- *Corresponding Author:
- S. D. Vachala
Department of Pharmaceutical Chemistry, R. R. College of Pharmacy, R.R. Institute of Technology, Bengaluru, Karnataka 560090, India
E-mail: geethsen@gmail.com
| Date of Received | 23 May 2023 |
| Date of Revision | 30 August 2024 |
| Date of Accepted | 31 October 2024 |
| Indian J Pharm Sci 2024;86(5):1852-1864 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Novel angularly fused pyrido[3',2':4,5]furo[3,2-d]pyrimidines were synthesized using 3-amino-4- methyl-N-phenyl-6-substituted furo[2,3-b]pyridine-2-carboxamide and various aromatic carboxylic acids using microwave irradiation. The compounds were characterized and tested for in silico, in vitro and in vivo anticancer activities. The synthesized compounds were tested for receptor binding with dihydrofolate reductase and thymidylate synthase enzymes. Compounds pyrido[3',2':4,5]furo[3,2-d] pyrimidine-5, pyrido[3',2':4,5]furo[3,2-d]pyrimidine-6 and pyrido[3',2':4,5]furo[3,2-d]pyrimidine-7 showed effective binding with the target receptors through hydrogen bonding interactions. The 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide assay results revealed the cytotoxicity effect of compounds on Henrietta Lacks cervical cancer cell line, human breast cancer cell line and rat glioma cell line. Compounds, pyrido[3',2':4,5]furo[3,2-d]pyrimidine-5, pyrido[3',2':4,5]furo[3,2-d] pyrimidine-6 and pyrido[3',2':4,5]furo[3,2-d]pyrimidine-7 showed better acitivty on those cell lines with lesser half maximal inhibitory concentration values. So, they were selected for the in vivo anticancer study and in vitro deoxyribonucleic acid ladder assay. Pyrido[3',2':4,5]furo[3,2-d]pyrimidine-5 and pyrido[3',2':4,5]furo[3,2-d]pyrimidine-7 exhibited total tumor volume as 4.9±0.01 and 3.95±0.25 ml/mouse, respectively. Pyrido[3',2':4,5]furo[3,2-d]pyrimidine-5 and pyrido[3',2':4,5]furo[3,2-d] pyrimidine-6 markedly increased the mean survival time levels with an enhanced % increase in life span to 70.83 and 29.16, respectively. From the above results, it is concluded that compounds containing phenyl ring substituted with electron withdrawing groups such as 3-fluoro, 4-trifluoromethyl and 3,5-dimethoxy substitutions at the C-2 of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidin-4(3H)-one could be drug like candidates for further structural modification to enhance their biological activities.
Keywords
Angularly fused pyrimidines, microwave irradiation, in silico screening, 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide assay, anticancer, deoxyribonucleic acid ladder assay
Newer methodologies such as high-throughput screening for faster development of promising biologically active molecules are readily available nowadays. So far, the important pharmacophores which have been identified in fused aromatic heterocycles are quinazolines, pyrido[2,3-d] pyrimidine and furano[3,2-d]pyrimidines. They are of current interest in the drug discovery processes. These privileged structures were extensively studied and represented in chemical compound libraries. Moreover, they have been studied widely, due to their specificity in the inhibition of Dihydrofolate Reductase (DHFR) and Thymidylate Synthase (TS). Angularly fused tricyclic heterocycles have not been studied as far as other fused heterocyclic systems. Roopa et al.[1] reported some naphtho[2,1-b]furan encompassing pyrimidines derivatives as potent antimicrobial agents. Fusion of furo[2,3-b]pyridine with pyrimidine was attempted to bring out a new scaffold with better pharmacological profile. 2-aryl- 4-morpholinopyrido[3',2':4,5]furo[3,2-d]pyrimidine derivative was reported for its antiproliferative activity. The isosteric replacement of sulphur moiety by oxygen showed approximately 400-fold greater potency than 4-morpholin-4-yl-pyrido[3',2':4,5] thieno[3,2-d]pyrimidine and also showed antiproliferative activity in various cell lines, including multi drug resistant multidrug-resistant breast adenocarcinoma (MCF7/ADR-res) cells[2]. There was a significant decrease in the tumor cell chemomigration and invasion being observed.
Further, the in vitro antiproliferative activity was exhibited in four different tumor cell types and in vivo in the MDA-MB-435 breast carcinoma and OVCAR-3 orthotopic ovarian carcinoma model[3]. In 2019, Sirakanyan et al.[4] reported the synthesis and antitumor activity of new amino derivatives of pyrido[3′,2′:4,5](furo)thieno[3,2-d]pyrimidines. And their results revealed that the compound activity depended mostly on the nature of the amine fragments[4]. These findings opened up an avenue for the synthesis of novel angularly fused pyrido[3',2':4,5]furo[3,2-d]pyrimidines to examine their biological properties.
Materials and Methods
Chemistry:
The melting points of the synthesized compounds were determined in open capillary tubes and were uncorrected. The characterization of the compounds was done by various spectral analyses. The IR spectra of the compounds were recorded in the range of 500- 4000 cm-1 on Shimadzu FT-IR 8310 using Potassium bromide (KBr) pellets. The proton Nuclear Magnetic Resonance (1H-NMR) spectra were recorded on Joel, model GSV-400 MHz spectrometer using deuterated chloroform (CDCl3)/Dimethylsulfoxide (DMSO)-d6 as solvent. The chemical shifts were reported as parts per million downfield from tetramethylsilane (Me4Si). Mass spectra were recorded on the Shimadzu GC-MS QP5050. The purity of the compounds were checked by thin layer chromatography on Silicon dioxide (SiO2) gel (HF254, 200 mesh) coated plates. The spots were visualized by Ultraviolet (UV) light and Retention factor (Rf) value was calculated for each compounds.
General procedure for the synthesis of 2-chloro-Nphenylacetamide:
To a solution of chloroacetyl chloride (1 mM) in chloroform, aniline (1.2 mM) and Triethylamine (TEA; 2 mM) were added in succession and the reaction mixture was stirred at room temperature for 30 min. Then it was diluted with Dichloromethane (DCM; 20 ml) and washed with water. The organic layer was dried over anhydrous sodium sulphate and concentrated to dryness under reduced pressure to yield a solid residue, which was crystallized to obtain 2-chloro-Nphenylacetamide[5].
General procedure for the synthesis of 3-amino-4- methyl-N-phenyl-6-substituted furo[2,3-b]pyridine- 2-carboxamide:
To a solution of 4,6-disubstituted-2-hydroxypyridine-3- carbonitrile in dry acetone, 2-chloro-N-phenylacetamide and anhydrous potassium carbonate (0.2 M) were added in equimolar ratio and the reaction mixture was refluxed on water bath for 8 h. The recovered potassium salt was filtered off and the subsequent trituration of the same with ethanol gave 3-amino-4-methyl-N-phenyl-6- substituted furo[2,3-b]pyridine-2-carboxamide[6].
