- *Corresponding Author:
- Tingting Li
Department of Stomatology, Tianjin Women’s and Children’s Health Center, Tianjin 300070, China
E-mail: tingtingli1221@163.com
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “81-87” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Based on the unclear pathological mechanism as well as adverse events following carboplatin treatment, in this study, we developed a comprehensive differential gene profile for oral squamous cell carcinoma patients treated with cisplatin. The messenger ribonucleic acid sequencing data as well as micro ribonucleic acid sequencing data of oral squamous cell carcinoma patients treated with/without carboplatin from two datasets (The Cancer Genome Atlas and hospital) were compared using differential expression analysis; the micro ribonucleic acid-messenger ribonucleic acid’s network was constructed; the functions of potential hub genes were verified with external clinical patient. Compared with untreated group, the carboplatin treated patients showed 123 differentially expressed messenger ribonucleic acids, including 96 up-regulated messenger ribonucleic acids and 27 down-regulated messenger ribonucleic acids. Meanwhile, 12 differential micro ribonucleic acids (Homo sapiens (human)-micro ribonucleic acid-514, human micro ribonucleic acid- 7974 as well as human-micro ribonucleic acid-509 as primary ones) were significantly different between two treatments. The functions and the pathological mechanism of carboplatin treatment were analyzed by quantitative real-time polymerase chain reaction and Western blotting methods. This integrated study hypothesized a micro ribonucleic acid dependent signaling pathway for carboplatin function and provided a beneficial reference for better evaluation of the popularly utilized chemotherapeutic drug.
Keywords
Oral squamous cell carcinoma, carboplatin, differential gene analysis, messenger ribonucleic acid
Oral Squamous Cell Carcinoma (OSCC) is characterized as the most malignancy of the oral neoplasm, which comprises for more than 90 % of all oral malignancies as well as approximately 38 % of head and neck tumors[1]. As approximately 500 000 new cases diagnosed every year with a fairly onerous prognosis, OSCC has been ranked as the 8th most common human cancer[2]. The worldwide incidence of OSCC is not homogeneous based on the variable prevalence of the risk factors[3]. Growing evidences based on retrospective assessments suggest the connection between OSCC and environmental risk factors including diabetes, alcoholism, smoking as well as dietary habits especially in developed regions[4-6]. Even with decades of study, the knowledge of the genetic basis for OSCC is still inferior[7,8].
Carboplatin or carboplatinum, is a well-accepted chemotherapeutic drug, which has been administrated for numerous human cancers including lung cancer, germ cell tumours, gynaecological cancers as well as head and neck cancer[9,10]. As cisplatin, carboplatin also has a broad spectrum of clinical activity in oncology. Cisplatin was first initiated in the mid-19th Century and also known as the name of Peyrone’s chloride. It functions as a crosslink with the purine bases on the Deoxyribonucleic Acid (DNA), interfering with DNA repair mechanisms, subsequently causing DNA damage and stimulating apoptosis in various cancer cells[11]. Cisplatin represents the oldest member of this family. Since then, the subsequent analogues including carboplatin, oxaliplatin, as well as satraplatin have become the golden standard chemotherapeutic administration for a broad range of malignancies[12,13]. Carboplatin refers to cis-diammine-cyclobutanedicarboxylatoplatinum (II), which was initially produced to be a great “substitute” chemotherapeutic drug for cisplatin. Although carboplatin has opened a window of hope for effective agents to treat various tumor types like OSCC, the overall outcomes are still far from satisfactions. Carboplatin has been reported to be associated with significant myelotoxicity (particularly thrombocytopenia) but less nephrotoxicity and neurological sequelae[14]. OSCC patients developed frequently undesirable adverse events with carboplatin treatment such as severe various allergic reactions, gastrointestinal disorders, kidney problems, decrease immunity to infections, as well as hemorrhage especially in younger patients[15,16]. Meanwhile, a number of OSCC patients were shown to develop drug resistance to carboplatin[17]. All of these largely limit the applications of carboplatin for OSCC patients. Based on the truth of unclear mechanisms behind the carboplatin, its function in OSCC has been greatly restricted. To address these issues, a systematically differential expression analysis was generated on OSCC patients treated with or without carboplatin. With an innovative combination of bioinformatics methods and clinical experimental verification, we comprehensively explored the potential hub genes (messenger Ribonucleic Acids (mRNAs) and microRNA (miRNAs)) as well as closely associated signaling pathways for carboplatin functions, which provided a beneficial guidance for the precise administration of carboplatin for OSCC patients.
