- *Corresponding Author:
- Xiaoyun Xu Department of General Surgery, The Second Affiliated Hospital of Shantou University Medical College, Shantou, Guangdong Province 515051, China E-mail: meiyangyang1304@126.com
| Date of Received | 18 December 2021 |
| Date of Revision | 05 October 2022 |
| Date of Acceptance | 17 August 2023 |
| Indian J Pharm Sci 2023;85(4):1118-1125 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the role of circ_0084927 in lung cancer cell sensitivity to cisplatin. Half-maximal inhibitory concentration value of cisplatin was detected by cell counting kit-8. circ_0084927 and microRNA-634 expression were determined by reverse transcription quantitative polymerase chain reaction. Cell counting kit-8, scratch healing, colony formation as well as Transwell experiments were used for analyzing cell viability, scratch healing rate, clone number and invasive ability, respectively. Dualluciferase reporter method was utilized to validate targeting relationship. The half-maximal inhibitory concentration value of cisplatin and circ_0084927 expression were increased in A549/DDP cells, whereas microRNA-634 expression displayed an opposite result in comparison with A549 cells (p<0.05). Relative to the DDP+si-NC group, the cell optical density value, scratch healing rate, clone numbers and invasion numbers were significantly reduced in the DDP+si-circ_0084927 group (p<0.05). Optical density value, scratch healing rate, clone numbers as well as invasion numbers in the DDP+microRNA-634 group were significantly reduced relative to the DDP+microRNA-NC group (p<0.05). Circ_0084927 directly bound to microRNA-634. Relative to the DDP+si-circ_0084927+anti-microRNA-NC group, optical density value, scratch healing rate, clone numbers as well as invasion numbers in the DDP+si-circ_0084927+antimicroRNA- 634 group were increased (p<0.05). Interference with circ_0084927 reduced the cisplatin resistance of lung cancer cells by promoting microRNA-634.
Keywords
Lung cancer, circ_0084927, cisplatin resistance, microRNA-634
In 2020, there were about 1.796 million deaths from lung cancer worldwide, making it the leading cause of cancer death worldwide[1]. Smoking remains the main risk factor. Non-small cell lung cancer, which grows and divides slowly and spreads, and metastasizes relatively late, is a major subtype of lung cancer and cisplatin is currently the main treatment agent for the disease. However, chemotherapy resistance severely limits the therapeutic effect of cisplatin, leading to poor prognosis. Studying the mechanism of cisplatin resistance may provide effective treatment strategies for lung cancer. Circular RNAs (circRNAs) are Ribonucleic Acids (RNA) molecules that commonly bind to microRNAs (miRNAs) to regulate gene expression[2]. circRNA has been differentially expressed in various cancers and participated in cell proliferation and cell cycle, thus mediating tumorigenesis and drug resistance[3]. circ_0084927 is involved in malignant pleural effusion associated with lung adenocarcinoma[4]. Other studies have indicated that the high expression of circ_0084927 promotes the formation and invasion of cervical cancer tumors, and circ_0084927 absence inhibits cervical cancer cell malignant phenotypes[5,6]. However, whether circ_0084927 controls cisplatin resistance in lung cancer remains unclear.
miR-634 is a tumor-related miRNA. miR-634 expression is downregulated in liver cancer as well as cervical cancer tissues, and its overexpression affects cell migration and cycle progression, and promotes cell apoptosis, thus exerting an antitumor activity[7,8]. miR-634 mediated cisplatin resistance in ovarian cancer and up-regulation of miR-634 can re-sensitize drug-resistant ovarian cancer cells to cisplatin[7]. Target prediction software has shown that there are binding sites between miR-634 and circ_0084927, but there are no relevant studies on the regulation of cisplatin resistance by circ_0084927 and miR-634 in lung cancer. This study analyzed circ_0084927 and miR-634 expression and investigated the effect of circ_0084927 and miR-634 under cisplatin treatment on A549/DDP cell phenotypes with the hope of providing an effective target for reversing cisplatin resistance in lung cancer.
