- *Corresponding Author:
- Wenfang Liu
Department of Thyroid and Breast Surgery, Cangzhou Central Hospital, Cangzhou, Hebei 061001, China
E-mail: 164442488@qq.com
| This article was originally published in a special issue, “Role of Biomedicine in Pharmaceutical Sciences” |
| Sci 2023:85(2) Spl Issue “159-165” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Breast cancer is the most common global malignancy and the leading cause of cancer deaths for women. Aromatase inhibitors are clinically recommended therapy for the disorders. However, resistance to aromatase inhibitors therapies still occurs and the underlined molecular mechanisms are still poorly understood. The tumour specimens consisted of aromatase inhibitor-sensitive and aromatase inhibitor-resistant breast cancer patients, who were compared and the differentially expressed genes for aromatase inhibitor-sensitive patients were generated and related genetic network was explored. The enriched cellular process and signaling cassettes for aromatase inhibitor-sensitive patients were also in-depth investigated. Compared with the aromatase inhibitor-resistant patients, 896 genes showed a significant differential expression. 621 were significantly up-regulated and 275 were remarkably downregulated. From the interaction network, the differentially expressed genes including signal recognition particle 72, B-cell lymphoma-2 associated transcription factor 1, cathelicidin antimicrobial peptide and defensin alpha 1B could be considered as the key target genes for aromatase inhibitors treatment in breast cancer patients. These differentially expressed genes were enriched in processes involving immunoglobulin complex, cell division and others while, drug metabolism and other signalling mechanisms were enriched in aromatase inhibitors treatment of breast cancer patients. The study speculated several novel gene targets for aromatase inhibitors therapeutic treatment of breast cancer patients and initiated a beneficial direction for future aromatase inhibitors drug resistance research.
Keywords
Breast cancer, aromatase inhibitors, differentially expressed genes, protein-protein interaction
Breast cancer is the leading cause of cancer death in women and the second most common cancer overall after lung cancer. It is also the fifth most common cause of cancer death[1], so it makes sense that breast cancer is one of the most studied malignancies in the world [2]. Many factors have been shown to be directly related to breast cancer, including those related to hormones, pregnancy, anthropometric indices, physical condition, diet, environmental exposures, etc.[3]. With currently available treatments, including surgery, chemotherapy, endocrine therapy, radiotherapy and targeted therapy, the clinical outcomes of breast cancer have improved over the past 20 y. However, the treatment of breast cancer remains challenging due to its significant heterogeneity[4]. In the clinic, breast cancer patients can be classified into a number of subtypes. To date, classifications have been based on different expression patterns of the Progesterone Receptor (PR), the Estrogen Receptor (ER) and the Human Epidermal Growth Factor Receptor-2 (HER-2)[5]. These breast cancers that have a significant number of receptors for either estrogen or progesterone are called Hormone Receptor (HR) positive breast cancers[6].
Aromatase is a Cytochrome P450 (CYP450) enzyme encoded by the Cytochrome P450 Family 19 Subfamily A Member 1 (CYP19A1) gene that can convert androgens to estrogens, specifically testosterone and androstenedione to the aromatic estrogens, estradiol and estrone[7]. Aromatase is expressed in many tissues including the placenta, central nervous system, bone, muscle, testes, prostate, adrenal glands and skin[8]. Aromatase is often overexpressed in breast endothelial cells and the surrounding stroma in HR+ breast cancer, leading to local estrogen production in the tumour microenvironment and promoting cancer progression by activating the ER[9]. Therefore, Aromatase Inhibitors (AIs) play a key role in the treatment of HR+ breast cancer. AIs include both steroidal and non-steroidal agents and have been shown to be useful in the treatment of breast cancer[10]. In clinical practice, AIs are the most commonly used first-line endocrine therapy options for postmenopausal women with ER-positive/PR- positive locally advanced or metastatic breast cancer, while tamoxifen or an AI plus ovarian suppression is the recommended therapy for premenopausal women[11]. However, resistance to AI therapies still occurs[12]. The molecular mechanism of resistance to AI therapy in breast cancer is still unclear.
Based on the uncertain molecular mechanism behind AI resistance in breast cancer patients, we compared tumour tissues between AI-sensitive and AI-resistant ER-positive breast cancer patients. At the same time, the potential target genes and genetic network were also investigated in detail. All these promising findings shed light on future breast cancer research.
Materials and Methods
Specimens and Differentially Expressed Gene (DEG) analysis:
The Ribonucleic Acid (RNA) sequencing data and complete clinical information in this study were obtained from the Gene Expression Omnibus (GEO) database with accession number GSE206199 (https://www.ncbi.nlm.nih.gov/geo/query/acc. cgi?acc=GSE206199). Of these, 14 tumour samples were obtained from ER-positive breast cancer patients receiving adjuvant AI treatment. The 14 tumour samples consisted of 7 AI-sensitive (S1-S7) and 7 AI-resistant (R1-R7) patients.