Materials and Methods
Data source:
The mRNA sequencing data, miRNA sequencing data as well as clinical information of OSCC patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://TCGA-data.nci.nih.gov/TCGA/). In the mRNA dataset, 16 patients with OSCC were treated with carboplatin and 93 patients were without carboplatin treatment. Meanwhile, in the miRNA dataset, 16 patients with OSCC were treated with carboplatin and 92 patients were without carboplatin treatment.
Differential gene analysis:
The limma package in R language was developed to analyze the differentially expressed mRNAs as well miRNAs between different groups, taking the absolute value of the log-transformed differential expression multiple Fold Change (Log2FC)>1 and p-value<0.05 as a standard for analysis.
Functional enrichment analysis:
For the obtained differentially expressed genes, we used the “clusterProfiler” function package in R language for enrichment analysis of Gene Ontology (GO) (including biological process, molecular function and cellular component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. When p-value<0.05, we considered the corresponding entries to be significantly enriched.
Prediction of miRNA target genes:
The target genes of miRNAs were predicted through the miRNA Target Prediction Database (miRDB) (http://mirdb.org/index.html, version 6.0) database[18]. Meanwhile, the Cytoscape (https://cytoscape.org/, version 3.7.2) was established to visualize the miRNA-mRNA regulatory network.
Patient tissue sample:
25 OSCC patients treated with carboplatin and 25 OSCC patients treated without any chemotherapeutic drug underwent routine resection in the Second Hospital of Tianjin Medical University. These tissues were snap-frozen and stored in liquid nitrogen. This study was given official approval by the ethics committees of the hospital.
Quantitative real-time Polymerase Chain Reaction (PCR):
The total RNA was extracted using RNAiso Plus (Takara, Beijing, China). The real-time PCR was performed with BrightGreen 2× quantitative PCR (qPCR) MasterMix-Low ROX (ABM, Vancouver, Canada). PCR conditions were set up as follows: One cycle at 95° for 5 min and 40 cycles at 95° for 15 s and followed by 60° for 40 s. The relative expression was processed with the 2-ΔΔCt method.
Western blotting analysis:
The cancerous tissues of OSCC patients treated with or without carboplatin were harvested and washed twice with Phosphate Buffered Saline (PBS). The cell lysis buffer contained 25 mM 4-(2-Hydroxyethyl)- 1-Piperazineethanesulfonic acid (HEPES) pH 7.5, 150 mM Sodium Chloride (NaCl), 0.5 % Nonyl Phenoxypolyethoxylethanol-40 (NP-40) and 0.1 % Sodium Dodecyl Sulfate (SDS) plus proteinase inhibitor cocktail (Sigma-Aldrich, St. Louis, Missouri). The protein concentration of cell lysate was determined using the Pierce Bicinchoninic Acid (BCA) protein assay kit (Thermo Fisher Scientific, Beijing, China). Then 30 mg of proteins were loaded on premade 8 %-15 % SDS polyacrylamide gel (Invitrogen) for separation and then electrotransferred onto a nitrocellulose membrane. The blots were incubated in PBS buffer with 5 % defat milk and 0.02 % Tween-20 at room temperature for 1 h before primary antibody incubation overnight at 4°. Horseradish Peroxidase (HRP)-conjugated secondary antibody (Jackson Immunoresearch Laboratory) incubation was conducted at room temperature for 1 h. Supersignal West Pico chemiluminescent substrate (Pierce; Thermo Scientific) was used for visualization of immunoreactive proteins.