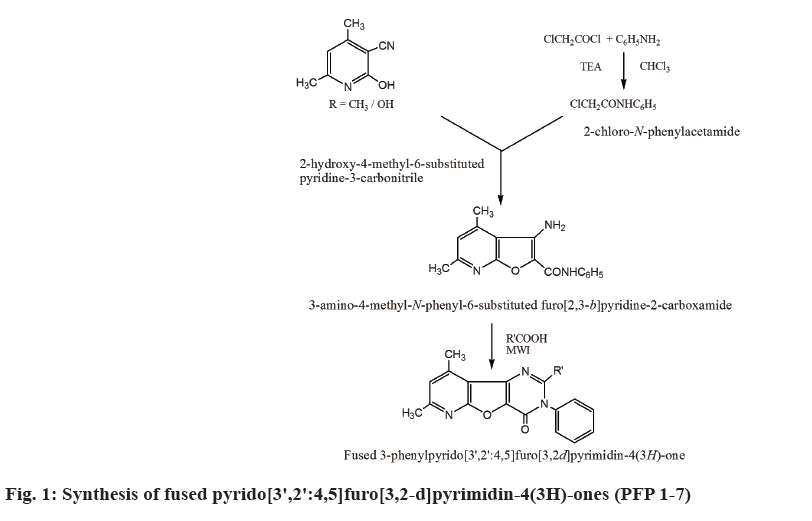
Procedure for the synthesis of fused pyrido[3',2':4,5] furo[3,2-d]pyrimidin-4(3H)-ones (PFP 1-7):
Equimolar amount of 3-amino-4-methyl-N-phenyl- 6-substituted furo[2,3-b]pyridine-2-carboxamide and various aromatic carboxylic acids (Table 1) were irradiated using microwave radiation (300 W) for 4-11 min to obtain fused pyrido[3',2':4,5]furo[3,2-d] pyrimidin-4(3H)-ones (Table 1).
| Compound code | R′ | Molecular formula |
|---|---|---|
| PFP-1 | 4-bromobenzyl | C24H18BrN3O2 |
| PFP-2 | 4-chlorobenzyl | C24H18ClN3O2 |
| PFP-3 | (4-chlorophenoxy)methyl | C24H18ClN3O3 |
| PFP-4 | 4-fluorophenyl | C23H16FN3O2 |
| PFP-5 | 3-fluorophenyl | C23H16FN3O2 |
| PFP-6 | 4-trifluoromethylphenyl | C24H16F3N3O2 |
| PFP-7 | 3,5-dimethoxyphenyl | C25H21N3O4 |
Table 1: Codes for the Synthes?zed 9-Methyl-3-Phenyl-2,7-D?subst?tuted Pyr?do[3',2':4,5]Furo[3,2-D]Pyr?m?d?n-4(3h)-Ones (PFP 1-7) Along w?th the?r Subst?tuents
Biological screening:
Docking studies on DHFR and TS: Protein Data Bank (PDB) files 1BOZ and 1IOO needed for docking studies were selected and subjected to structure validation procedures. The incorrect amino acid residues and their formal charges were adjusted at this step. The high-resolution X-ray structure of TS complex with tomudex (Raltitrexed; RTX) and DHFR complex with Methotrexate (MTX) were imported into Molegro Virtual Docker (MVD) and the ligands and water molecules were extracted. Missing bond order, hybridization states and hydrogen atoms were added, charges were assigned and flexible torsions of ligands were detected. A grid volume that was big enough to cover the entire surface of the protein was used for the docking calculation, while other parameters were at default.
Molecular structures and optimization: A library of molecules was generated by performing structural modifications in fused pyrimidines. These modifications were carried out on fused pyrimidines, viz., from benzopyrimidine, pyridopyrimidine and tricyclic angularly fused pyrimidines in order to arrive molecules with better in silico inhibition molecular docking (MolDock) score as compared with RTX and MTX. The structures of all the fused pyrimidines (ligands) were constructed using advanced chemistry development ChemSketch (ACD) software version 12.01 installed in Intel Pentium 4 machine. The ligands were saved in molecular database (MOL) format and then converted to 3D energy minimized conformations. Finally, the ligands were saved in a single molecular design limited Structure data file (MDL SDF), followed by selection of the same with lower energy conformation for our further studies.
Molecular docking: Molecular docking of fused pyrimidines on the active site of TS and DHFR were carried out using MVD version 2011.4.3. The default settings, including a grid resolution of 0.30 Å for grid generation and a 15 Å radius from the template as the binding site were used. Maximum numbers of poses were generated from the default value of 5. MVD essentially works based on evolutionary algorithm. In a normal situation, consecutive docking runs do not give exactly the same fitness scores. To address this issue, ten successive runs were allowed and then the ligands with the best conformational and MolDock scores were selected for further experimental tests. MVD was used to calculate the interaction energy between ligands and macromolecular systems from the 3D structures of the protein and ligands. The MolDock Score energy, Escore, was defined by the following equation[7].
Escore = Einter+Eintra
Where, Einter was the ligand-protein interaction energy and Eintra was the internal energy of the ligand.
In vitro anticancer activity by 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay:
MTT assay is one of in vitro assays which was developed to determine the sensitivity of specific tumors and to individualize therapy. It measures mitochondrial dehydrogenase activity as a reflection of cell viability[8]. Cancerous cells were grown in T-25 cm2 tissue culture flasks containing Minimum Essential Medium (MEM) media supplemented with 10 % Fetal Bovine Serum (FBS), 1 % L-glutamine and 50 μg/ml gentamicin sulphate at 37° in Carbon dioxide (CO2) incubator in an atmosphere of humidified 5 % CO2 and 95 % air. The cells were maintained by routine subculturing. Single cell suspension was obtained from a monolayer culture. Cells were dislodged from the culture flasks by trypsinization. Exponentially growing HeLa, T47D and C6 cells were harvested from T-25cm2 tissue culture flasks and a stock cell suspension (5×104 cells/ml) was prepared. A sterile 96-well flat bottom tissue culture plate was seeded with 5×103 cells in 0.1 ml of MEM supplemented with 10 % FBS and allowed to attach for 24 h. Cells were treated with different concentrations of test compound (12.5-100 μM) in triplicates and incubated for 24 h. The control group cells were treated with only the medium containing 0.1 % DMSO (DMSO control). Drug containing media was removed and washed with 100 μl of PBS and then 100 μl of MTT reagent (1 mg/ml) was added and incubated for 3 h at 37°. After 3 h of incubation, MTT was removed by draining on tissue paper and the formazan crystals formed in each well were dissolved in 100 μl of DMSO. The absorbance was measured by an ELISA plate reader at 540 nm and the percentage cytotoxicity was calculated.
DNA ladder assay:
DNA ladder assay demonstrates the chromosomal DNA fragmentation, which is the biological hallmark of apoptosis; whereas, in necrosis, random digestion of DNA occurs and it shows ladder pattern in agarose gel electrophoresis[9]. The monolayer cell culture was trypsinized and cell count was adjusted to 4×105 cells/ ml using MEM containing 10 % FBS. To each one of the wells, 2 ml of diluted cell suspension was added and incubated at 37° for 24 h in 5 % CO2 incubator. After 24 h, when a partial monolayer was formed, the supernatant was discarded and 2 ml of diluted test solution was added to each well in duplicate and kept for incubation at 37° in 5 % CO2 incubator. After 24 h of exposure, cells were harvested by trypsinization and centrifuged for 5 min at 1000 rpm. The supernatant liquid was then discarded and the cell pellet was dissolved in 1 ml Nicoletti digestion buffer and then incubated at 45° overnight. Thus, the genomic DNA was extracted. Further, ribonuclease (RNAase; 40 μg/ml, prepared in distilled water) was added in the aqueous phase and was kept at room temperature for 2 h. The genomic DNA extracted was further extracted with chloroform and the successive treatment with ethanol precipitates the same in presence of 0.3 M sodium acetate. The DNA was pelleted by centrifugation at 10 000 rpm for 5 min at 4° and it was suspended in 100 μl of 1×Tris-EDTA buffer. 10 μg of DNA was subjected to 2 % agarose gel electrophoresis. The gel was run at 65 V for 1 h or until the tracking dye moved to the bottom of gel. The gel was then visualized by placing under the UV transilluminator and the fluorescence of DNA fragments was observed. The photographs were then taken from gel documentation system.
