- *Corresponding Author:
- Yi Xu
Department of Otolaryngology,
Hwamei Hospital,
University of Chinese Academy of Sciences,
Ningbo,
Zhejiang 315000,
China
E-mail: xuyidoctorxuyi@163.com
| This article was originally published in a special issue, “RecentDevelopments in Biomedical Research and PharmaceuticalSciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “121-126” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To clarify the underlying mechanism of radioresistance nasopharyngeal carcinoma, we identified correlated gene modules and key genes through analyzing the gene expression omnibus dataset with the weighted gene co-expression network analysis. GSE32389 dataset were obtained from the gene expression omnibus. The weighted gene co-expression network analysis was introduced to establish a gene co-expression network and mine important gene modules and the key genes. Gene ontology enrichment was performed with the genes in modules which were related to radioresistance nasopharyngeal carcinoma. Then, we established a proteinprotein interaction network with the genes in such module and identified hub genes in this protein-protein interaction network using molecular complex detection. Finally, we analyzed these hub genes overall survival using gene expression profiling interactive analysis database. Thirty five gene co-expression modules were obtained through weighted gene co-expression network analysis, genes in module had similar expression pattern. We found that module light cyan was the most correlated module to radioresistance nasopharyngeal carcinoma. Genes in this module were mainly related to cytokine production, regulation of cell activation and immune effector process. Five hub genes were discovered, namely cluster of differentiation 2 molecule, interleukin 10 receptor subunit alpha, cluster of differentiation 38 molecule, interleukin 2 receptor subunit beta and C-C motif chemokine ligand 19. In this research, we found five hub genes through analyzing the gene module which was mostly related to radioresistance nasopharyngeal carcinoma. They were involved in multiple cellular processes including cytokine production, regulation of cell activation and immune effector process. All these may improve our basic understanding of the molecular mechanisms underlying radioresistance nasopharyngeal carcinoma.
Keywords
Gene ontology, nasopharyngeal carcinoma, interleukin, radiotherapy
Nasopharyngeal Carcinoma (NPC) is one of the most common head and neck malignancies in China. According to the Global Cancer Statistics in 2018, there would be more than 129 000 new cases and approximately 73 000 cancer-related deaths of NPC [1]. With the development of diagnosis and therapy, especially radiotherapy, the overall prognosis of NPC has improved significantly[2]. Currently, the standard of treatment for NPC is the Intensity-Modulated Radiotherapy (IMRT) with good tumor coverage and healthy tissue protection[3]. However, radioresistance is the key factor of NPC treatment failure. The local control rate, progression-free survival and 3 y Overall Survival (OS) of locally advanced recurrent NPC patients are quite low, with only 44.3 %, 17.5 % and 47.2 %[4]. There are many reasons for the occurrence of radiotherapy resistance, such as abnormal gene expression, epigenetic factor, abnormal activation of some signal pathways and change of tumor microenvironment[5]. A deeper understanding of the mechanism of radioresistance NPC will help to inhibit tumor progression, reduce the necessary radiation dose and improve the survival rate of patients. It is very important to identify key factors and potential mechanism for radioresistance NPC.
With the tremendous development of next generation sequencing, together with the development of network analysis in bioinformatics field, the study of co- expression[6] genes related to clinical trait has increased our understanding of the pathological mechanism of radioresistance NPC. Besides, Weighted Gene Co-Expression Network Analysis (WGCNA) is widely used to analyze large-volume gene expression data and to discover key gene modules which are correlated to clinical trait[7]. In this study, we established a co- expression network using WGCNA and clustered similar expressed genes into the same gene modules. The modules which were most related to the radioresistance NPC were obtained. The genes in light cyan module which were mostly correlated to NPC patient’s radioresistance were mainly enriched in regulation of cytokine production.