Results and Discussion
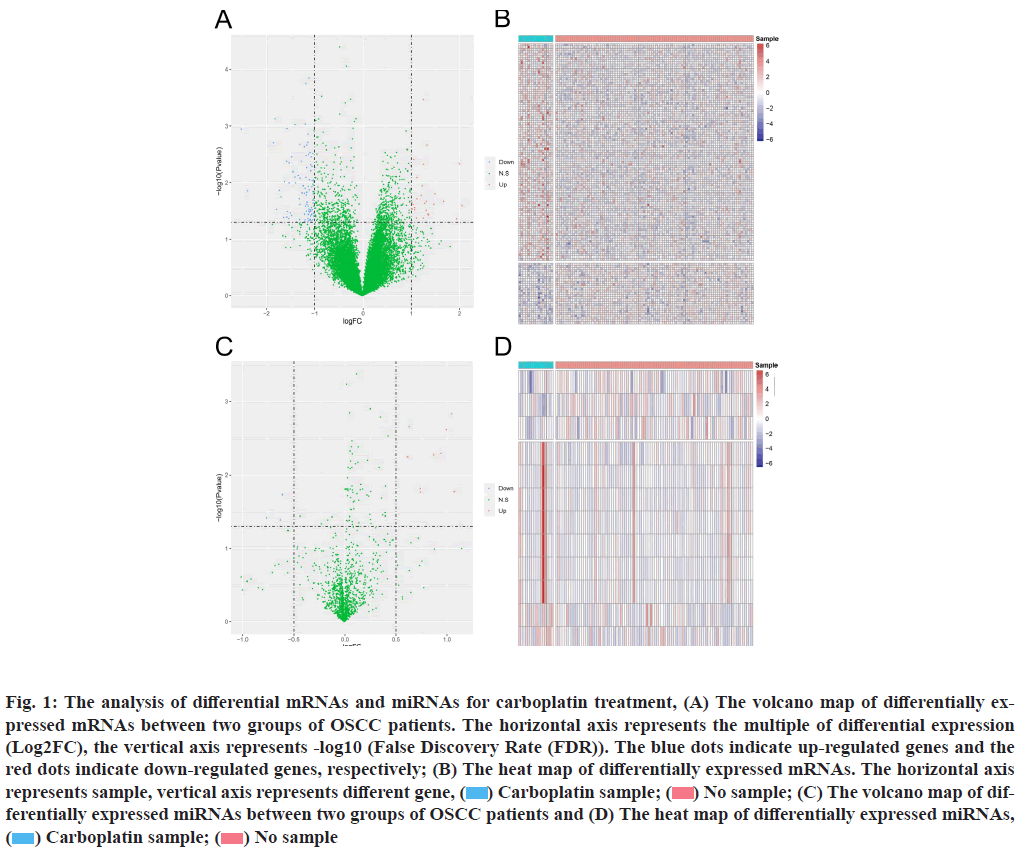
Differentially expressed analysis of mRNA and miRNA is explained below. We compared the mRNA expression of OSCC patients treated/untreated with carboplatin. Compared with control group (no carboplatin treatment), OSCC patients treated with carboplatin alone demonstrated 123 differentially expressed mRNAs, of which 96 mRNAs were up-regulated and 27 mRNAs were down-regulated (fig. 1A).
Fig. 1: The analysis of differential mRNAs and miRNAs for carboplatin treatment, (A) The volcano map of differentially expressed mRNAs between two groups of OSCC patients. The horizontal axis represents the multiple of differential expression (Log2FC), the vertical axis represents -log10 (False Discovery Rate (FDR)). The blue dots indicate up-regulated genes and the red dots indicate down-regulated genes, respectively; (B) The heat map of differentially expressed mRNAs. The horizontal axis represents sample, vertical axis represents different gene, ( ) Carboplatin sample; (
) Carboplatin sample; ( ) No sample; (C) The volcano map of differentially expressed miRNAs between two groups of OSCC patients and (D) The heat map of differentially expressed miRNAs, (
) No sample; (C) The volcano map of differentially expressed miRNAs between two groups of OSCC patients and (D) The heat map of differentially expressed miRNAs, ( ) Carboplatin sample; (
) Carboplatin sample; ( ) No sample
) No sample
The differentially expressed mRNAs were significantly diverse among the two groups (fig. 1B). At the same time, 12 different miRNAs displayed specific expression pattern between two groups. Compared with untreated OSCC patients, carboplatin treatment initiated 3 upregulated miRNAs and 9 down-regulated miRNAs (fig. 1C and fig. 1D). With further investigation, Olfactory Receptor Family 6 Subfamily C Member 2 (OR6C2), Transmembrane Protein 225B (TMEM225B), Villin 1 (VIL1), Tumor Suppressing Subtransferable Candidate 2 (Pseudogene) (TSSC2), C-C Motif Chemokine Ligand 25 (CCL25), Alkaline Phosphatase, Placental- Like 2 (ALPPL2), Cerebellin 2 Precursor (CBLN2), Potassium Voltage-Gated Channel Interacting Protein 3 (KCNIP3), Mitochondrial Ribosomal Protein L23 (MRPL23) and Gamma-Aminobutyric Acid Type A Receptor Subunit Gamma 3 (GABRG3) were primary hub mRNAs (with most significant difference compared the two groups). The Homo sapiens (human) (has)- miR-514, hsa-miR-7974 as well as hsa-miR-509 were hub miRNAs.
