ_0003645, pcDNA, miR-335 mimics, miR-NC, and Wild-Type/Mutant (WT/MUT)- hsa_circ_0003645 plasmids. Rabbit anti-human E-cadherin and N-cadherin were acquired by Santa Cruz Biotechnologies (Santa Cruz, CA, United States of America (USA)). Beyotime (Shanghai, China) provided dual-luciferase activity detection Kit.
Method:
Cell treatment and transfection: Referring to the previous description[16], 0.5, 1.0, 1.5 mg/ml MTE were employed to respectively stimulate CAL-27 cells (5.0×105 cells/well, in 6-well plates) in RPMI 1640 medium for 48 h, generated MTE-L/M/H groups. Synchronously, control group was normal cultured CAL-27 cells. According to lipofectamine method, we knock-downed hsa_circ_0003645 by transfecting si-NC or si-hsa_circ_0003645 into un-treated CAL-27 cells, marked si-NC or si-hsa_circ_0003645 group. Besides, pcDNA or pcDNA-hsa_circ_0003645 were transfected into CAL-27 cells, and incubated with 1.5 mg/ml MTE, recorded as MTE+pcDNA or MTE+pcDNA-hsa_ circ_0003645 group.
CCK-8 assay: In 96-well plates, transfected CAL- 27 cells (2.5×104 cells/well) were cultured for 24 h, followed by mixture with 10 μl, CCK-8 reagent. After being cultured for another 2 h, an enzyme meter was applied to assess the Optical Density (OD) values in different groups.
Colony formation assay: After being harvested and trypsinized, 1000 un-treated or treated cells in 6-well plates were cultured for 14 d and the medium was changed every 2 d. The culture was terminated when cell colonies were visible. After washing, cells were sequentially subjected to 4 % paraformaldehyde fixture and crystal violet staining. Under a microscope, colony number was counted (≥50 cells were regarded as a colony).
Flow cytometry: After being collected and washed with Phosphate Buffer Solution (PBS), 5.0×104 cells in 6-well plates were re-suspended in 500 μl binding buffer. Then, 5 μl annexin V-Fluorescein Isothiocyanate (FITC) and 5 μl Propidium Iodide (PI) were added into the cell the cells suspension, followed by fully mixture and incubation for 15 min at room temperature. At last, a flow cytometry was utilized to analyze cell apoptosis within 1 h.
Wound healing assay: Cell migration was measured in this experiment. In short, 5.0×104 cells in 6-well plates were maintained for 24 h. After that, a scratch was created using a sterile pipette tip in cell monolayer and scratch spacing was measured and denoted d0 h. After washing the floating cells, the cells were cultured with serum- free medium for 24 h and the intracellular spacing was examined and denoted d24 h. Finally, scratch healing rate (%)=(d0 h-d24 h)/d0 h×100 %.
Western blot: Based on Radio- Immunoprecipitation Assay (RIPA) lysis buffer, total CAL-27 cell proteins were prepared. After BCA method determination, the corresponding protein samples were appended with the loading buffer and denatured, followed by separation with Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE). Following shifted onto membranes and blocked for 1 h, the membranes were subjected to overnight incubation with primary antibodies: E-cadherin (1:1000), N-cadherin (1:1000), and Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) (1:2000). After being soaked in secondary antibody (1:2000) for 2 h, the band was analyzed and quantified.
Reverse Transcription quantitative-PCR (RT- qPCR): After extracted with Trizol reagent, the obtained total RNAs were reversely transcribed in complementary DNA (cDNA), which was adopted to RT-qPCR reaction. Amplification was initially carried out at 95° for 10 s for 35 cycles (95° 10 s, 58° 30 s, 72° 30 s). Primer was displayed as follows: hsa_circ_0003645: Forward: 5'-CACAGTGGCCTTGTTCCCT-3'; Reserved: 5'-TTCCCAAGACAGAGTTTTGCT-3'; miR-335: Forward: 5'-TCAAGAGCA ATAACGAAAA ATG T-3'; Reserved: 5'-GCTGTCAACGATACGCTACGT-3'; GAPDH: Forward: 5'-GTCAAGGCTGAGAACGGGAA-3'; Reserved: 5'-AAATGAGCCCCAGCCTTCTC-3'; U6: Forward: 5'-GCGATACAGAAGCACGAGAG-3' and Reserved: 5'-CGATACAGAGAGCGCGACTAC GAG-3'. Finally, GAPDH or U6 was respectively internal reference for hsa_circ_0003645 or miR-335, and results were assessed with 2-ΔΔCt method.