Materials and Methods
Cells and reagents:
A549 and A549/DDP (American Type Culture Collection, Manassas, Virginia, United States of America), cisplatin (P4394; Sigma Aldrich, Shanghai, China), Trizol and first strand complementary Deoxyribonucleic Acid (cDNA) synthesis kit (FastKing RT Kit) (Biolab, Beijing, China), SYBR Premix Ex TaqII (TaRaKa, Dalian, China), Cell Counting Kit-8 (CCK-8) kit, anti- Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) antibody (AF1186), anti-E cadherin antibody (AF6759), goat anti-rabbit IgG (A0208), rabbit anti-N-cadherin antibody (AF0243) (Beyotime, Shanghai, China), recombinant luciferase reporter plasmid, plasmid cloning DNA (pcDNA), miRNA mimics, anti-miRNA and small interfering RNA (si-RNA) (Synbio technologies, Suzhou, China) and Transwell chambers (Costar, Shanghai, China).
Methods:
circ_0084927 and miR-634 expression analysis: Total Ribonucleic Acid (RNA) was isolated with Trizol reagent, and reverse transcription was conducted using FastKing RT Kit, followed by Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR) based on the SYBR Premix Ex TaqII Kit. circ_0084927 and miR-634 expression were calculated through the 2-ΔΔCt method. circ_0084927 5-CTGGTGCAGCAAGATGGAAC-3 and 5-CTGCACCTCCCTTGGCAATA-3, GAPDH 5-GAAGAGAGAGACCCTCACGCTG-3 and 5-ACTGTGAGGAGGGGAGATTCAGT- 3 , miR-634 5-CAGTCTCAAACCAGCACC-3 and 5-TATGGTTGTTCACGACTCCTTCAC-3, U6 5-CTCGCTTCGGCAGCACA-3’ and 5’-AACGCTTCACGAATTTGCGT-3’.
Cell culture: A549 and A549/DDP cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) medium and inoculated into 24-well plates. Cells at 60 % confluence were introduced with plasmid or oligonucleotide according to Lipofectamine™ reagent instructions. The A549/ DDP cells were transfected with pcDNAcirc_ 0084927, si-circ_0084927 and controls. A549/DDP cells transfected with si-circ_0084927, miR-634 mimics, si-circ_0084927+anti-miR-634 and controls were treated with 2.5 μg/ml cisplatin for 24 h.
Detection of cell viability: Cells were inoculated into 96-well petri dishes and administrated with DDP (1.25, 2.5, 5, 10, 20, 40 μg/ml) for 24 h[9]. After that, cells were incubated with CCK-8. The Optical Density (OD) of each well was detected with a microprocessor, and proliferation inhibition rate was finally determined. In addition, A549/ DDP cells transfected with oligonucleotides alone or jointly were inoculated in 96-well plates and added with cisplatin. Then, samples were analyzed based on the above method.
Colony formation experiment: 5×102 cells were inoculated in petri dishes for 12-14 d until cell colonies appeared. After fixation and staining, the number of clones greater than 50 cells was determined.
Cell migration analysis by scratch healing assay: After the transfected cells were treated with cisplatin, 1×104 cells each group were inoculated incubated at 37° to 80 % confluence in 6-well plates, and a straight line was drawn on the surface of single layer cells with the tip of pipetting gun. Cell debris was removed, and cells were incubated in an incubator at 37°. The 6-well plates were placed under a microscope and photographed to determine the width of scratches.
Cell invasion analysis: Transfected A549/DDP cells were added to the top chamber pre-coated with Matrigel for invasion analysis. After incubating at 37° for 24 h, the cells in the lower compartment were fixed and stained. The number of invasive cells was determined under a microscope.