Finally, we discovered five hub genes (Cluster of Differentiation 2 (CD2), Interleukin 10 Receptor Subunit Alpha (IL10RA), Cluster of Differentiation 38 (CD38), Interleukin 2 Receptor Subunit Beta (IL2RB) and Chemokine (C-C motif) Ligand 19 (CCL19)) that could predict the radiotherapy resistance of NPC. Also, we combined Gene Expression Profiling Interactive Analysis (GEPIA) databases to verify if these hub genes be able to predict the prognosis of NPC.
Materials and Methods
Data acquisition and processing:
Gene Expression Omnibus (GEO) database contains a large number of disease-related gene expression data sets, which is a commonly used research database[8]. In our study, GSE32389 dataset was introduced to construct co-expression networks of radioresistance NPC. This dataset included 8 radiosensitive NPC patients and 12 radioresistance NPC patients. We downloaded the original data of the GSE32389 dataset and normalized the data with the limma package. In addition, we also annotated the genes with the ID conversion tool.
Construction of co-expression network:
WGCNA is a commonly used tool for constructing gene co-expression networks. It can cluster genes with similar expression patterns in data sets into a module and associate the module with clinical characteristics[9]. In this study, the co-expression network was constructed based on GSE32389 dataset using the R package “WGCNA”. The soft-thresholding power of the co- expression network we set was 7 and 0.8 was used as the correlation coefficient threshold. The minimum number of genes in modules was set as 30. To merge possible similar modules, we defined 0.2 as the threshold for cut height. In addition, eigengene and gene significance methods[10] were introduced to discover modules which were correlated with radiotherapy activity of NPC patients. The association between module eigengenes and clinical trait was used to identify the significant
clinical module.
Functional enrichment analysis:
In order to achieve deeper understanding of the biological function of the genes in the interested module related to radioresistance NPC, we introduced the Gene Ontology (GO) analysis using the powerful online bioinformatics tool Database for Annotation, Visualization and Integrated Discovery (DAVID) database[11] (https://david.ncifcrf.gov/home.jsp/) and p<0.05 was set as the cut-off.
Identification of the hub genes:
Gene module with highest connectivity to radiotherapy resistance was identified by the WGCNA algorithm. The Protein-Protein Interaction (PPI) network was constructed with the genes in such module using Cytoscape v3.7.0[12]. Genes located at network nodes are the key nodes of the pathway. The network was analyzed by Molecular Complex Detection (MCODE) and the network is clustered to find closely connected regions and the most significant cluster with 10 nodes was visualized. And the hub genes were discovered using the cytoHubba in cytoscape with Mathews Correlation Coefficient (MCC) algorithms[13].
Prognostic analysis of the hub genes:
GEPIA is a web-based powerful tool for cancer bioinformatics study based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data[14] offering several key functions including differential expression analysis and patient survival analysis. Since there is no record of NPC in TCGA database, we analyzed the Head and Neck Squamous Cell Carcinoma (HNSCC) data set in TCGA database as a substitute. In this study, we performed prognostic analysis using GEPIA to investigate the relationship between hub genes expression level and HNSCC patient’s prognosis with OS.