Differential expression analysis:
The edgeR package in the R language was used to analyze DEGs between AI-sensitive (S1-S7) and AI-resistant (R1-R7) patients. The absolute values of logarithmically transformed differential expression multiples (Log2Fold Change (FC))>1 and p-value<0.05 were used as screening criteria[13].
Functional enrichment analysis:
The clusterProfiler package in R language was processed for Gene Ontology (GO) analysis (including Biological Process (BP), Molecular Function (MF) and Cellular Component (CC)) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis[14], p-value<0.05 was considered statistically significant.
Protein-Protein Interaction (PPI) networks:
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database is a database that analyses and predicts functional protein-protein connections. We used STRING (https://string- db.org/, version 11.0) to analyze the functional connections and interactions of candidate proteins[15], from which the interaction pairs with a combined score greater than or equal to 0.4 (confidence score≥0.4) were retained. Cytoscape (https:// cytoscape.org/, version 3.7.2) was used to visualize the PPI network[16]. The network modules reserve specific biological meanings, which are usually the core of protein networks. We process the Molecular Complex Detection (MCODE) method plug-in in Cytoscape software to identify significant clustering modules, using MCODE score>2 as a threshold.
Statistical analysis:
The GraphPad Prism 9 was established for data collection and analysis in this study. The student t-test was used to compare the differences between two groups. The p-value<0.05 was considered as a statistical significant.
Results and Discussion
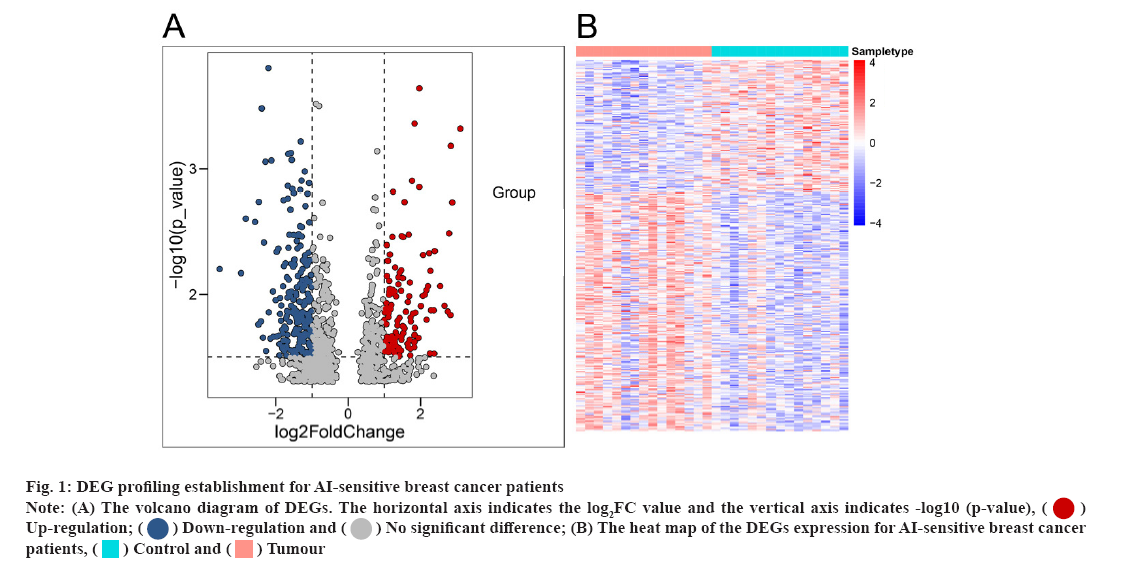
DEG profiling establishment for AI-sensitive breast cancer patients were shown here. In the study, we first analysed the differentially expressed messenger RNAs (mRNAs) using the online GEO chip GSEGSE206199, which contains RNA sequencing information for 7 AI-sensitive (S1-S7) and AI- resistant (R1-R7) breast cancer patients. Compared to the AI-resistant patients, 896 genes showed a significant differential expression (fig. 1A and fig. 1B).