Dual-luciferase reporter gene assay: Based on LipofectamineTM 2000, WT/MUT-hsa_ circ_0003645 was co-transfected into 5.0×104 CAL-27 cells in 6-well plates with miR-NC or miR- 335 mimics for 6 h. After changing the medium, cells were cultured for another 24 h. Then, cells were harvested and lysed for the detection of luciferase activity.
Statistical analysis:
Data with normal distribution were processed based on Statistical Package for the Social Sciences (SPSS) 21.0 and reported as (x͞ ±s). The comparisons of two-group and multiple groups were employed with student’s t-test or one-way Analysis of Variance (ANOVA). Meanwhile, Least Significant Difference (LSD)-t test was used for pairwise comparison between groups. Difference was deemed statistically significant at p<0.05.
Results and Discussion
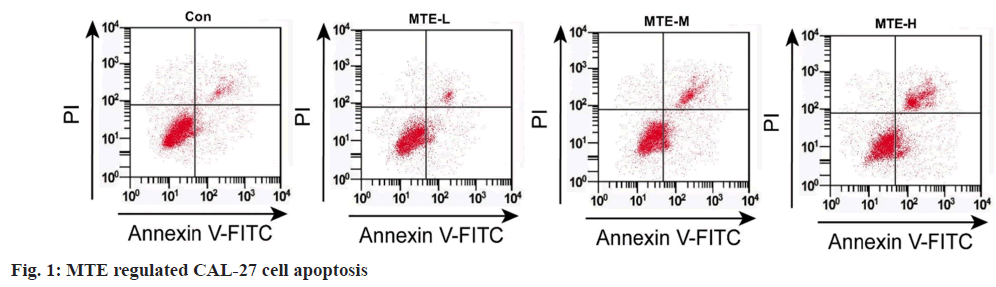
Referring to data displayed in fig. 1 and Table 1, MTE exposure might hinder cell OD value and colony formation number, and induce apoptosis rate in a dose-dependent manner (p<0.05).
| Groups | OD value | Colony formation number | Apoptosis rate % |
|---|---|---|---|
| Control | 1.16±0.09 | 118.67±5.73 | 6.81±0.37 |
| MTE-L | 1.16±0.08 | 117.33±6.34 | 6.81±0.24 |
| MTE-M | 0.98±0.06ab | 86.67±3.86ab | 13.00±0.67ab |
| MTE-H | 0.63±0.03abc | 64.67±2.49abc | 21.27±1.08abc |
| F | 39.426 | 86.466 | 311.200 |
| p | 0.000 | 0.000 | 0.000 |
Note: ap<0.05, bp<0.05, and cp<0.05 relative to control, MTE-L, and MTE-M group, respectively
Table 1: Effects of MTE on CAL-27 Cell Proliferation and Apoptosis (x͞ ±s, n=3)
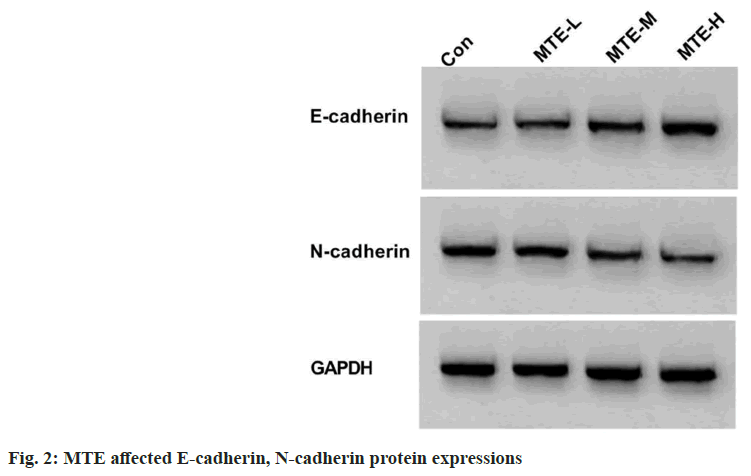
Based on the results exhibited in fig. 2 and Table 2, MTE treatment apparently reduced CAL-27 cell scratch healing rate and N-cadherin expression, and enhanced E-cadherin expression in a concentration-dependent way (p<0.05).