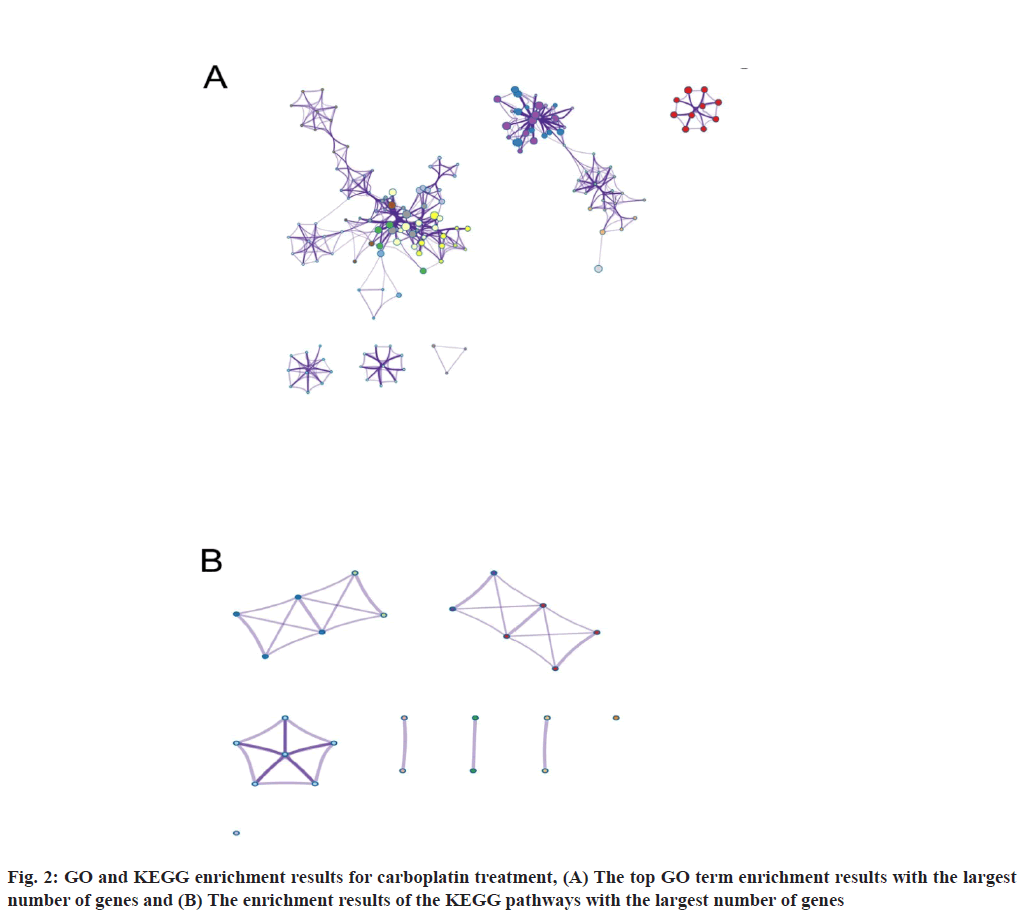
By performing GO and KEGG enrichment analysis on these 123 differentially expressed genes, we demonstrated that these differentially expressed genes were enriched in GO terms including such as cell cycle phase transition and cell division etc. (fig. 2A). In the figure, the horizontal axis represents the number of enriched genes and the vertical axis represents the name of each GO term respectively, ( ) Cell cycle phase transition; (
) Cell cycle phase transition; ( ) Cell division; (
) Cell division; ( ) Sensory organ morphogenesis; (
) Sensory organ morphogenesis; ( ) Skeletal system development; (
) Skeletal system development; ( ) Regulation of chromosome segregation; (
) Regulation of chromosome segregation; ( ) Tissue morphogenesis; (
) Tissue morphogenesis; ( ) Spingolipid translocation; (
) Spingolipid translocation; ( ) Embryonic morphogenesis; (
) Embryonic morphogenesis; ( ) Sex differentiation; (
) Sex differentiation; ( ) Cargo receptor activity; (
) Cargo receptor activity; ( ) DNA-binding transcription activator activity RNA polymerase II; (
) DNA-binding transcription activator activity RNA polymerase II; ( ) DNA dependent DNA replication; (
) DNA dependent DNA replication; ( ) Response to bleomycin; (
) Response to bleomycin; ( ) Cerebellar Purkinje cell layer structural organization; (
) Cerebellar Purkinje cell layer structural organization; ( ) Chromatin binding; (
) Chromatin binding; ( ) Autonomic nervous system development; (
) Autonomic nervous system development; ( ) IgM Immunoglobulin complex.