Western blot: After being treated with cisplatin for 24 h, transfected A549/DDP cells were precooled. After quantification, 30 μg of protein samples was transferred to polyvinylidene fluoride membrane by Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE). Anti-E-cadherin, anti-N-cadherin and anti-GAPDH were used to incubate membranes at 4°. After the membrane was fully washed using washing buffer, detection of antigen-antibody complex on membrane was performed by chemiluminescent reagent. Quantity One® software was used to quantify protein expression.
Dual-luciferase reporter assay: Wild-Type (WT) and Mutant Type (MUT) sequences of circ_0084927 were amplified and inserted into the PGL4 vector respectively to generate recombinant plasmids WT-circ_0084927 and MUT-circ_0084927. The above recombinant plasmids were transfected into A549/DDP cells with miR-634 mimics or miR-NC.
Relative luciferase activity was analyzed using dual luciferase detection system.
Statistical methods:
Each experiment was repeated three times with three multiple wells each time. Measurement data are expressed by (x̄ ±s). Independent sample t test, univariate analysis of variance and LSD-t test were used to compare the differences. p<0.05 indicated statistically significant.
Results and Discussion
As shown in Table 1, the proliferation inhibition rate of A549/DDP cells was significantly lower than that of A549 cells at the same concentration of cisplatin (1.25, 2.5, 5, 10, 20 or 40 μg/ ml) (p<0.05). The Half-Maximal Inhibitory Concentration (IC50) value of A549/DDP cells was significantly higher than that of A549 cells (31.04±5.60 vs. 8.72±1.64, p<0.05).
| Group | Concentration of cisplatin (μg/ml) | IC50 (μg/ml) | |||||
|---|---|---|---|---|---|---|---|
| 1.25 | 2.5 | 5 | 10 | 20 | 40 | ||
| A549 | 8.75±0.67 | 26.04±2.19 | 41.02±3.95 | 54.41±5.06 | 66.43±5.69 | 80.78±6.84 | 8.72±1.64 |
| A549/DDP | 5.11±0.44* | 8.58±0.62* | 16.81±1.33* | 24.21±2.18* | 40.24±3.91* | 57.05±4.36* | 31.04±5.60* |
| t | 13.623 | 23.013 | 17.426 | 16.444 | 11.38 | 8.777 | 11.475 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with A549 group, *p<0.05.
Table 1: Analysis Of Proliferation Inhibition Rate ( ,N=9).
,N=9).
As shown in Table 2, circ_0084937 expression was increased in A549/DDP cells relative to A549 cells (4.15±0.34 vs. 1.00±0.00, p<0.05), while miR-634 expression was lower (0.42±0.04 vs. 1.00±0.00, p<0.05).
| Group | circ_0084937 | miR-634 |
|---|---|---|
| A549 | 1.00±0.00 | 1.00±0.00 |
| A549/DDP | 4.15±0.34* | 0.42±0.04* |
| t | 27.794 | 43.5 |
| p | 0.000 | 0.000 |
Note: Compared with A549, *p<0.05
Table 2: circ_0084927 AND miR-634 Expression In Cisplatin-Resistant Lung Cancer Cells ( , n=9).
, n=9).