| Groups | Scratch healing rate (%) | E-cadherin | N-cadherin |
|---|---|---|---|
| Control | 59.92±2.14 | 0.20±0.02 | 0.74±0.06 |
| MTE-L | 59.81±2.28 | 0.22±0.02 | 0.72±0.06 |
| MTE-M | 49.00±2.15ab | 0.39±0.03ab | 0.47±0.04ab |
| MTE-H | 33.31±1.73abc | 0.71±0.05abc | 0.26±0.02abc |
| F | 108.813 | 159.048 | 67.598 |
| p | 0.000 | 0.000 | 0.000 |
Note: ap<0.05, bp<0.05, and cp<0.05 compared with control, MTE-L, and MTE-M group, respectively
Table 2: Effects of MTE on CAL-27 Cell Migration (x͞ ±s, n=3)
As shown in Table 3, hsa_circ_0003645 content was gradually reduced with increasing dose of MTE, but miR-335 expression was improved (p<0.05).
| Group | hsa_circ_0003645 | miR-335 |
|---|---|---|
| Control | 1.00±0.00 | 1.00±0.00 |
| MTE-L | 0.98±0.02 | 1.06±0.04 |
| MTE-M | 0.66±0.04ab | 1.57±0.09ab |
| MTE-H | 0.21±0.02abc | 3.40±0.14abc |
| F | 682.458 | 517.853 |
| p | 0.000 | 0.000 |
Note: ap<0.05, bp<0.05, and cp<0.05 vs. control, MTE-L, and MTE-M group, respectively
Table 3: Effects of MTE on HSA_circ_0003645 and miR-335 (x͞ ±s, n=3)
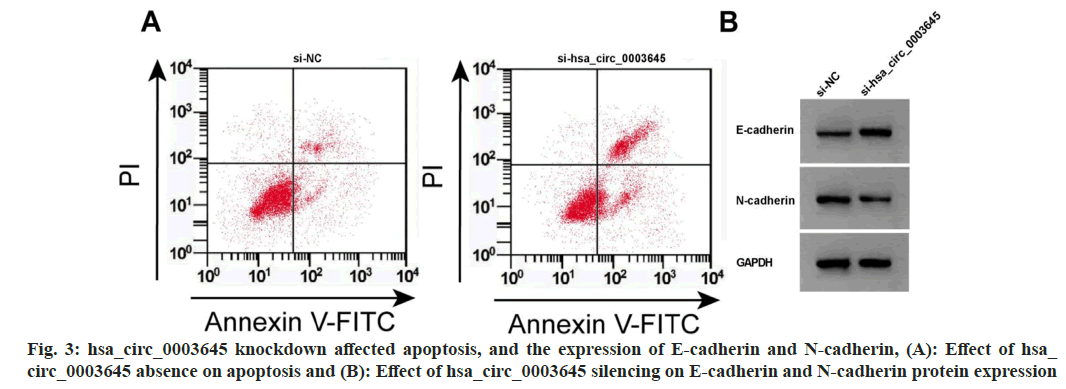
Data from fig. 3 and Table 4 displayed that hsa_circ_0003645 expression, cell OD value, colony formation number, scratch healing rate, and N-cadherin were obviously repressed after si-hsa_circ_0003645 introduction, while miR-335 expression, apoptosis rate, and E-cadherin expression were improved (p<0.05).