In vivo anticancer activity:
In vivo anticancer activity of the selected compounds was determined by liquid tumour model using Ehrlich Ascites Carcinoma (EAC) cells, originated from breast carcinoma by spontaneous passaging. The EAC cells were maintained in Swiss albino mice, by intraperitoneal (i.p.) transplantation on every 9th d. Swiss albino male mice (20±2 g) of 8-10 w old, were selected for in vivo anticancer study and maintained in the animal house at controlled temperature, humidty and light (23±2°,50±5 %, 14 and 10 h of light and dark, respectively). Animals were provided with sterile food and water ad libitum. The acute toxicity study of the compounds was evaluated in the mice by the up and down staircase method[10]. The dose was selected based on the acute toxicity study reports. 1/20th of the tolerable dose was selected for this study. The mice were weighed and divided into 6 groups (Table 2).
| Group | Treatment |
|---|---|
| I | Normal control |
| II | Tumor induced animals+2 % w/v Acacia (Tumor control) |
| III | Tumor induced animals+PFP-5 (50 mg/kg body weight, i.p.) |
| 1V | Tumor induced animals+PFP-6 (50 mg/kg body weight, i.p.) |
| V | Tumor induced animals+PFP-7 (50 mg/kg body weight, i.p.) |
| VI | Tumor induced animals+cisplatin (3.5 mg/kg body weight, i.p.) |
Table 2: Groups Under In V?vo Ant?cancer Act?v?ty
ach group consisted of 6 animals. EAC cells (1.5×105 cells/mouse) were injected by i.p. route to each mouse of each group except the normal control group (Group I). This was taken as Day 0. Group II served as a tumor control. Test compounds and reference drug treatment were continued for 1, 3, 5, 7, 9, 11, 13th d starting from Day 1. On the 14th d, 24 h after the last dose, half the number of animals (n=6) from each group were sacrificed and the rest were kept for the life span study of the tumor hosts. After sacrificing the animals, blood sample was collected to estimate the parameters.
Parameters monitored:
The effect of test compounds carried out on tumor growth and host’s survival time were examined by studying the parameters such as, tumor volume, packed cell volume, viable tumor cell count, non-viable tumor cell count, haematological cell count, Mean Survival Time (MST) and Percentage Increase in Life Span (% ILS) [11-14]. The values were recorded as mean±Standard Error of the Mean (S.E.M).
Tumor volume and packed cell volume:
The mice were dissected and ascitic fluid from peritoneal cavity was carefully collected to measure the total tumor volume. The fluid was centrifuged to determine the packed cell volume at 1000 rpm for 5 min.
Viable and non-viable tumor cell count:
To count the viable and non-viable cells, the ascitic fluid was diluted 20 times. This cell suspension was mixed with tryphan blue (0.4 % in normal saline) dye and allowed to stand for few minutes at room temperature. After staining, it was placed on the Neubauer counting chamber and the numbers of cells in the 64 small squares were counted. The cells that did not take up the dye were viable and those that took the stain were non-viable.
Haematological studies:
The blood sample was collected from mice by retroorbital puncture under anesthesia and the estimation of haematological parameters such as, Red Blood Cells (RBC), White Blood Cells (WBC) and Haemoglobin (Hgb) were performed using Veterinary blood cell counter. Monocyte and lymphocyte cells were also counted.
MST and % ILS:
The sets of animal groups allotted for the survival observations were maintained separately. These animals were observed for mortality daily for 6 w or till the death. The effect of compounds on life span was measured by calculating the MST and the % ILS. An enhancement of life span by 25 % or more was considered as an effective antitumor response.
MST=[Day of 1st death+Day of last death]/2
% ILS=[(Mean survival of treated group/Mean survival of control group)-1]×100
Results and Discussion
The synthesis of 9-methyl-3-phenyl-2,7-disubstituted pyrido[3',2':4,5]furo[3,2-d]pyrimidin-4(3H)-ones (PFP 1-7) were carried out as outlined in fig. 1. In presence of TEA, chloroacetyl chloride reacted with aniline to give 2-chloro-N-phenylacetamide which on cyclization with 4,6-disubstituted-2-hydroxypyridine- 3-carbonitrile yielded 3-amino-4-methyl-N-phenyl- 6-substituted furo[2,3-b]pyridine-2-carboxamide (Table 1). Further, the microwave irradiation of the above compound with appropriate aromatic acids resulted in the formation of compounds PFP 1-7. The physicochemical properties of compounds PFP 1-7 were determined were shown in Table 3.
| Compound code | Molecular weight (Da) | Yield (%) | MWI Reaction time (min) | Melting point (°) | Rf | UV λmax (nm) |
|---|---|---|---|---|---|---|
| PFP-1 | 459 | 95 | 11 | 97 | 0.87 | 201 |
| PFP-2 | 415 | 96 | 7 | 110 | 0.79 | 202 |
| PFP-3 | 431 | 71 | 8 | 123 | 0.8 | 205 |
| PFP-4 | 385 | 68 | 8 | 129 | 0.75 | 207 |
| PFP-5 | 385 | 70 | 7 | 105 | 0.61 | 203 |
| PFP-6 | 435 | 71 | 7 | 152 | 0.72 | 210 |
| PFP-7 | 427 | 75 | 6 | 172 | 0.78 | 204 |
Table 3: Phys?co-Chem?cal Propert?es of Compounds PFP 1-7.
Structure of the compounds was confirmed by spectral analysis. Spectral data of 2-(3,5-dimethoxyphenyl)- 7,9-dimethyl-3-phenylpyrido[3',2':4,5]furo[3,2-d] pyrimidin-4(3H)-one (PFP-7) revealed the following details, IR (KBr) (cm-1): 3007 (aromatic -CH-, Str.), 1676 (C=O, Str.), 2953 (-CH3-, -CH- Str.), 2837 (OCH3, CH Str.), 1600 (ring C=N), 1550 (ring C=C Str.), 1454 (ring C-N Str.), 1211 (unsymmetrical C-O-C Str.). 1H-NMR (CDCl3): 8.3-8.5 (pyridyl -CH-, 1H, singlet), 7.1-7.8 (aromatic protons, 5H, multiplet), 6.7 - 6.2 (dimethoxyphenyl ring -CH-, 3H, multiplet), 4.3-4.2 (singlet, 3H, -OCH3), 3.8-3.9 (singlet, 3H, -OCH3), 3.1 (singlet, 3H, CH3), 2.4 (singlet, 3H, CH3). MS: 428 (M+1), 397 (M+-C2H6), 290 (M+-C8H9O2), 145 (C9H7NoO), 137 (C8H9O2), 121 (C7H7NO), 108 (C6H4O2), 77 (C6H5). Anal. Calc. for C25H21N3O4: C, 70.25; H, 4.95; N, 9.83; O, 14.97. Found: C, 70.30; H, 4.60; N, 9.75; O, 14.85.
Chemically induced cytotoxicity is one of the major approaches to produce antitumor activity. Reports are available on the drugs being listed as cytotoxic agents[15]. MTT assay measures mitochondrial dehydrogenase activity as a reflection of cell viability and is very useful as one of the most extensively used techniques in the screening and evaluation of new anticancer agents[8].