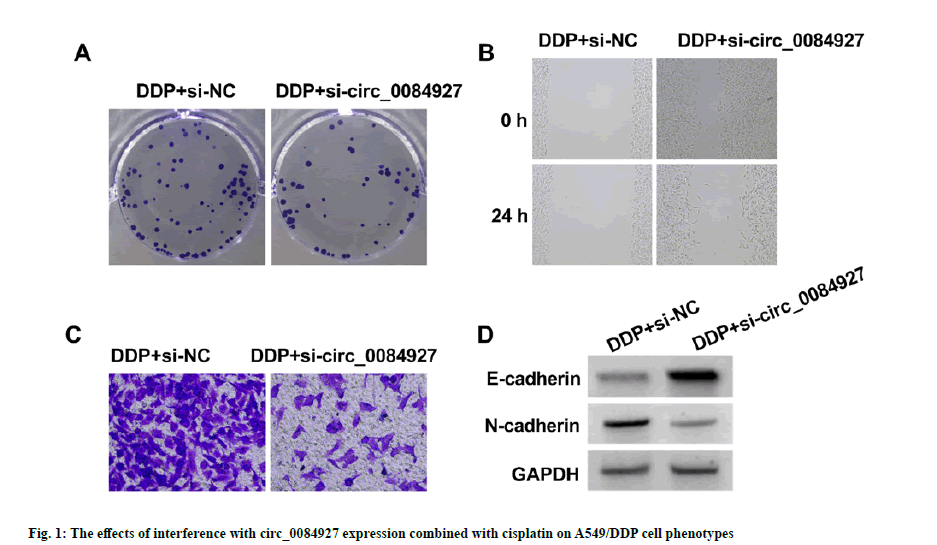
As shown in fig. 1 and Table 3, relative to DDP+si-NC group, circ_0084927 expression (0.34±0.03 vs. 1.00±0.00), OD value (0.36±0.04 vs. 0.75±0.05), number of cell colonies (39.91±4.09 vs. 87.45±6.51), scratch healing rate (24.31±2.22 vs. 65.36±5.05), invasion number (46.82±4.19 vs. 107.94±11.12) and N-cadherin production (0.31±0.03 vs. 0.77±0.06) of A549/ DDP cells in DDP+si-circ_0084927 group were decreased (p<0.05), but E-cadherin production was significantly increased (0.63±0.05 vs. 0.23±0.02, p<0.05).
| Group | circ_0084927 | OD value (450 nm) | Number of colonies | Scratch healing rate (%) | Number of invasion | E-cadherin protein | N-cadherin protein |
|---|---|---|---|---|---|---|---|
| DDP+si-NC | 1.00±0.00 | 0.75±0.05 | 87.45±6.51 | 65.36±5.05 | 107.94±11.12 | 0.23±0.02 | 0.77±0.06 |
| DDP+si-circ_0084927 | 0.34±0.03* | 0.36±0.04* | 39.91±4.09* | 24.31±2.22* | 46.82±4.19* | 0.63±0.05* | 0.31±0.03* |
| t | 66.000 | 18.272 | 18.551 | 22.324 | 15.43 | 22.283 | 20.572 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with DDP+si-NC group, *p<0.05.
Table 3: The Effects Of Interference With Circ_0084927 Expression Combined With Cisplatin On A549/Ddp Cell Phenotypes (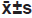 , N=9).
, N=9).
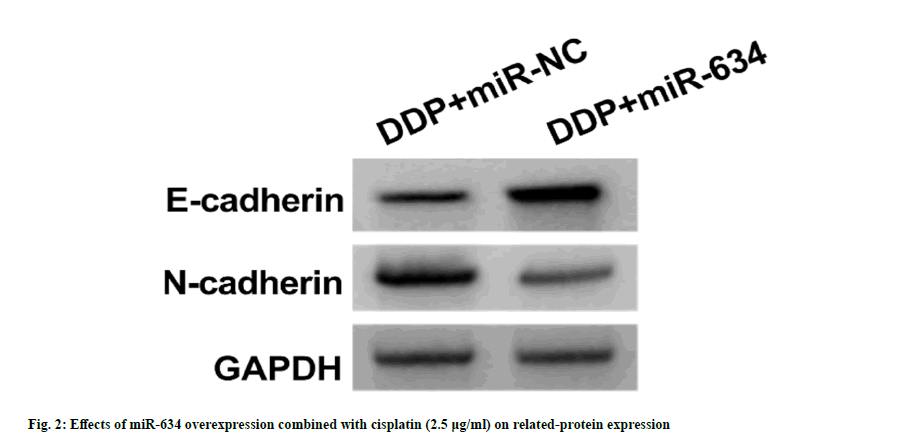
As shown in fig. 2 and Table 4, relative to DDP+miR-NC group, miR-634 (3.12±0.24 vs. 1.00±0.00) and E-cadherin protein levels (0.55±0.04 vs. 0.22±0.02) in A549/DDP cells in DDP+miR-634 group were increased (p<0.05), but OD value (0.40±0.04 vs. 0.77±0.05), the number of cell colonies (42.92±4.08 vs. 89.28±6.58), scratch healing rate (31.57±3.16 vs. 66.83±4.84), number of invasion (53.37±5.09 vs. 109.71±11.64) and N-cadherin production (0.40±0.04 vs. 0.79±0.06) were significantly decreased (p<0.05).