| Group | hsa_circ_0003645 | miR-335 | OD value | Colony formation number | Apoptosis rate (%) | Scratch healing rate (%) | E-cadherin | N-cadherin |
|---|---|---|---|---|---|---|---|---|
| si-NC | 1.00±0.0 | 1.00±0.00 | 1.16±0.09 | 118.33±5.73 | 6.87±0.38 | 60.17±2.14 | 0.20±0.02 | 0.73±0.07 |
| si-hsa_circ_0003645 | 0.11±0.01a | 4.55±0.16a | 0.54±0.03a | 55.67±2.05a | 23.14±1.27a | 31.36±1.27a | 0.88±0.06a | 0.19±0.02a |
| t | 154.153 | 37.889 | 11.320 | 17.834 | 21.258 | 20.053 | 18.623 | 12.847 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: ap<0.05 vs. si-NC
Table 4: HSA_circ_0003645 Downregulation Regulated Proliferation, Apoptosis and Migration (x̄±s, n=3)
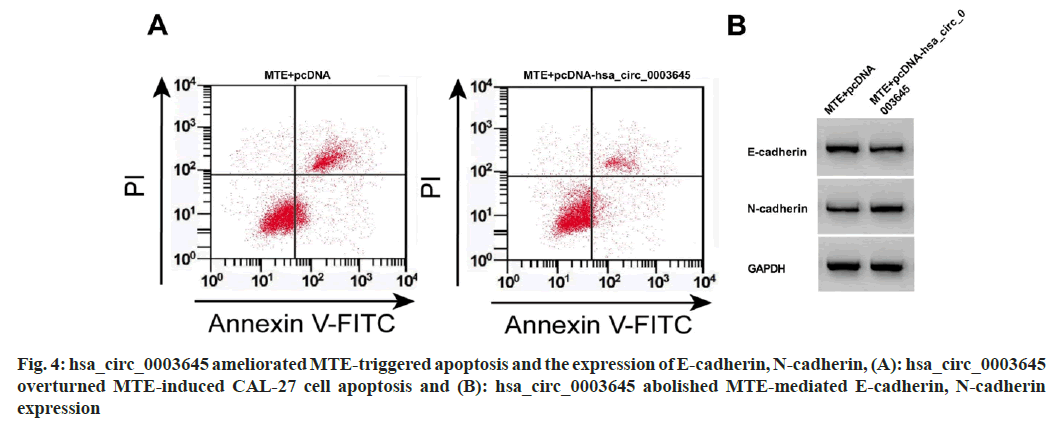
Compared with MTE+pcDNA, hsa_circ_0003645 level, OD value, colony formation number, migration, and N-cadherin were clearly increased in the MTE+pcDNA-hsa_circ_0003645 group, however, miR-335 expression, apoptosis, and E-cadherin were blocked (fig. 4 and Table 5).