Results and Discussion
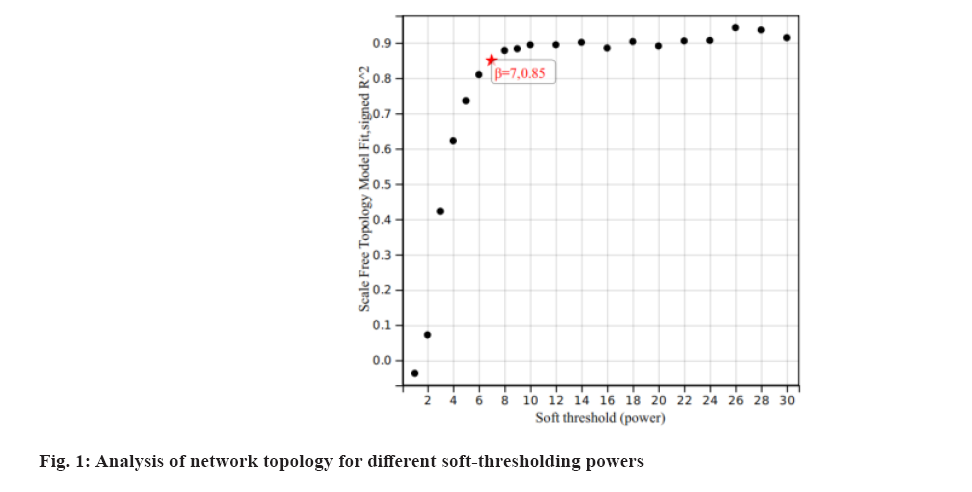
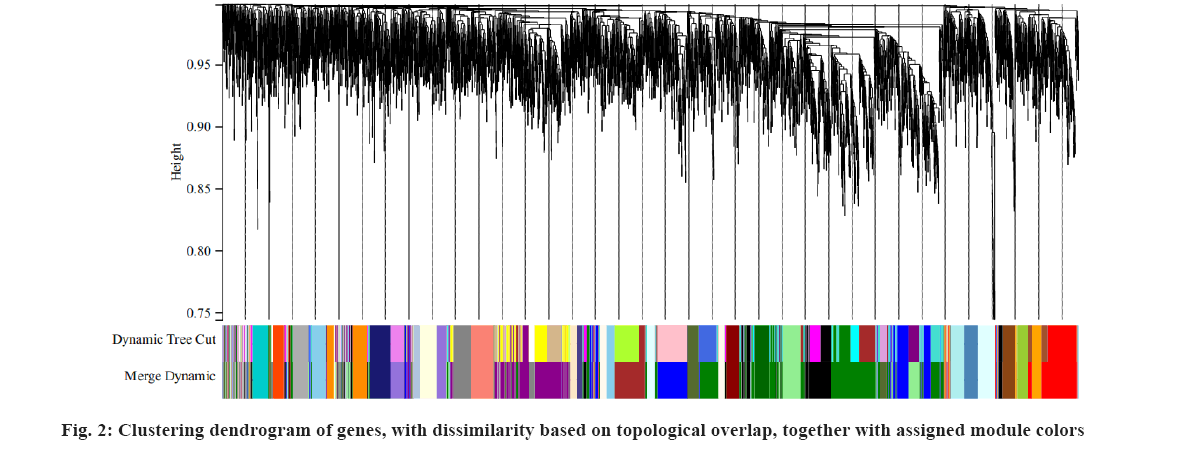
We downloaded the dataset GSE32389 of NPC tissues together with its clinical trait data from the GEO database. The GSE32389 dataset was GPL4381 platform. There were 8 radiosensitive NPC patients and 12 radioresistance NPC samples in GSE32389. The raw data of GSE32389 were normalized with the Robust Multichip Average (RMA) method in R-project[15]. In this study, to construct a WGCNA network, the soft thresholding power beta (β) was set at 7 and the scale independence reached 0.85 (fig. 1). Then, we used the one-step network to construct the functional to