circRNA interactive predicted the presence of continuous binding sites between miR-634 and circ_0084927 (fig. 3). Relative luciferase activity of miR-634 mimics and WT-circ_0084927 cotransfected cells was lower than that of miRNC and WT-circ_0084927 co-transfected cells (0.36±0.03 vs. 1.01±0.07, t=25.605, p<0.05). But, the relative luciferase activity of miR- 634 mimics and MUT-Circ_0084927 group was similar with that of miR-NC and MUTCirc_ 0084927 co-transfected cells (1.00±0.07 vs. 1.03±0.06, t=0.976, p=0.344). miR-634 expression in the pcDNA-circ_0084927 group (0.45±0.04) was lower than that of the pcDNA group (1.00±0.00) (p<0.05). miR-634 expression in the si-circ_0084927 group (3.22±0.34) was higher than that of the si-NC group (1.02±0.06) (p<0.05), as shown in Table 5.
Relative to DDP+si-circ_0084927+anti-miRNC group, the expression levels of miR-634 (0.24±0.03 vs. 1.00±0.00) and E-cadherin protein (0.32±0.03 vs. 0.65±0.05) in the DDP+sicirc_ 0084927+anti-miR-634 group were decreased (p<0.05), while OD value (0.66±0.04 vs. 0.35±0.03), the number of cell colonies (79.04±5.75 vs. 36.93±3.09), scratch healing rate (57.08±5.17 vs. 22.45±2.11), invasion number (89.24±7.86 vs. 43.73±4.09) and N-cadherin protein level (0.68±0.06 vs. 0.30±0.03) were significantly increased (p<0.05), as shown in fig. 4 and Table 6.
Cisplatin is a common anticancer agent widely utilized in multiple malignancies including lung cancer, but patients often develop resistance after several cycles of cisplatin therapy, which limits the clinical efficacy of cisplatin and can lead to tumor recurrence. Recent studies have shown that dysregulation of non-coding RNAs including circRNAs is related to cisplatin resistance. Previous evidence pointed out that circ_103809 was increased in cisplatin-resistant lung cancer cells, and its silencing enhanced the repressive influence of cisplatin on lung cancer cell vitality[10]. circ_102272 was significantly elevated in liver cancer and promoted cisplatin resistance[11]. Sun et al.[12] reported that circ- MCTP2 sensitized gastric cancer cells to cisplatin via increasing MTMR3 production through association with miR-99a-5p. Therefore, exogenous alteration of circRNA expression may be a novel therapeutic strategy against cisplatin resistance.
circ_0084927 is increased in cervical cancer, breast cancer, colorectal cancer and other cancers, and promote cancer progression by inhibiting cell apoptosis and inducing cell proliferation and migration[13-15]. In this study, circ_0084927 content in A549/DDP cells was increased when compared with control, suggesting that abnormal circ_0084927 expression is involved in lung cancer cell sensitivity to DDP. Functional analysis revealed interference with circ_0084927 expression decreased A549/DDP cell proliferation and motility under cisplatin treatment, suggesting that interference with circ_0084927 expression could improve cisplatin sensitivity in lung cancer cells. Abnormal EMT contributes to the highly malignant characteristics of cancer cells and chemotherapy resistance[16,17]. Activation of Epithelial-Mesenchymal Transition (EMT) is closely related to chemotherapy resistance of lung cancer, and targeting EMT increases chemotherapy sensitivity and inhibits lung cancer progression[18,19]. This study revealed circ_0084927 knockdown combined with cisplatin significantly down-regulated N-cadherin expression and up-regulated E-cadherin, suggesting that interference with circ_0084927 may overcome cisplatin resistance by inhibiting EMT in lung cancer cells.