| Groups | hsa_circ_0003645 | miR-335 | OD value | Colony formation number | Apoptosis rate (%) | Scratch healing rate (%) | E-cadherin | N-cadherin |
|---|---|---|---|---|---|---|---|---|
| MTE +pcDNA | 0.20±0.02 | 3.40±0.16 | 0.63±0.05 | 64.67±2.87 | 21.38±1.11 | 33.38±1.74 | 0.71±0.06 | 0.25±0.02 |
| MTE +pcDNA-hsa_circ_0003645 | 0.86±0.06a | 1.26±0.06a | 1.04±0.09a | 101.33±5.44a | 9.20±0.60a | 55.22±2.02a | 0.26±0.02a | 0.68±0.05a |
| t | 18.075 | 21.691 | 6.897 | 10.324 | 16.719 | 14.189 | 12.324 | 13.83 |
| p | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: ap<0.05 compared with MTE+pcDNA
Table 5: HSA_circ_0003645 Reversed the Influence of MTE on CAL-27 Cell Malignant Behaviors (x͞ ±s, n=3)
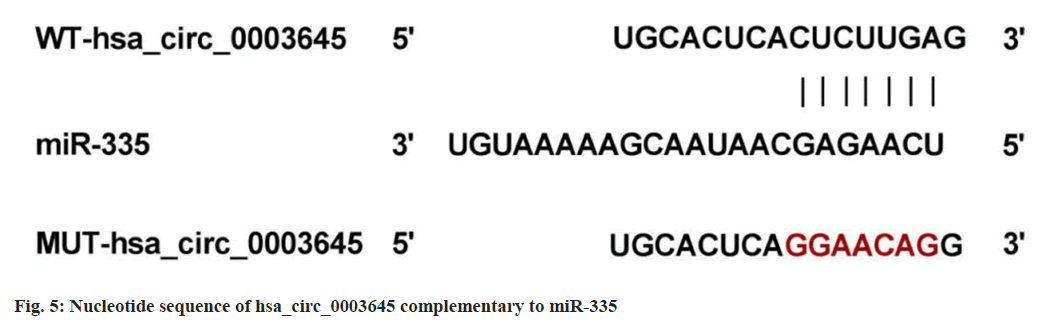
Based on circRNA interactive prediction, existence of complementary sequences between hsa_circ_0003645 and miR-335 was found (fig. 5). In addition, miR-335 upregulation led to an apparent enhancement in the luciferase activity of WT-hsa_circ_0003645, rather than the MUT group (Table 6).
| Group | WT-hsa_circ_0003645 | MUT-hsa_circ_0003645 |
|---|---|---|
| miR-NC | 0.95±0.06 | 0.95±0.05 |
| miR-335 | 0.22±0.02a | 0.96±0.06 |
| t | 19.992 | 0.222 |
| p | 0.000 | 0.835 |
Note: ap<0.05 relative to miR-NC
Table 6: Results of Dual Luciferase Reporter Experiments (x͞ ±s, n=3)
As a TCM, M. tenacissima is rich in alkaloids, organic acids and polysaccharides, and other active ingredients, with anti-inflammatory, anti- tumor, and other pharmacological activities. Of note, convincing evidence has suggested that MTE exerts a potent potential tumor-suppressor effect in various human cancers. It has been reported that MTE might repress hematological tumor cell proliferation by boosting Poly ADP-Ribose Polymerases (PARP) expression and repressing p-Protein Kinase B (AKT) expression[17]. Beyond that, MTE might induce non-small cell lung cancer cell apoptosis via improving caspase-3 activity[18]. In addition, it has been reported that MTE might retard melanoma cell growth through regulating Phosphoinositide 3-Kinase (PI3K)/ AKT/mammalian Target of Rapamycin (mTOR) pathway[19]. Previous studies have described that excessive cell proliferation and impaired apoptosis are principal reasons for tumor development[20].
Herein, our data found that MTE exposure might effectively impede OSCC cell proliferative ability and boost apoptosis in a dose-dependent manner, suggesting that MTE has the underlying value of repressing OSCC development. Furthermore, tumor cell migration has been reported as the main cause of tumor recurrence and metastasis[21]. Tumor cells undergo the Epithelial-Mesenchymal Transition (EMT) process responsible for cytoskeleton alteration and intercellular adhesion reduction, which makes tumor cells easy for migrate[22]. In the current work, applying MTE decline scratch healing rate and N-cadherin and enhance E-cadherin, supporting the repression of MTE on OSCC cell migration ability.
It has been widely accepted that circRNAs might target miRNAs to control OSCC cell growth and metastasis. For example, circFNDC3B might accelerate OSCC migration and invasion by droving EMT[23]. Moreover, circCDR1as overexpression might elevate OSCC cell autophagy, proliferation, motility, and decrease apoptosis[24]. Herein, hsa_circ_0003645 absence might hinder OSCC cell proliferation, migration, and facilitate apoptosis, verifying the suppressive role of hsa_circ_0003645 silencing on OSCC development and that hsa_circ_0003645 might be used as a target for OSCC treatment. Consistent with lncRNA[7], our data validated that MTE exposure might block hsa_circ_0003645 expression in OSCC cells. Functional experiments presented that hsa_circ_0003645 knockdown-mediated OSCC cell proliferation and migration inhibition and apoptosis promotion were partly abrogated after MTE treatment, validating that applying MTE might retard OSCC progression via modulating hsa_circ_0003645.