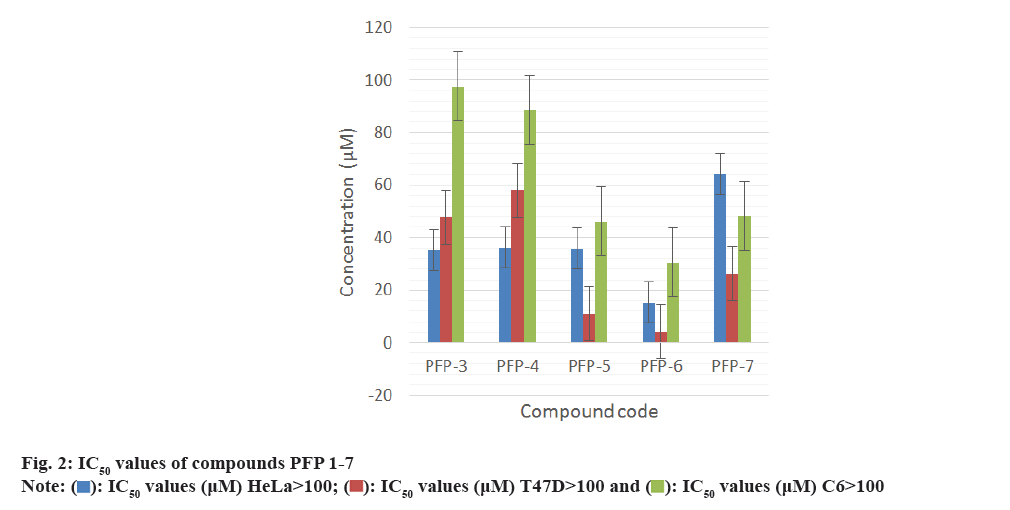
All the synthesized compounds were evaluated against three tumor cells; HeLa, T47D and C6 in 24 h drug exposure assay. In this study, the dose response curve was plotted to determine their IC50 values (fig. 2-fig. 5).
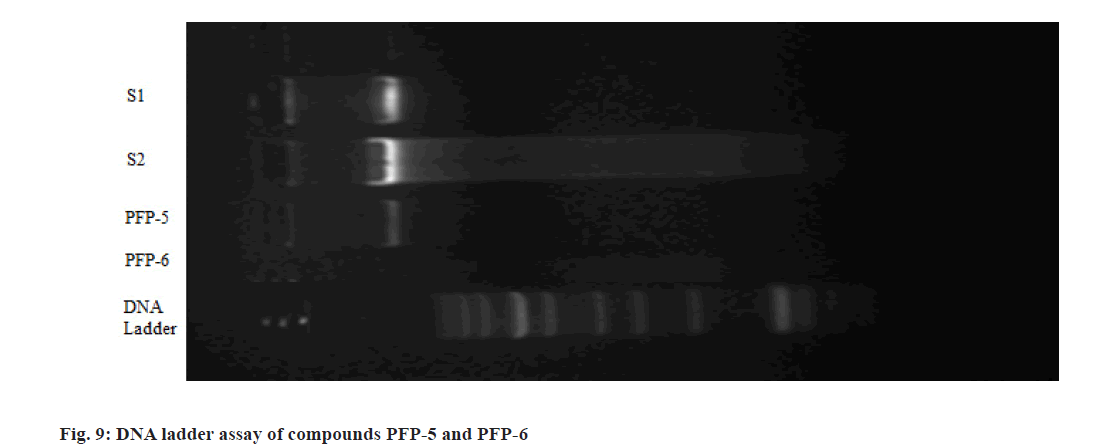
The MTT assay performed on the angularly fused pyrimidin-4(3H)-ones revealed that, compounds PFP- 5 and PFP-6 exhibited 50 % growth inhibition of T47D cells at 11.0 and 4.2 μM, respectively and indicated that, they could be good systems for predicting the antiproliferative activity. DNA fragmentation is the most widely studied event in programmed cell death (Apoptosis). Hence, they were selected for DNA ladder assay. The T47D cells were exposed to the test compounds for 24 h and from this; the DNA was isolated and subjected for electrophoresis using 2 % agarose gel. Photographs were then taken from the gel documentation system (fig. 6). Cisplatin (standard-1) and doxorubicin (standard-2) were used as standards. Here, the medium was treated as the negative control and the solvent control was 0.05 % DMSO. The results suggested that, compounds PFP-5 and PFP-6 showed their cytotoxicity not by means of the internucleosomal DNA fragmentation.
The compounds were found to be safe at a single oral dose of 500 and 1000 mg/kg body weight. Thus, the 1/20th of the higher tolerable dose were selected for the in vivo anticancer study. Parameters such as, total tumor volume, VC, non-viable cell count, packed cell volume, MST and % ILS were assessed. The substance which shows increase in the % ILS could be (≥25 %) considered as a sign for an effective anticancer agent.
Additionally, mean increase in the body weight could be a useful parameter to evaluate the anticancer activity. All the selected compounds showed significant reduction in the total tumor volume, as shown in Table 4. Similarly, RBC, WBC, differential counts (monocytes, lymphocytes) and haemoglobin levels in drug-treated animals were also shown in Table 5. Further, the anticancer efficacy of these compounds the % ILS and MST were shown in fig. 6. The mean increase in body weight of compounds treated group and tumor control are given in fig. 7.
| Groups | Total volume (ml) | Packed cell volume (ml) | Viable cells (106 cells/mouse) | Non-viable cells (106 cells/mouse) |
|---|---|---|---|---|
| Group II (Tumor control) | 13.0±0.70 | 7.0±0.15 | 62.1±0.12 | 1.8±0.16 |
| Group III (PFP-5) | 4.9±0.01*** | 1.95±0.15*** | 25.18±0.01** | 2.3±0.08 |
| Group IV (PFP-6) | 8.0±0.17*** | 1.6±0.11*** | 31.2±0.35** | 3.3±0.41** |
| Group V (PFP-7) | 3.95±0.25*** | 1.4±0.20*** | 20.7±0.18*** | 2.7±0.09 |
| Group VI (Cisplatin) | 2.0±0.02*** | 1.5±0.12 | 21.8±0.19*** | 3.7±0.43*** |
Note: * p<0.05; ** p<0.01; *** p<0.001 as compared to tumor control (group II)
Table 4: Effect of PFP-5, 6 and 7 on Total Volume, Packed Cell Volume, V?able and Non-V?able Tumor Cell Count of the EAC Cells Bear?ng M?ce
| Groups | Haematological Parameters | ||||
|---|---|---|---|---|---|
| RBC (106/μl) | WBC (103/μl) | Hgb (g/l) | Monocyte (103/μl) | Lymphocyte (103/μl) | |
| Group I | 9.00±0.01 | 5.10±0.28*** | 12.0±0.16*** | 0.10±0.32*** | 5.00±0.15*** |
| Group II | 4.23±0.68c | 92.9±0.45c | 5.5±0.53c | 5.4±0.15c | 44.2±0.23c |
| Group III (PFP-5) | 7.54±0.06*** | 40.7±0.10c*** | 9.55±0.55c*** | 3.15±0.12c*** | 31.05±0.48c*** |
| Group IV (PFP-6) | 8.20±0.16*** | 84.6±0.04c*** | 11.3±0.16*** | 6.2±0.23c | 43.4±0.17c |
| Group V (PFP-7) | 7.61±0.01*** | 39.1±0.18c*** | 9.4±0.17c*** | 3.2±0.16c*** | 32.5±0.25c*** |
| Group VI (Cisplatin) | 6.12±0.51c* | 44.2±0.11c*** | 7.7±0.18c*** | 3.4±0.29c*** | 10.20±0.53c*** |
Note: * p<0.05; **p<0.01; ***p<0.001 as compared to tumor control (group II); a<0.05; b<0.01; c<0.001 as compared to normal control (group I)
Table 5: Effect of Compounds PFP-5, PFP-6 and PFP-7 on the Haematolog?cal Parameters of the EAC Cells Bear?ng Sw?ss Alb?no M?ce (Mean±Sem, N=6)
The synthesis of 9-methyl-3-phenyl-2,7-disubstituted pyrido[3',2':4,5]furo[3,2-d]pyrimidin-4(3H)-ones (PFP 1-7) were carried out using 3-amino-4-methyl- N-phenyl-6-substituted furo[2,3-b]pyridine-2- carboxamide on microwave irradiation with different aromatic carboxylic acids (PFP 1-7).