The carcinogenic effect of circ_0084927 in cervical cancer and breast cancer is related to the regulation of miRNA expression[13,14]. To determine the mechanism of circ_0084927 in DDP resistance in lung cancer, this study conducted dual-luciferase report analysis to confirm miR- 634 was the target of circ_0084927. Recovery of miR-634 expression sensitized drug-resistant cells to temozolomide[20]. Long noncoding RNA ZXF1 inhibited lung cancer cell growth via upregulating the miRNA[21]. miR-634 inhibited radiation resistance in breast cancer cells through downregulating STAT3[22]. In this study, miR- 634 content was reduced in A549/DDP cells. Increasing miR-634 expression under cisplatin treatment resulted in decreased A549/DDP cell colony-forming, invasion and migration ability, reduced N-cadherin production as well as increased E-cadherin production. These results indicated that overexpression of miR- 634 reversed cisplatin resistance in lung cancer cells. Interference with circ_0084927 expression and miR-634 mimics in lung cancer had the same effect on enhancing cisplatin sensitivity in lung cancer, and that circ_0084927 negatively regulated miR-634 expression, suggesting that circ_0084927/miR-634 molecular axis regulates cisplatin sensitivity in lung cancer. Under cisplatin treatment, downregulation of miR-634 relieved the effects of circ_0084927 absence on A549/DDP cell phenotypes, further confirming that circ_0084927 targeted miR-634 to promote cisplatin resistance. However, the downstream target of miR-634 remains to be further explored.
In conclusion, interference with circ_0084927 inhibited lung cancer cell proliferation and motility, and increased cisplatin sensitivity by promoting miR-634. Therefore, interference with circ_0084927/miR-634 molecular axis may be an important strategy to overcome cisplatin resistance in lung cancer.
Conflict of interests:
The authors declared no conflict of interests.
References
- Liu ZC, Li ZX, Zhang Y. Interpretation on the report of Global Cancer Statistics 2020. J Multidisciplinary Cancer Manag 2021;7(2):1-13.
- Verduci L, Strano S, Yarden Y, Blandino G. The circRNA–micro RNA code: Emerging implications for cancer diagnosis and treatment. Mol Oncol 2019;13(4):669-80.
[Crossref] [Google Scholar] [PubMed]
- Tianxiang CH, Yunhai YA. Role of circular RNA in diagnosis, development and durg resistance of lung cancer. Zhongguo Fei Ai Za Zhi 2019;22(8):532-6.
[Crossref] [Google Scholar] [PubMed]
- Wen Y, Wang Y, Xing Z, Liu Z, Hou Z. Microarray expression profile and analysis of circular RNA regulatory network in malignant pleural effusion. Cell Cycle 2018;17(24):2819-32.
- Chen L, Zhang X, Wang S, Lin X, Xu L. Circ_0084927 facilitates cervical cancer development via sponging miR-142-3p and upregulating ARL2. Cancer Manag Res 2020;20(1):9271-83.
[Crossref] [Google Scholar] [PubMed]
- Qu X, Zhu L, Song L, Liu S. circ_0084927 promotes cervical carcinogenesis by sponging miR-1179 that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int 2020;20(1):333-43.