) IgM Immunoglobulin complex.
At the same time, the DNA polymerase complex related pathways were significantly enriched in KEGG analysis (fig. 2B). The horizontal axis in the figure indicates the number of genes enriched and the vertical axis indicates the name of each KEGG pathway respectively, ( ) DNA polymerase zeta complex; (
) DNA polymerase zeta complex; ( ) RPA complex; (
) RPA complex; ( ) Melatonin biosynthesis, tryptophan≥serotonin≥melatonin; (
) Melatonin biosynthesis, tryptophan≥serotonin≥melatonin; ( ) Wnt signaling pathway; (
) Wnt signaling pathway; ( ) Hippo signaling pathway; (
) Hippo signaling pathway; ( ) COPII complex; (
) COPII complex; ( ) Glycosphingolipid biosynthesis-globo and isoglobo; (
) Glycosphingolipid biosynthesis-globo and isoglobo; ( ) Complement and coagulation cascades and (
) Complement and coagulation cascades and ( ) BMP signaling.
) BMP signaling.
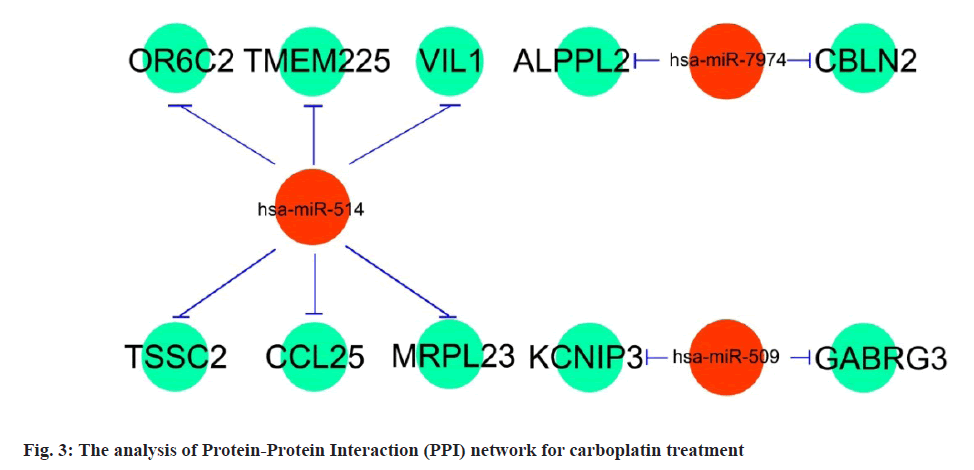
miRNA-mRNA regulatory network is shown below. The 3 candidate miRNAs (hsa-miR-514, hsa-miR-7974 as well as hsa-miR-509) and 10 potential mRNAs (OR6C2, TMEM225B, VIL1, TSSC2, CCL25, ALPPL2, CBLN2, KCNIP3, MRPL23 and GABRG3) were further explored for a regulatory network establishment visualized with Cytoscape software. Among the three miRNAs, hsa-miR-514 manipulated the largest number of target genes (6), which were OR6C2, TMEM225B, VIL1, TSSC2, CCL25 and MRPL23. Compared with hsa-miR-514, hsa-miR-7974 and hsa-miR-509 regulated 2 target genes, as ALPPL2 and CBLN2 for hsa-miR-7974, KCNIP3 and GABRG3 for hsa-miR-509 respectively (fig. 3). Based on these outcomes, the 3 miRNAs (hsa-miR-514, hsa-miR-7974 and hsa-miR-509) especially hsa-miR-514 were supposed as the primary carboplatin targeted miRNAs for OSCC treatment.