Structure of the compounds was confirmed by spectral analysis. In the IR spectra of compounds PFP 1-7, the carbonyl group absorption was observed in the region of 1670-1690 cm-1, whereas, the hetero aromatic -C N- stretching was observed around 1556-1600 cm- 1. Further, the compounds containing -OH and -CH3 groups produced absorption band in the range of 3180- 3381 cm-1 and 2922-2953 cm-1, respectively. In the 1H NMR spectra of compound PFP-7, the methyl group in all the compounds showed a singlet between δ 2.4- 3.5 ppm. In addition, compounds containing -OCH3 protons exhibited a singlet in the region of 3.8-4.2 ppm. GCMS spectral data of compounds were also in agreement with the proposed structure of compounds PFP 1-7.
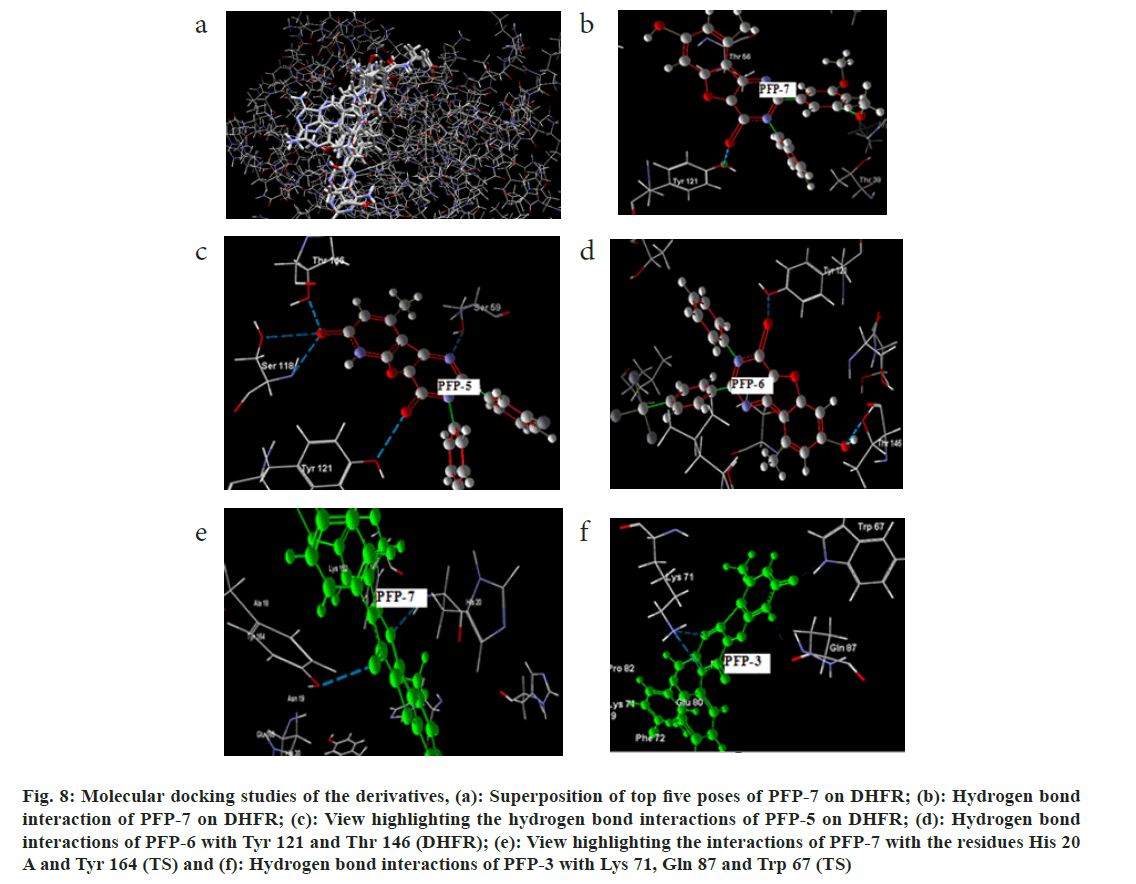
The synthesized 7-hydroxy-9-methyl-3-phenyl-2- substituted pyrido[3',2':4,5]furo[3,2-d]pyrimidin- 4(3H)-one (PFP 1-7) derivatives were docked into DHFR and TS enzymes. The docking results of the derivatives are summarized in Table 6 and Table 7. The pyrido[3',2':4,5]furo[3,2-d]pyrimidin-4(3H)- one system (PFP 1-7) showed interactions with the residues such as, Ser 59, Thr 146, Thr 121, Lys 55 and Gly 20. Among this series, compound PFP-7 showed the lowest binding energy and the MolDock score was found to be -155.83 kcal/M. Further, the best pose of this compound showed the hydrogen bond interaction with the -OH group of Tyr 121 residue, as shown in fig. 8b. The superposition of the top 5 poses of the ligand, PFP-7 in the bound form on DHFR is given fig. 8a. Further, the effective hydrogen bond interactions were also shown by the compound PFP-6 and its MolDock score and the number of poses obtained for this compound were registered as -147.812 kcal/M and 13, respectively. The hydrogen bond interactions of this compound were seen with the -OH group of Thr 146 and Tyr 121 residues, as shown in fig. 8d and the total bond energy was found to be -1.5966 kcal/M. Interestingly, compound PFP- 5, having the 3-fluorophenyl substitution at the 2nd position exhibited maximum of five hydrogen bond interactions with the enzyme and its MolDock score was -133.58 kcal/M (fig. 8c).