[Crossref] [Google Scholar] [PubMed]
- Cong J, Liu R, Wang X, Jiang H, Zhang Y. miR-634 decreases cell proliferation and induces apoptosis by targeting mTOR signaling pathway in cervical cancer cells. Artif Cells Nanomed Biotechnol 2016;44(7):1694-701.
[Crossref] [Google Scholar] [PubMed]
- Zhang CZ, Cao Y, Fu J, Yun JP, Zhang MF. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol 2016;10(10):1532-41.
[Crossref] [Google Scholar] [PubMed]
- Lv J, Zhang F, Zhai C, Wang G, Qu Y. Bag-1 silence sensitizes non-small cell lung cancer cells to cisplatin through multiple gene pathways. Onco Targets Ther 2019;12(1):8977-89.
[Crossref] [Google Scholar] [PubMed]
- Zhu X, Han J, Lan H, Lin Q, Wang Y, Sun X. A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer 2020;20(1):1190-200.
- Guan Y, Zhang Y, Hao L, Nie Z. CircRNA_102272 promotes cisplatin-resistance in hepatocellular carcinoma by decreasing miR-326 targeting of RUNX2. Cancer Manag Res 2020;12(1):12527-34.
[Crossref] [Google Scholar] [PubMed]
- Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res 2020;39(1):246-56.
[Crossref] [Google Scholar] [PubMed]
- Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S. Hsa_circ_0084927 regulates cervical cancer advancement via regulation of the miR-634/TPD52 axis. Cancer Manag Res 2020;12(1):9435-48.
[Crossref] [Google Scholar] [PubMed]
- Gong G, She J, Fu D, Zhen D, Zhang B. Circular RNA circ_0084927 regulates proliferation, apoptosis and invasion of breast cancer cells via miR-142-3p/ERC1 pathway. Am J Transl Res 2021;13(5):4120.
- Liu F, Xiao XL, Liu YJ, Xu RH, Zhou WJ, Xu HC, et al. CircRNA_0084927 promotes colorectal cancer progression by regulating miRNA-20b-3p/glutathione S-transferase mu 5 axis. World J Gastroenterol 2021;27(36):6064-78.
[Crossref] [Google Scholar] [PubMed]
- Du B, Shim JS. Targeting epithelial–mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 2016;21(7):965.
[Crossref] [Google Scholar] [PubMed]
- Zhang YF. Research progress on the relationship between epithelial-mesenchymal transition and targeted drug resistance in non-small cell lung cancer. Oncol Prog 2016;14(3):220-2.
- Tao L, Shu-Ling W, Jing-Bo H, Ying Z, Rong H, Xiang-Qun L, et al. miR-451a attenuates doxorubicin resistance in lung cancer via suppressing epithelial mesenchymal transition (EMT) through targeting c-Myc. Biomed Pharmacother 2020;125:109962-72.
[Crossref] [Google Scholar] [PubMed]
- Sun H, Zhou X, Bao Y, Xiong G, Cui Y, Zhou H. miR-103 targets PTEN to promote dasatinib resistance in lung cancer A549 cells via activating PI3K/AKT pathway. Chin J Cancer Biother 2019;26(3):266-72.
- Tan Z, Zhao J, Jiang Y. miR-634 sensitizes glioma cells to temozolomide by targeting CYR 61 through Raf-ERK signaling pathway. Cancer Med 2018;7(3):913-21.
[Crossref] [Google Scholar] [PubMed]
- Ren X, Xu N, Zhang Y, Wang T. Downregulation of long non-coding RNA ZXF1 restricts cell survival by targeting miR-634-GRB2 in lung adenocarcinoma. Acta Biochim Pol 2020;67(1):31-9.
[Crossref] [Google Scholar] [PubMed]
- Yang B, Kuai F, Chen Z, Fu D, Liu J, Wu Y, et al. miR-634 decreases the radioresistance of human breast cancer cells by targeting STAT3. Cancer Biother Radiopharm 2020;35(3):241-8.
[Crossref] [Google Scholar] [PubMed]