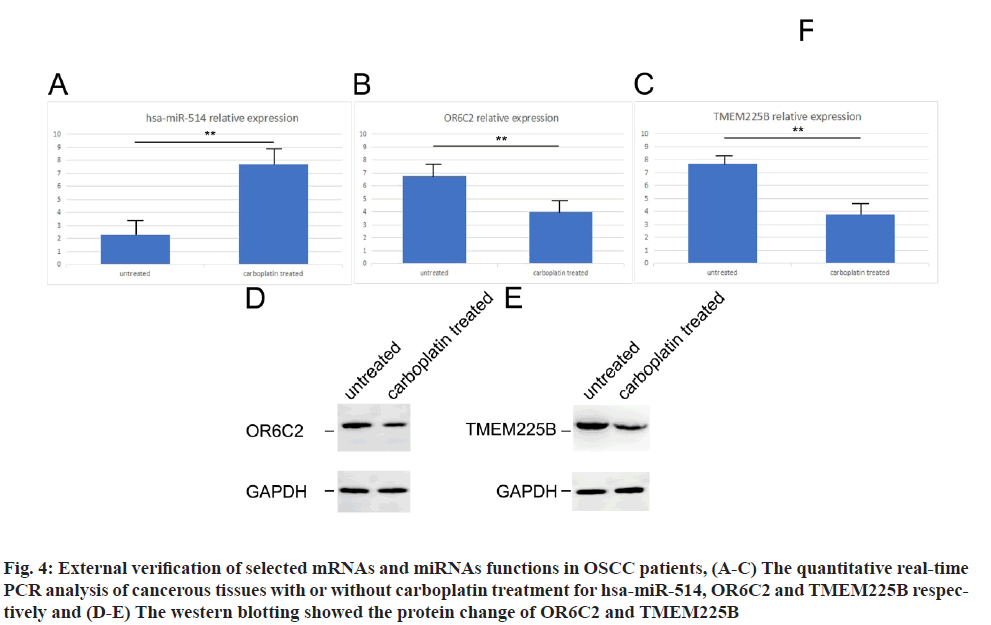
From the results above, we have implicated the potential mRNAs as well as miRNAs candidates for the carboplatin functions in OSCC treatment. Next, we sought to verify the selected potential mRNAs and miRNAs using the clinical patient’s specimen. Using the cancerous tissues from carboplatin treated/untreated OSCC patients, the gene expression of has-miR-514 increased while the gene expression of OR6C2 and TMEM225B decreased in carboplatin treated patients tissues compared with untreated patients by quantitative real-time PCR (fig. 4A-fig. 4C). With respect to protein expression, both OR6C2 and TMEM225B dropped significantly in carboplatin treated OSCC patients using western blotting examination (fig. 4D and fig. 4E). These outcomes were in line with the differentially expression results.
Fig. 4: External verification of selected mRNAs and miRNAs functions in OSCC patients, (A-C) The quantitative real-time PCR analysis of cancerous tissues with or without carboplatin treatment for hsa-miR-514, OR6C2 and TMEM225B respectively and (D-E) The western blotting showed the protein change of OR6C2 and TMEM225B
The aggressiveness and complexity of OSCC have brought many obstacles to clinical treatment. The discovery of chemotherapeutic drug for OSCC patients is still a very strenuous journey. Despite decades of hard work for the studies, only few drugs have come out with promising results, including cisplatin and carboplatin[19]. The cisplatin first came out in the 1800s, which was originally applied as a chemical compound for inorganic synthesis[20]. As popularly applied in tumor treatment, cisplatin is so called the “penicillin of cancer” subsequently[21]. Carboplatin, as an advanced analogue in the family with cisplatin, is functional via the similar molecular mechanism involving tumour cell apoptosis. It has been reported that cisplatin is effective in treating specific tumor types while carboplatin is more favourable toxicity profiles[12]. Recently, a randomised non-inferiority trial comparing the use of single agent carboplatin against cisplatin as a radiosensitizer suggested the similar survival outcomes for OSCC patients, as the 3 y Overall Survival (OS) was 79.2 % and 77.7 % respectively, (p=0.9884; Hazard Ratio (HR): 0.83; 95 % Confidence Interval (CI): 0.613-1.010)[22]. Another study implied that cisplatin was significantly superior to carboplatin with respect to adjuvant chemoradiotherapy for locally advanced oropharynx and oral cavity, in which the 3 y local control rate of 85 % against 62 % and 3 y OS of 78 % against 51 % respectively[23].