identification module using WGCNA in R, 35 gene co- expression modules were finally achieved that genes in each module had similar expression pattern as shown in fig. 2
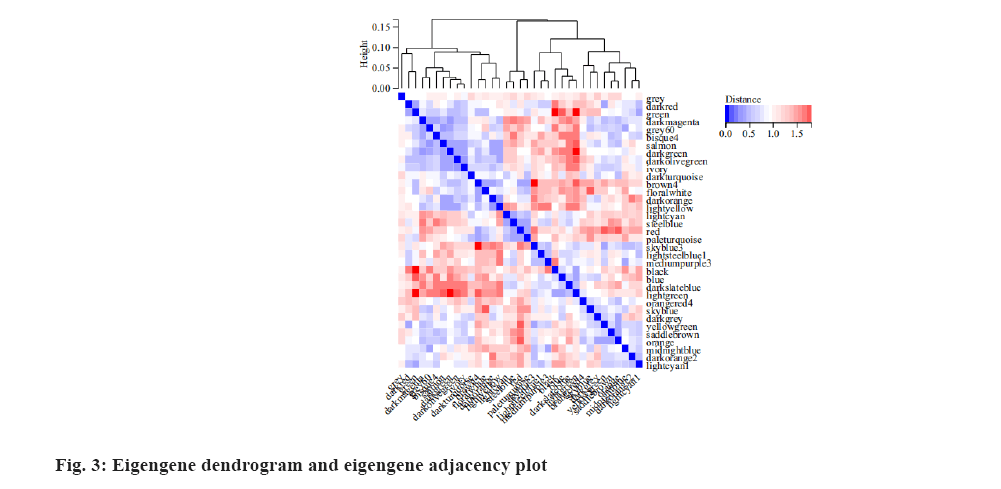
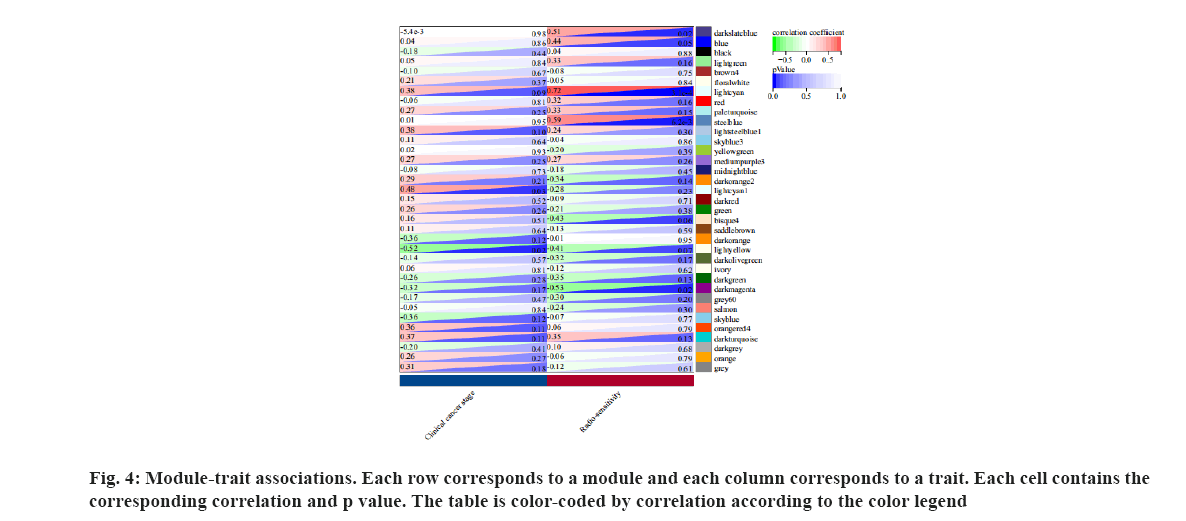
We analyzed the correlation of eigengenes between the gene co-expression modules. The eigengenes were clustered into several modules. Finally, thirty five modules were clustered into two clusters (fig. 3). We associate the gene module with the radiotherapy effect and identify the most significant correlation. The module light cyan was most significantly correlated with radiotherapy resistance as shown in fig. 4.