| Code | MolDock score (grid) | MolDock score | Total no. of poses | No. of H-bond | Hydrogen bond interactions | |
|---|---|---|---|---|---|---|
| H-bond energy | Interacting amino acid | |||||
| PFP-1 | -142.41 | -142.61 | 25 | 1 | -0.43 | Ser 59 |
| PFP-2 | -139.38 | -134.6 | 24 | 2 | -2.13 | Gly 20, Lys 55 |
| PFP-3 | -135.19 | -129.11 | 25 | 5 | -4.19 | 2Ser 59, Ser 118, Thr 146, Tyr 121 |
| PFP-4 | -134.78 | -129.91 | 23 | 2 | -2.55 | 2Ser 59 |
| PFP-5 | -139.25 | -133.58 | 26 | 5 | -4.35 | 2Ser 118, Thr 146, Ser 59, Tyr 121 |
| PFP-6 | -148.82 | -147.81 | 13 | 2 | -1.59 | Tyr 121, Thr 146 |
| PFP-7 | -157.03 | -155.83 | 21 | 1 | 0 | Tyr 121 |
Table 6: B?nd?ng Data for the Lowest Energy Conformer of Compounds 7-Hydroxy-9-Methyl-3-Phenyl-2-Subst?tuted Pyr?do[3',2':4,5]Furo[3,2-D]Pyr?m?d?n-4(3h)-One (Pfp 1-7) Docked on the DHFR Act?ve S?te and the?r H-Bond Interact?ons w?th Am?no Ac?d Res?dues
| Code | Grid score | MolDock score | No. of poses | Hydrogen bond Interactions | |||
|---|---|---|---|---|---|---|---|
| H-bond energy | No. of H-bond | Interaction with TS (chain) | Interacting amino acid | ||||
| PFP-1 | -131.1 | -126.9 | 25 | -4.67 | 3 | B | Lys 71, Leu 70, Gln 87 |
| PFP-2 | -127.3 | -121.8 | 38 | -2.13 | 2 | B | Lys 71 |
| PFP-3 | -127.6 | -121.7 | 28 | -3.48 | 4 | B | Lys 71, Gln 87, Trp 67 |
| PFP-4 | -118 | -112.3 | 26 | -2.68 | 3 | B | Ser 83, 2Gln 87 |
| PFP-5 | -130.8 | -129.5 | 34 | -1.4 | 2 | B | Lys 71, His 32 |
| PFP-6 | -109.4 | -105.7 | 26 | -2.09 | 3 | B | Lys 71, Gln 87, Trp 67 |
| PFP-7 | -131.7 | -130.9 | 25 | -2.32 | 2 | A+B | His20,Tyr 164 |
Table 7: B?nd?ng Data for the Lowest Energy Conformer of Compounds PFP 1-7 Docked on the TS Act?ve S?te and the?r H-Bond Interact?ons w?th Am?no Ac?d Res?dues
Fig. 8: Molecular docking studies of the derivatives, (a): Superposition of top five poses of PFP-7 on DHFR; (b): Hydrogen bond interaction of PFP-7 on DHFR; (c): View highlighting the hydrogen bond interactions of PFP-5 on DHFR; (d): Hydrogen bond interactions of PFP-6 with TYR 121 and THR 146 (DHFR); (e): View highlighting the interactions of PFP-7 with the residues HIS 20 A and TYR 164 (TS) and (f): Hydrogen bond interactions of PFP-3 with LYS 71, GLN 87 and TRP 67 (TS)
Among the compounds tested (PFP 1-7), for their binding affinity with TS, compound having 3,5-dimethoxyphenyl substitution at the 2nd position in the pyrido[3',2':4,5]furo[3,2-d]pyrimidin-4(3H)-one system (PFP-7) showed remarkable hydrogen bond interactions with chain ‘A’ and chain ‘B’ of 1IOO and the binding energy was found to be -130 kcal/M. The hydrogen bond interactions of compounds PFP-7 and PFP-3 are depicted in fig. 8e and fig. 8f, respectively. The above results revealed that the pyrido[3',2':4,5] furo[3,2-d]pyrimidin-4(3H)-one scaffold could be sustained for the effective enzyme binding.
All the synthesized compounds were evaluated for their cytotoxicity studies on three different tumor cell lines namely, HeLa, T47D and C6 in 24 h drug exposure assay. The effect of substituted phenyl group at the 2nd position and methyl group at the 7th and 9th positions were studied. In this study, the dose response curve was plotted to determine their IC50 values (fig. 2-fig. 5). From the results, it was understood that, the cell proliferation decreased with increase in the dose.
Among the seven compounds tested, PFP-1 and PFP-2 having 4-bromobenzyl and 4-chlorobenzyl substitution respectively at the 2nd position, did not effectively inhibit the growth on any cell lines and their IC50 values were found to be >100 μM. However, other compounds in this series showed encouraging cytotoxicity on these three cell lines. Among them, compound PFP-6, bearing 4-(trifluoromethyl)phenyl substitution at the 2nd position exhibited a greater cytotoxicity on HeLa, T47D and C6 cells with the 50 % inhibition of the growth at 15.2, 4.26 and 30.5 μM, respectively. Compounds PFP- 5 and PFP-7 also were nearly equivalent in exhibiting their antiproliferative activity.
DNA fragmentation is widely studied in apoptosis study. From the MTT study results, it was observed that the compounds PFP-5 and PFP-6 were found be active on T47D cells with less inhibitory concentration of 11.0 and 4.2 μM, respectively. It indicated that, they could be good systems for predicting the antiproliferative activity. The T47D cells were exposed to the test compounds for 24 h and from this; the DNA was isolated and subjected for electrophoresis using 2 % agarose gel. Photographs were then taken from the gel documentation system. Cisplatin (standard-1) and doxorubicin (standard-2) were used as standards. Here, the medium was treated as the negative control and the solvent control was 0.05 % DMSO. No DNA fragmentation was observed for compounds PFP-5 and PFP-6 (fig. 9). Thus the results suggested that, compounds PFP-5 and PFP-6 showed their cytotoxicity not by means of the internucleosomal DNA fragmentation.
Acute toxicity studies were carried out to determine the safer dose of test compounds on mice. The compounds were found to be safe at a single oral dose of 500 and 1000 mg/kg body weight. Blood parameters, MST, % ILS and mean increases in the body weight were evaluated. Cells get accumulated in the peritoneal cavity in tumor bearing mice and the ascitic fluid present in the peritoneal cavity is very important for tumor growth, due to its nutritional value. Generally, ascitic tumor implantation promotes local inflammatory reactions and provides an increase in the vascular permeability, which results in the formation of oedema, cellular migration and progressive ascitic fluid formation.
The reduction in the peritoneal fluid volume, packed cell volume and the viable cell count in tumor bearing mice indicate the anticancer activity. The other attributes, which provides anticancer potential could be an increase in the non-viable cell count[16,17]. Anemia and myelosuppression are the main problems encountered in cancer chemotherapy[18]. Anemia induced in tumor bearing mice is mainly due to the reduction of RBC or haemoglobin percentage and this may occur either, due to iron deficiency or haemolytic or myelopathic conditions[19]. It is noteworthy to mention here that, tumorigenesis and its progression are accompanied by the following changes compared with normal cytogenesis; gradual decrease in the haemoglobin content and erythrocyte count, gradual increase in leukocytes and thrombocytes.
Further, to support the anticancer potential, the improvement in the blood parameters such as, increase in haemoglobin level, RBC count and decrease in WBC count, and differential cell count towards the normal value would help. Thus, the improvements in haematological parameters also will enable to evaluate the potency of the test compound. Based on the MTT assay results, from the synthesized compounds, three compounds were selected and tested for their in vivo anticancer activity[20].
All the selected compounds showed significant reduction in the total tumor volume, as shown in Table 4. Particularly, compounds PFP-5 and PFP-7 exhibited the total tumor volume at 4.9±0.01 and 3.95±0.25 ml/ mouse, respectively. The total peritoneal fluid volume measured in the cisplatin treated group was found to be at 2.0±0.02 ml/mouse. Similarly, the packed cell volume was also decreased significantly by the test compounds.
However, the maximum reduction in the packed cell volume was exhibited by compound PFP-7 (1.4±0.20 ml/mouse). Further, the fluid was tested to find out the presence of the viable and non-viable tumor cells by tryphan blue assay. All the four compounds significantly decreased the viable cell count. Particularly, it was reduced to a great extent for compound PFP-7 (20.71±0.18×106 cells/mouse). Further, increase in non-viable cell count was observed (Table 4) in all the treatment groups. However, treatment by compound PFP-6 exhibited the presence of non-viable cells at 3.3 ± 0.41×106, respectively.
RBC, WBC, differential counts (monocytes, lymphocytes) and Hb levels in drug-treated animals were also restored significantly when compared to that of the EAC control values. The effects of compounds on haematological parameters are shown in Table 5. The RBC count and Hgb level were significantly increased by the treatment with compound PFP-6 when compared with that of the standard. Other two compounds also showed improvement in the RBC count and Hgb levels. The decrease in the WBC cell count also supported to measure the anticancer potential of a compound. Furthermore, the monocytes and lymphocytes counts were also brought down towards near normal levels as, shown in Table 5. Thus, the results obtained were supportive.