In this integrated study, we comprehensively compared differentially expressed mRNAs as well as miRNAs between carboplatin treated and non-treated OSCC patients. The primary 10 mRNAs and 3 miRNAs were proposed as hub genes for carboplatin functions in OSCC treatment. Interestingly, compared with hub genes for cisplatin treatment, no overlapping mRNAs or miRNAs could be identified for these two drugs. These results suggested that even with similar pathological process, the signaling axis was not the same between the two chemotherapeutic drugs for OSCC treatment. Here with the differential expression analysis, 3 potential miRNAs (hsa-miR-514, hsamiR- 7974 and hsa-miR-509) displayed significant differences between two treatments, which were all upregulated (shown as fig. 1C, fig. 3 and fig. 4). Since miRNAs in cancerous tissues are coordinated with de novo repression of downstream target genes. It was reasonable to predict the repressed level of target mRNAs (OR6C2, TMEM225B, VIL1, TSSC2, CCL25, ALPPL2, CBLN2, KCNIP3, MRPL23 and GABRG3) (shown as fig. 1A, fig. 3 and fig. 4). Previously, low expression of miR-514 was shown to be related to poor prognosis in ovarian cancer patients and miR-514 repressed proliferation in ovarian cancer cells through targeting Adenosine Triphosphate (ATP) binding cassette subfamily[24]. The miR-7974 was reported to be involved in the formation of lung adenocarcinoma and lung squamous cell carcinoma[25]. A study by Liang et al. claimed that miR-509 functionally interacted with NONHSAT112228.2, a sense-overlapping long non-coding RNA (lncRNA) transcribed by a noncode gene overlapping with p21 gene, to release p21 expression, which inversely inhibited the proliferation and migration of lung cancer cells[26]. Except for these published articles, the association between selected hub genes from this study and OSCC has not been fully explored yet, which call for a great point for the future study.
In summary, in the light of the fact that the molecular mechanism underline carboplatin treatment for OSCC is still under controversial, we summarized the differential expression profile for mRNAs and miRNAs from two databases. With functional enrichment analysis as well as external clinical experiments, the potential associated signaling pathway had been established, which initiated new insights into the management as well as evaluation of chemotherapeutic drug and provided a beneficial reference for future clinical research.
Author’s contributions:
Zhe Han and Bo Zhang contributed equally to this work. Zhe Han designed the work and performed the experiments; Bo Zhang analyzed the results and wrote the article and Tingting Li supervised the work.
Funding:
This work was supported by the Tianjin Municipal Science and Technology Plan Project (No: 19KPXMRC00020).
Conflict of interests:
The authors declare no competing interests.
References
- Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol 2015;8(9):11884-94.
[Google Scholar] [PubMed]
- Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, et al. Oesophageal cancer. Nat Rev Dis Primers 2017;3(1):1-21.
[Crossref] [Google Scholar] [PubMed]
- Afzali P, Ward BB. Management of the neck in oral squamous cell carcinoma: Background, classification, and current philosophy. Oral Maxillofac Surg Clin North Am 2019;31(1):69-84.
[Crossref] [Google Scholar] [PubMed]
- Alves AM, Diel LF, Lamers ML. Macrophages and prognosis of oral squamous cell carcinoma: A systematic review. J Oral Pathol Med 2018;47(5):460-7.
[Crossref] [Google Scholar] [PubMed]
- Chakraborty D, Natarajan C, Mukherjee A. Advances in oral cancer detection. Adv Clin Chem 2019;91:181-200.
[Crossref] [Google Scholar] [PubMed]
- Sasahira T, Kirita T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int J Mol Sci 2018;19(8):2413.
[Crossref] [Google Scholar] [PubMed]
- Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 2016;8(9):1-35.
[Crossref] [Google Scholar] [PubMed]
- Kengkarn S, Petmitr S, Reamtong O, Poomsawat S, Sanguansin S. Identification of novel candidate biomarkers for oral squamous cell carcinoma based on whole gene expression profiling. Pathol Oncol Res 2020;26(4):2315-25.