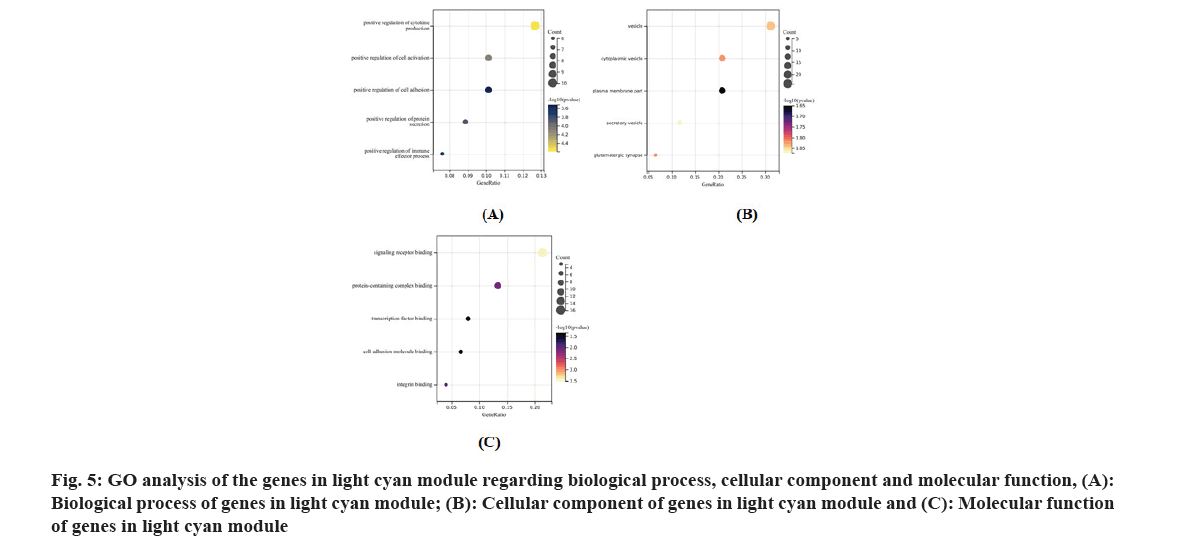
We performed GO analysis[16] for genes in the module light cyan. Regarding the biological process, the genes in this module were enriched in cytokine production, regulation of cell activation and immune effector process. As for the cellular component, these genes were enriched in vesicle, plasma membrane, secretory vesicle and glutamatergic synapse. Besides, regarding molecular function, these genes were enriched in signaling receptor binding, protein-containing complex binding, transcription factor binding, cell adhesion molecule binding and integrin binding as shown in fig. 5.
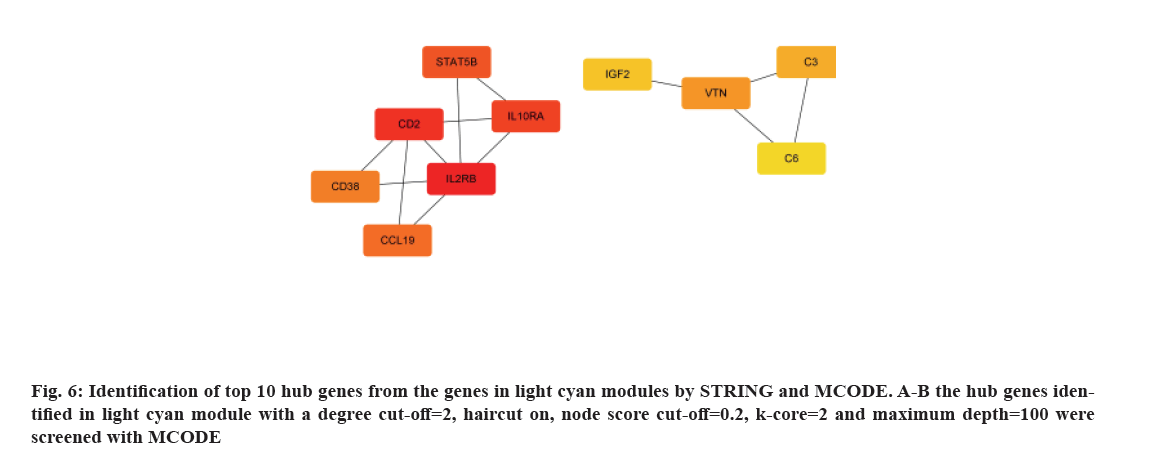
To achieve a deeper understanding of the associations between genes in the gene modules, a PPI network[17] with the module genes was constructed using Cytoscape software. As the result shown in fig. 6, top ten hub genes were identified in the light cyan module by of the PPI network using cytoHubba package, including C3, C6, CCL19, CD2, CD38, IGF3, IL2RB, IL10RA, Signal Transducer and Activator of Transcription 5B (STAT5B) and Vitronectin (VTN).