Further, the anticancer efficacy of these compounds was supported by the enhanced survival time of the EAC bearing mouse. Among these four test compounds, PFP-5 and PFP-6 markedly increased the MST levels at 20.5 and 15.5 d, respectively. The enhancement of ILS was observed at 70.83 % and 29.16 %, respectively. However, compound PFP-7 did not increase the life span considerably and it showed the percentage ILS only at around 4.16. Cisplatin at a dose of 3.5 mg/kg (i.p.) increased the % ILS value to be at 79.16, as shown in fig. 6.
The mean increase in body weight of compounds treated group and tumor control are given in fig. 7 and it was observed that the compounds showed a significant decrease in the increased body weight than that of the tumor control. Among the four derivatives, compound PFP-6 exhibited significant reduction in the mean increase in the body weight. The other compounds also reduced the increase in the body weight. After treatment with the compound PFP-5, 6 and 7 under investigation, all the depleted haematological parameters were reverted back towards normal. Moreover they enhanced the MST and % ILS. This results disclosed that, the compounds containing phenyl ring substituted with electron withdrawing trifluoromethyl substitution at the 2nd position of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidin- 4(3H)-one exhibited better anticancer activity.
In the present work, a simple and efficient practical method for the synthesis of angularly fused tricyclic pyrimidines was achieved by using MWI technique. The recovery of the product was much easier as compared to the conventional procedures. Further, it contributed to the development of clean, cost effective and eco-friendly procedure with good yield. All the compounds showed good antioxidant activity. Further, compound PFP-6 containing 4-(trifluoromethyl) phenyl substitution at the 2nd position showed promising anti-inflammatory and anticancer activities. Hence, it is concluded that, the PFP scaffold with carbonyl and 4-trifluoromethyl phenyl functions at the 4th and 2nd positions, respectively supported the biological activities. Results of biological activities with substitutions in other positions may widen our understanding on the structure activity relationship remarkably. Optimization of the activity of pyrido[3',2':4,5]furo[3,2-d]pyrimidin-4(3H)-one scaffold using more number of cancer cell lines would be a possible suggestion for future studies.
Acknowledgement
The authors are thankful to AICTE, New Delhi for providing the research grant and also we express our gratitude to the Management of Manipal College of Pharmaceutical Sciences, Manipal University, for providing all the facilities to carry out this work. The corresponding author also thank to the Principal, RRCOP, for providing the facilities to publish this work.
Conflict of interests:
The authors of this manuscript declare that there is no conflict of interest.
References
- Roopa DL, Basavarajaiah SM, Punarva HB. Design, synthesis, spectral analysis, drug likeness prediction, and molecular docking investigations of new Naphtho [2, 1-b] furan encompassing pyrimidines as potential antimicrobial agents. Polycycl Aromat Compd 2024;44(9):6042-63.
- Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Yamano M, et al. Synthesis and biological evaluation of pyrido [3', 2': 4, 5] furo [3, 2-d] pyrimidine derivatives as novel PI3 kinase p110a inhibitors. Bioorg Med Chem Lett 2007;17(9):2438-42.
[Crossref] [Google Scholar] [PubMed]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell 2006;9(5):341-9.
[Crossref] [Google Scholar] [PubMed]
- Sirakanyan SN, Spinelli D, Geronikaki A, Hakobyan EK, Sahakyan H, Arabyan E, et al. Synthesis, antitumor activity, and docking analysis of new pyrido [3', 2': 4, 5] furo (thieno)[3, 2-d] pyrimidin-8-amines. Molecules 2019;24(21):3952.
[Crossref] [Google Scholar] [PubMed]
- Ghosh S, Tiwari P, Pandey S, Misra AK, Chaturvedi V, Gaikwad A, et al. Synthesis and evaluation of antitubercular activity of glycosyl thio-and sulfonyl acetamide derivatives. Bioorg Med Chem Lett 2008;18(14):4002-5.
[Crossref] [Google Scholar] [PubMed]
- Mahadevan KM, Vaidya VP, Vagdevi HM. Synthesis of novel naphtho [2, 1-b] furopyrimidine derivatives. Ind J Chem 2003;42B:1931-7.
- Thomsen R, Christensen MH. MolDock: A new technique for high-accuracy molecular docking. J Med Chem 2006;49(11):3315-21.
[Crossref] [Google Scholar] [PubMed]
- Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res 1988;48(3):589-601.
[Google Scholar] [PubMed]
- Zhu N, Wang Z. An assay for DNA fragmentation in apoptosis without phenol/chloroform extraction and ethanol precipitation. Anal Biochem 1997;246(1):155-8.
[Crossref] [Google Scholar] [PubMed]
- Ghosh MI. Fundamentals of experimental pharmacology. Scientific Book Agency: Calcutta; 1984.
- Zarafoenetis CJ. Methods for the examination of the blood. Approved laboratory techniques. Appleton-Century-Crofts, New York. 1969:39-126.
- Al-Harbi MM, Qureshi S, Raza M, Ahmed MM, Giangreco AB, Shah AH. Influence of anethole treatment on the tumour induced by Ehrlich ascites carcinoma cells in paw of Swiss albino mice. Eur J Cancer Prev 1995;4(4):307-18.
[Crossref] [Google Scholar] [PubMed]
- Al-Harbi MM, Qureshi S, Raza M, Ahmed MM, Giangreco AB, Shah AH. Influence of anethole treatment on the tumour induced by Ehrlich ascites carcinoma cells in paw of Swiss albino mice. Eur J Cancer Prev 1995;4(4):307-18.
[Crossref] [Google Scholar] [PubMed]
- Qureshi S, Al-Shabanah OA, Al-Harbi MM, Al-Bekairi AM, Raza M. Boric acid enhances in vivo Ehrlich ascites carcinoma cell proliferation in Swiss albino mice. Toxicology 2001;165(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Suffness M, Pezzuto JM. Methods in plant Biochemistry. New York, Academic Press 1991; pp. 71.
- Bala A, Kar B, Haldar PK, Mazumder UK, Bera S. Evaluation of anticancer activity of Cleome gynandra on Ehrlich's Ascites Carcinoma treated mice. J Ethnopharmacol 2010;129(1):131-4.
[Crossref] [Google Scholar] [PubMed]
- Dongre SH, Badami S, Godavarthi A. Antitumor activity of Hypericum hookerianum against DLA induced tumor in mice and its possible mechanism of action. Phytother Res 2008;22(1):23-9.
[Crossref] [Google Scholar] [PubMed]
- Hirsch J. An anniversary for cancer chemotherapy. JAMA 2006;296(12):1518-20.
[Crossref] [Google Scholar] [PubMed]
- Fenninger LD, Mider GB. Energy and nitrogen metabolism in cancer. Adv Cancer Res 1954;2:229-53.
[Crossref] [Google Scholar] [PubMed]
- Segura JA, Barbero LG, Márquez J. Ehrlich ascites tumour unbalances splenic cell populations and reduces responsiveness of T cells to Staphylococcus aureus enterotoxin B stimulation. Immunol Lett 2000;74(2):111-5.