[Crossref] [Google Scholar] [PubMed]
- Fox LE. Carboplatin. J Am Anim Hosp Assoc 2000;36(1):13-4.
[Crossref] [Google Scholar] [PubMed]
- Xiang M, Colevas AD, Holsinger FC, Le QT, Beadle BM. Survival after definitive chemoradiotherapy with concurrent cisplatin or carboplatin for head and neck cancer. J Natl Compr Canc Netw 2019;17(9):1065-73.
[Crossref] [Google Scholar] [PubMed]
- Moncharmont C, Auberdiac P, Mélis A, Afqir S, Pacaut C, Chargari C, et al. Cisplatin or carboplatin, that is the question. Bull Cancer 2011;98(2):164-75.
[Crossref] [Google Scholar] [PubMed]
- Ho GY, Woodward N, Coward JI. Cisplatin versus carboplatin: Comparative review of therapeutic management in solid malignancies. Crit Rev Oncol Hematol 2016;102:37-46.
[Crossref] [Google Scholar] [PubMed]
- Pujade-Lauraine E, Guastalla JP, Vincent P. Carboplatin in cancer of the ovary. Bull Cancer 2000;87:40-7.
[Google Scholar] [PubMed]
- Hall MD, Telma KA, Chang KE, Lee TD, Madigan JP, Lloyd JR, et al. Say no to DMSO: Dimethylsulfoxide inactivates cisplatin, carboplatin, and other platinum complexes. Cancer Res 2014;74(14):3913-22.
[Crossref] [Google Scholar] [PubMed]
- Stordal B, Davey M. Understanding cisplatin resistance using cellular models. IUBMB Life 2007;59(11):696-9.
[Crossref] [Google Scholar] [PubMed]
- Oun R, Rowan E. Cisplatin induced arrhythmia; electrolyte imbalance or disturbance of the SA node? Eur J Pharmacol 2017;811:125-8.
[Crossref] [Google Scholar] [PubMed]
- Wouda RM, Hocker SE, Higginbotham ML. Safety evaluation of combination carboplatin and toceranib phosphate (Palladia) in tumour?bearing dogs: A phase I dose finding study. Vet Comp Oncol 2018;16(1):E52-60.
[Crossref] [Google Scholar] [PubMed]
- Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47.
- Moskovitz J, Moy J, Ferris RL. Immunotherapy for head and neck squamous cell carcinoma. Curr Oncol Rep 2018;20(2):1-7.
[Crossref] [Google Scholar] [PubMed]
- Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol 2019;16(11):669-83.
[Crossref] [Google Scholar] [PubMed]
- Rivera C, Oliveira AK, Costa RA, de Rossi T, Leme AF. Prognostic biomarkers in oral squamous cell carcinoma: A systematic review. Oral Oncol 2017;72:38-47.
[Crossref] [Google Scholar] [PubMed]
- Chitapanarux I, Lorvidhaya V, Kamnerdsupaphon P, Sumitsawan Y, Tharavichitkul E, Sukthomya V, et al. Chemoradiation comparing cisplatin versus carboplatin in locally advanced nasopharyngeal cancer: Randomised, non-inferiority, open trial. Eur J Cancer 2007;43(9):1399-406.
[Crossref] [Google Scholar] [PubMed]
- Rades D, Ulbricht T, Hakim SG, Schild SE. Cisplatin superior to carboplatin in adjuvant radiochemotherapy for locally advanced cancers of the oropharynx and oral cavity. Strahlenther Onkol 2012;188(1):42-8.
[Crossref] [Google Scholar] [PubMed]
- Xiao S, Zhang M, Liu C, Wang D. MiR-514 attenuates proliferation and increases chemoresistance by targeting ATP binding cassette subfamily in ovarian cancer. Mol Genet Genomics 2018;293(5):1159-67.
[Crossref] [Google Scholar] [PubMed]
- Dong R, Liu J, Sun W, Ping W. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in lung adenocarcinoma and lung squamous cell carcinoma. Pathol Oncol Res 2020;26(3):1935-45.
[Crossref] [Google Scholar] [PubMed]
- Liang JJ, Wang JY, Zhang TJ, An GS, Ni JH, Li SY, et al. MiR-509-3-5p-NONHSAT112228. 2 Axis Regulates p21 and suppresses proliferation and migration of lung cancer cells. Curr Top Med Chem 2020;20(10):835-46.