In terms of molecular mechanisms, our data discovered that hsa_circ_0003645 directly targeted miR-335. Several researches have indicated that miR-335 acted as a tumor-suppressor role by dampening cell proliferation and migration in different tumors[25-27]. Meanwhile, it has been confirmed that Platycodin D (PD) might diminish bladder cancer cell growth, invasion, and EMT[28]. In the present work, hsa_circ_0003645 upregulation might partially counteract MTE treatment-evoked miR-335 content enhancement in OSCC cells, further supporting MTE exposure might dwindle OSCC cell malignant phenotypes via targeting hsa_circ_0003645/miR-335.
In summary, applying MTE suppressed OSCC progression via decreasing hsa_circ_0003645 and increasing miR-335, contributing to the theoretical basis for MTE against OSCC.
Conflict of interests:
The authors declared no conflict of interests.
References
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74(1):12-49.
[Crossref] [Google Scholar] [PubMed]
- Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK, et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol 2021;121:105451.
[Crossref] [Google Scholar] [PubMed]
- Vos JL, Elbers JB, Krijgsman O, Traets JJ, Qiao X, van der Leun AM, et al. Neoadjuvant immunotherapy with nivolumab and ipilimumab induces major pathological responses in patients with head and neck squamous cell carcinoma. Nat Commun 2021;12(1):7348.
[Crossref] [Google Scholar] [PubMed]
- Wang P, Yang J, Zhu Z, Zhang X. Marsdenia tenacissima: A review of traditional uses, phytochemistry and pharmacology. Am J Chin Med 2018;46(07):1449-80.
[Crossref] [Google Scholar] [PubMed]
- Pan Y, Liao X, Yang L, Zhang C, Wang J, Zheng P, et al. Extract of Marsdenia tenacissima (Roxb.) moon (apocynaceae) suppresses hepatocellular carcinoma by inhibiting angiogenesis. Front Pharmacol 2022;13:900128.
[Crossref] [Google Scholar] [PubMed]
- Yuan Y, Guo Y, Guo ZW, Hao HF, Jiao YN, Deng XX, et al. Marsdenia tenacissima extract induces endoplasmic reticulum stress-associated immunogenic cell death in non-small cell lung cancer cells through targeting AXL. J Ethnopharmacol 2023;314:116620.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Gong X, Huang M. Marsdenia tenacissima extract prevents the malignant progression of glioma through upregulating lncRNA MEG3 and SFRP1-dependent inhibition of Wnt/β-catenin pathway. CNS Neurosci Ther 2023;29(5):1272-89.
[Crossref] [Google Scholar] [PubMed]
- Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 2022;19(3):188-206.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 2021;505:49-57.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Ke S, Zheng W, Zhu Z, Wu Y. Hsa_circ_0003645 promotes breast cancer progression by regulating miR-139-3p/HMGB1 axis. Onco Targets Ther 2020;13:10361-72.
[Crossref] [Google Scholar] [PubMed]
- Lin D, Wang Y, Lei L, Lin C. Circ_0003645 serves as miR-335-5p sponge to promote the biological process of diffuse large B-cell lymphoma by upregulating NFIB. Autoimmunity 2022;55(2):127-35.
[Crossref] [Google Scholar] [PubMed]
- Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 2020;6(4):319-36.
[Crossref] [Google Scholar] [PubMed]
- Song G, Ma Y, Ma Y, Liu P, Hou L, Xu Z, et al. miR-335-5p targets SDC1 to regulate the progression of breast cancer. Crit Rev Eukaryot Gene Expr 2022;32(6):21-31.