[Crossref] [Google Scholar] [PubMed]
 ): IC50 values (µM) HeLa>100; (
): IC50 values (µM) HeLa>100; (  ): IC50 values (µM) T47D>100 and (
): IC50 values (µM) T47D>100 and ( ): IC50 values (µM) C6>100
): IC50 values (µM) C6>100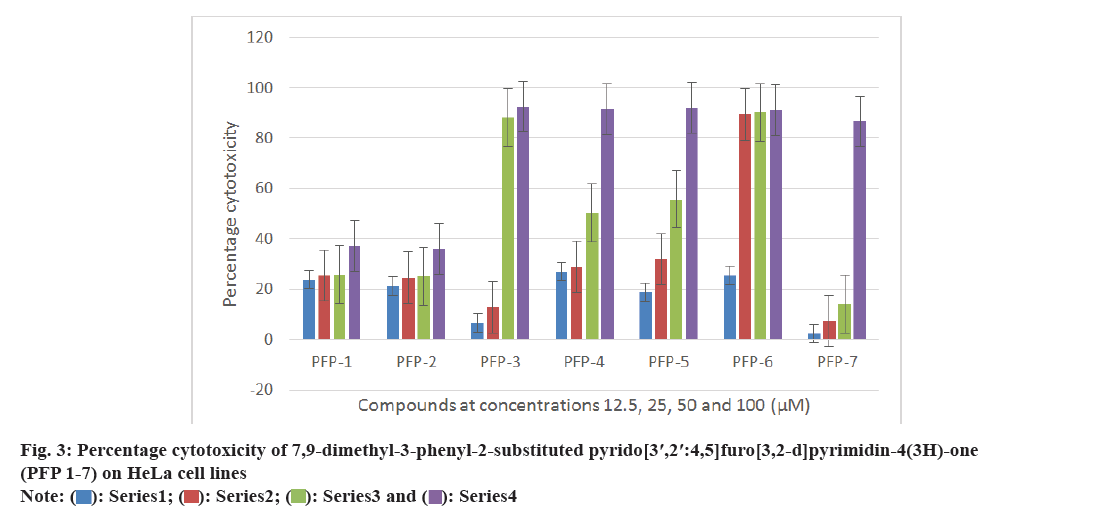
 ): Series1; (
): Series1; ( ): Series2; (
): Series2; (  ): Series3 and (
): Series3 and ( ): Series4
): Series4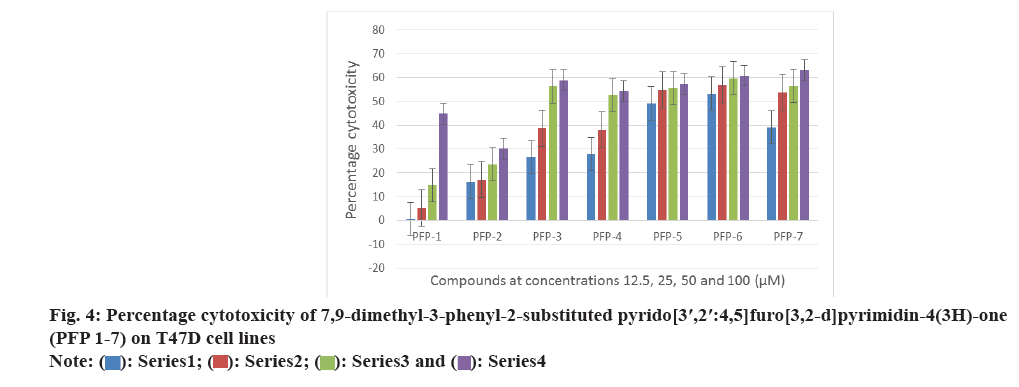
 ): Series1; (
): Series1; ( ): Series2; (
): Series2; ( ): Series3 and (
): Series3 and ( ): Series4
): Series4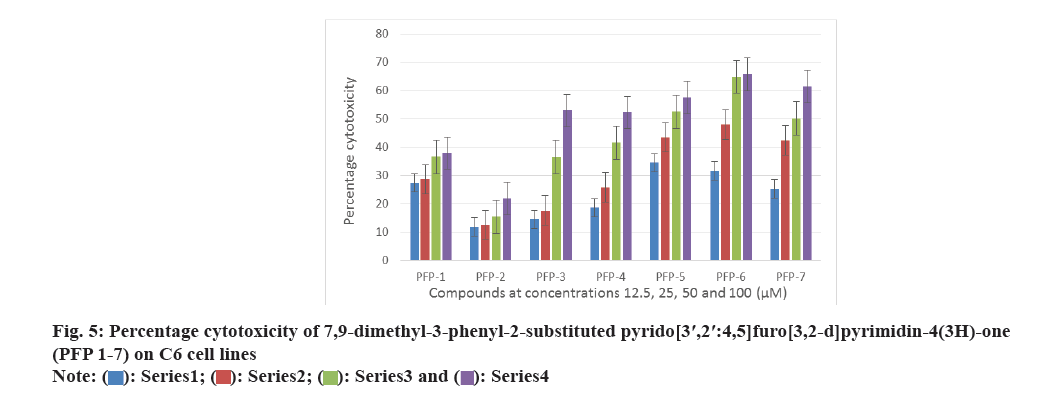
 ): Series1; (
): Series1; ( ): Series2; (
): Series2; ( ): Series3 and (
): Series3 and ( ): Series4
): Series4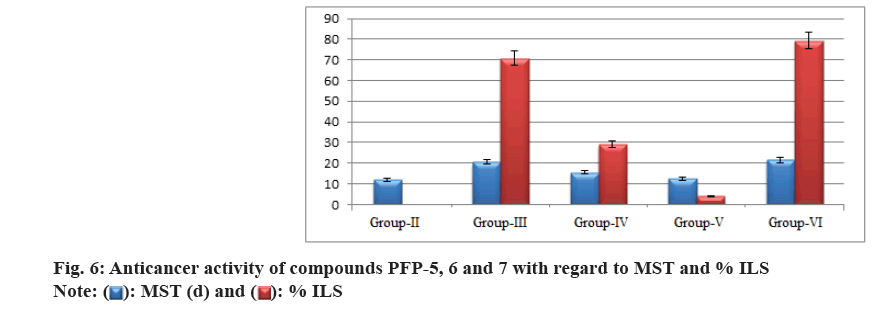
 ): MST (d) and (
): MST (d) and ( ): % ILS
): % ILS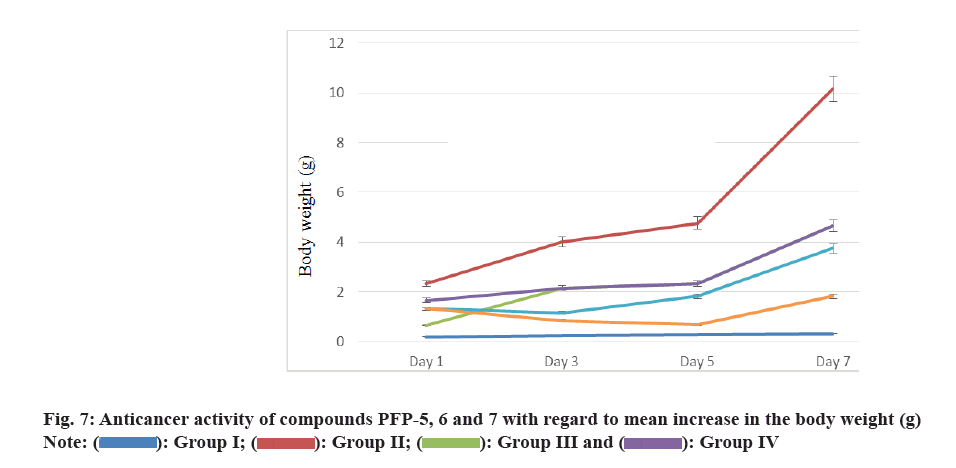
 ): Group I; (
): Group I; ( ): Group II; (
): Group II; ( ): Group III and (
): Group III and ( ): Group IV
): Group IV