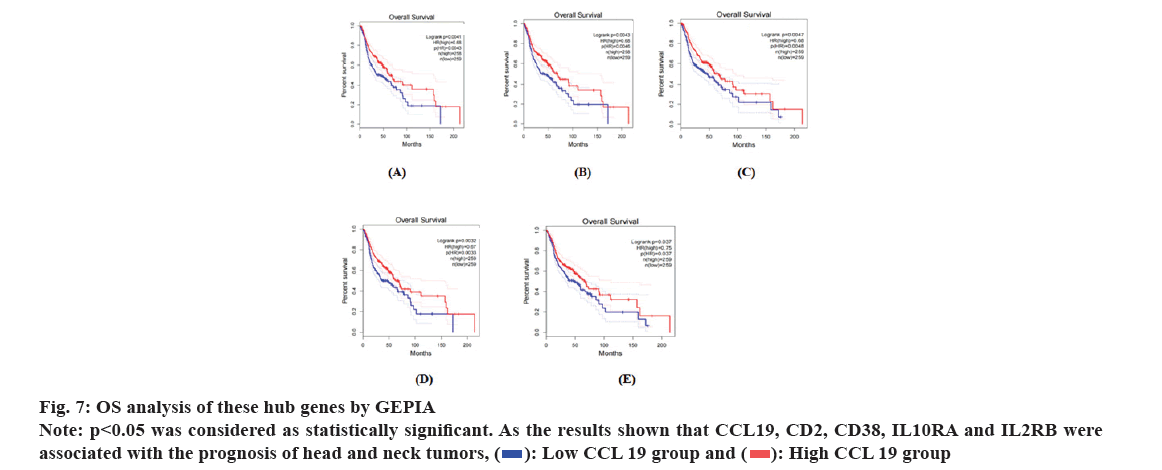
We performed the OS analysis of these ten key genes in survival plotting module in HNSC using GEPIA. The results showed that CD2, IL10RA, CD38, IL2RB and CCL19 could cause poor OS in NPC patients as shown in fig. 7.
NPC, one of the most common head and neck malignancies in China, has been becoming an important health problem that threatens the health and life of human beings[17]. IMRT is the current standard of treatment for NPC with its good curative effect[18]. However, radioresistance is the key factor of NPC treatment failure[5]. A deep understanding of the underlying mechanism of radioresistance NPC will help to inhibit tumor progression, reduce the necessary radiation dose and improve the survival rate of patients. Therefore, exploring susceptibility modules and genes for radioresistance patients is important.
We established the co-expression network by WGCNA using the published data of radioresistance NPC. All the genes in this dataset were included in the network. We obtained 35 gene modules that genes in each module have similar expression pattern. Then, we identified the light cyan module as the mostly correlated gene modules with radioresistance NPC patients. And genes in this key module were enriched in cytokine production, regulation of cell activation and immune effector process. Besides, we constructed a PPI network with genes in this key module; we identified 5 hub genes which were related to NPC radiotherapy effect and prognosis, namely CD2, IL10RA, CD38, IL2RB and CCL19.
WGCNA is widely used to analyze large-volume gene expression data and to discover gene modules which are highly correlated to clinical trait[9]. By analyzing the GSE32389 dataset, we constructed a co- expression network and found the light cyan module was most relevant to radioresistance in patients with NPC. GO analyses show that cytokine production, regulation of cell activation and immune effector process was activated during the radioresistance in NPC patients. Moreover, we identified 5 hub genes which were highest correlated to radioresistance NPC and prognosis, including CD2, IL10RA, CD38, IL2RB and CCL19. It was reported that CD38 regulated the metabolic-associated signaling pathways and serve a carcinogenic role in NPC[19]. Yu shown that IL-10 promoter genes including 1082 A/G, 819 T/C and 592 A/C are associated with NPC by promoting tumor progression[20]. Besides, Ju had reported that Atypical Chemokine Receptor 4 (ACKR4) CCL21 mediated NPC development[21]. These researches are consistent with the results of this study. We found several hub genes that play important role in radioresistance NPC, which may improve our knowledge of the molecular mechanisms underlying radioresistance NPC.