[Crossref] [Google Scholar] [PubMed]
- Wu YH, Huang YF, Chang TH, Wu PY, Hsieh TY, Hsiao SY, et al. MiR-335 restrains the aggressive phenotypes of ovarian cancer cells by inhibiting COL11A1. Cancers 2021;13(24):6257.
[Crossref] [Google Scholar] [PubMed]
- Ou D, Wu Y, Liu J, Lao X, Zhang S, Liao G. MiRNA‑335 and miRNA‑182 affect the occurrence of tongue squamous cell carcinoma by targeting survivin. Oncol Lett 2016;12(4):2531-7.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Chen YY, Lin C. Effects of extract of Fructus radix on tumor growth, metastasis and expression of cyclooxygenase-2 in MKN-45 rat model of human gastric cancer in situ. Chin J Gerontol 2015;35(11):2939-41.
- Huang X, Chen XY. Study on the inhibitory effect and mechanism of the extract of Wugu Teng on hematological tumor cells. Beijing Tradit Chin Med 2019;38(7):650-3.
- Jiao YN, Wu LN, Xue D, Liu XJ, Tian ZH, Jiang ST, et al. Marsdenia tenacissima extract induces apoptosis and suppresses autophagy through ERK activation in lung cancer cells. Cancer Cell Int 2018;18(5):149-60.
[Crossref] [Google Scholar] [PubMed]
- He XW, Guo GL, Chen JX. Extract of Phyllanthus sinensis inhibits the viability and induces apoptosis of melanoma cells by regulating PI3K/AKT/mTOR signaling pathway. Chin J Physiol 2018;34(12):2180-5.
- Liu X, Wang H, Tao GL, Chu TB, Wang YX, Liu L. LncRNA-TMPO-AS1 promotes apoptosis of osteosarcoma cells by targeting miR-329 and regulating E2F1. Eur Rev Med Pharmacol Sci 2020;24(21):11006-15.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Wang XF, Qin YC, Gong YB, Wang L, Li NC. Downregulation of long non-coding RNA DUXAP10 inhibits proliferation, migration, and invasion of renal cell carcinoma. Eur Rev Med Pharmacol Sci 2020;24(21):11041-51.
[Crossref] [Google Scholar] [PubMed]
- Wang JS, Wang MJ, Lu X, Zhang J, Liu QX, Zhou D, et al. Artesunate inhibits epithelial-mesenchymal transition in Non-Small-Cell Lung Cancer (NSCLC) cells by down-regulating the expression of BTBD7. Bioengineered 2020;11(1):1197-207.
[Crossref] [Google Scholar] [PubMed]
- Li X, Wang C, Zhang H, Li Y, Hou D, Liu D, et al. circFNDC3B accelerates vasculature formation and metastasis in oral squamous cell carcinoma. Cancer Res 2023;83(9):1459-75.
[Crossref] [Google Scholar] [PubMed]
- Cui L, Huang C, Zhou D. Overexpression of circCDR1as drives oral squamous cell carcinoma progression. Oral Dis 2023;29(3):957-67.
[Crossref] [Google Scholar] [PubMed]
- Huo W, Zhang M, Li C, Wang X, Zhang X, Yang X, et al. Correlation of microRNA-335 expression level with clinical significance and prognosis in non-small cell lung cancer. Medicine 2020;99(34):e21369.
- Wang S, Li Y, Sun S, Cai J, Cao J. Sp1 promotes ovarian cancer cell migration through repressing miR-335 expression. Biochem Biophys Res Commun 2020;524(1):211-6.
[Crossref] [Google Scholar] [PubMed]
- Li Q, Wang XJ, Jin JH. SOX2-induced upregulation of lncRNA LINC01510 promotes papillary thyroid carcinoma progression by modulating miR-335/SHH and activating Hedgehog pathway. Biochem Biophys Res Commun 2019;520(2):277-83.
[Crossref] [Google Scholar] [PubMed]
- Chen D, Chen T, Guo Y, Wang C, Dong L, Lu C. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp Cell Res 2020;396(1):112281.
[Crossref] [Google Scholar] [PubMed]