Conflict of interests:
The authors declared no conflict of interests.
References
- Chen YP, Chan AT, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet 2019;394(10192):64-80.
[Crossref] [Google Scholar] [PubMed]
- Guo R, Mao YP, Tang LL, Chen L, Sun Y, Ma J. The evolution of nasopharyngeal carcinoma staging. Br J Radiol 2019;92(1102):20190244.
- Liu YP, Wen YH, Tang J, Wei Y, You R, Zhu XL, et al. Endoscopic surgery compared with intensity-modulated radiotherapy in resectable locally recurrent nasopharyngeal carcinoma: A multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol 2021;22(3):381-90.
[Crossref] [Google Scholar] [PubMed]
- Zhan Y, Fan S. Multiple mechanisms involving in radioresistance of nasopharyngeal carcinoma. J Cancer 2020;11(14):4193-204.
- Lee AW, Ng WT, Chan JY, Corry J, Mäkitie A, Mendenhall WM, et al. Management of locally recurrent nasopharyngeal carcinoma. Cancer Treat Rev 2019;79:101890.
[Crossref] [Google Scholar] [PubMed]
- Kakati T, Bhattacharyya DK, Barah P, Kalita JK. Comparison of methods for differential co-expression analysis for disease biomarker prediction. Comput Biol Med 2019;113:103380.
[Crossref] [Google Scholar] [PubMed]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene co-expression network analysis: State of the art. J Biopharm Stat 2010;20(2):281-300.
[Crossref] [Google Scholar] [PubMed]
- Barrett T, Edgar R. Gene expression omnibus: Microarray data storage, submission, retrieval and analysis. Methods Enzymol 2006;411:352-69.
[Crossref] [Google Scholar] [PubMed]
- Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatcs 2008;9(1):559.
- Montenegro JD. Gene co-expression network analysis. Methods Mol Biol 2022;2443:387-404.
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization and integrated discovery. Genome Biol 2003;4(9):1.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8(4):S4-11.
[Crossref] [Google Scholar] [PubMed]
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45(W1):W98-102.
- Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47.
- Thomas PD, Mi H, Lewis S. Ontology annotation: Mapping genomic regions to biological function. Curr Opin Chem Biol 2007;11(1):4-11.
[Crossref] [Google Scholar] [PubMed]
- Chang ET, Ye W, Zeng YX, Adami HO. The evolving epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2021;30(6):1035-47.
[Crossref] [Google Scholar] [PubMed]
- Ng WT, Soong YL, Ahn YC, AlHussain H, Choi HC, Corry J, et al. International recommendations on reirradiation by intensity modulated radiation therapy for locally recurrent nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 2021;110(3):682-95.
[Crossref] [Google Scholar] [PubMed]
- Ge Y, Long Y, Xiao S, Liang L, He Z, Yue C, et al. CD38 affects the biological behavior and energy metabolism of nasopharyngeal carcinoma cells. Int J Oncol 2019;54(2):585-99.
[Crossref] [Google Scholar] [PubMed]
- Yu YF, Han ZG, Guo WB, Zhang GJ, Yang JK, Wu FL, et al. Interleukin-10 polymorphisms and nasopharyngeal carcinoma risk: A meta-analysis. Genet Mol Res 2015;14:18945-57.
- Ju Y, Sun C, Wang X. Loss of atypical chemokine receptor 4 facilitates C-C motif chemokine ligand 21-mediated tumor growth and invasion in nasopharyngeal carcinoma. Exp Ther Med 2019;17(1):613-20.
[Crossref] [Google Scholar] [PubMed]