Fig 1: DEG profiling establishment for AI-sensitive breast cancer patients
Note: (A) The volcano diagram of DEGs. The horizontal axis indicates the log2FC value and the vertical axis indicates -log10 (p-value), Up-regulation;
Up-regulation; Down-regulation and
Down-regulation and No significant difference; (B) The heat map of the DEGs expression for AI-sensitive breast cancer patients,
No significant difference; (B) The heat map of the DEGs expression for AI-sensitive breast cancer patients,  Control and
Control and  Tumour
Tumour
Of the 896 DEGs, 621 were significantly up-regulated (Table 1). At the same time, 275 were remarkably downregulated in 7 AI-sensitive (S1-S7) breast cancer patients compared to AI-resistant patients (Table 2).
| Gene name | Results | Adjusted p value | p value | logFC |
|---|---|---|---|---|
| SRP72 | 31670_s_at | 6.71E-16 | 1.98E-18 | 5.73919 |
| BCLAF1 | 38050_at | 4.94E-21 | 2.09E-24 | 4.664181 |
| LUC7L3 | 34397_at | 9.47E-12 | 6.80E-14 | 4.536923 |
| DNTTIP2 | 37938_at | 2.31E-18 | 1.95E-21 | 4.416025 |
| TRAPPC8 | 36002_at | 2.36E-09 | 3.54E-11 | 4.310544 |
| CLEC2B | 40698_at | 0.0125 | 0.00233 | 4.251532 |
| EXOSC8 | 36968_s_at | 4.14E-10 | 5.08E-12 | 4.191484 |
| RECQL | 34684_at | 1.09E-12 | 6.21E-15 | 4.027176 |
| RBM6 | 40869_at | 7.22E-10 | 9.15E-12 | 3.81235 |
| PRPF40A | 37506_at | 2.33E-17 | 3.94E-20 | 3.764995 |
| SRSF11 | 32183_at | 6.93E-18 | 8.62E-21 | 3.703981 |
| ITGA4 | 2061_at | 7.46E-08 | 1.81E-09 | 3.675505 |
| ZFR | 40610_at | 2.79E-09 | 4.42E-11 | 3.634965 |
| SMARCA5 | 39132_at | 1.54E-14 | 6.52E-17 | 3.592693 |
| ZC3H15 | 35750_at | 2.39E-21 | 5.05E-25 | 3.554028 |
| RPS6KB1 | 2037_s_at | 1.07E-10 | 1.04E-12 | 3.505803 |
| SECISBP2L | 41634_at | 1.43E-09 | 1.97E-11 | 3.474673 |
| SMC4 | 34878_at | 8.15E-09 | 1.53E-10 | 3.462946 |
| PPIG | 37385_at | 1.59E-18 | 1.01E-21 | 3.443338 |
| KTN1 | 32846_s_at | 6.93E-18 | 8.78E-21 | 3.396649 |
Table 1: Top 20 Gene List of Enhanced Expression for AI-Sensitive Breast Cancer Patients Compared With AI-Resistant Breast Cancer Patients
| Gene name | Results | Adjusted p value | p value | logFC |
|---|---|---|---|---|
| NR2F1 | 39294_at | 0.0518 | 0.0144 | -1.00729 |
| KAZN | 39615_at | 0.00221 | 0.00028 | -1.00736 |
| PTPRU | 33750_at | 0.0897 | 0.0303 | -1.00861 |
| HIST2H2BE | 33352_at | 0.207 | 0.0937 | -1.01303 |
| NOP2 | 1979_s_at | 0.00966 | 0.0017 | -1.01368 |
| IGHMBP2 | 31861_at | 0.00544 | 0.000835 | -1.01482 |
| LBR | 288_s_at | 0.0326 | 0.00774 | -1.02196 |
| GNA11 | 564_at | 0.00278 | 0.000368 | -1.02255 |
| FAM214B | 33841_at | 0.00259 | 0.000337 | -1.02311 |
| ARL2-SNX15 | 37570_at | 0.00327 | 0.000451 | -1.02462 |
| CDH6 | 37784_at | 0.124 | 0.0465 | -1.02556 |
| CCND3 | 1794_at | 0.0293 | 0.00684 | -1.03604 |
| SOD3 | 692_s_at | 0.0421 | 0.0108 | -1.03861 |
| KRT31 | 36310_at | 0.0111 | 0.00201 | -1.03882 |
| AURKB | 33266_at | 0.00838 | 0.00141 | -1.03884 |
| HLA-F | 37420_i_at | 0.116 | 0.0428 | -1.04081 |
| TGFA | 160025_at | 0.215 | 0.0988 | -1.044 |
| HEXA | 39340_at | 0.0598 | 0.0173 | -1.04657 |
| CDKN2D | 1797_at | 0.00879 | 0.00151 | -1.05586 |
| CXCR4 | 649_s_at | 0.165 | 0.0687 | -1.06204 |
Table 2: Top 20 Gene List of Decreased Expression For AI-Sensitive Breast Cancer Patients Compared with AI-Resistant Breast Cancer Patients
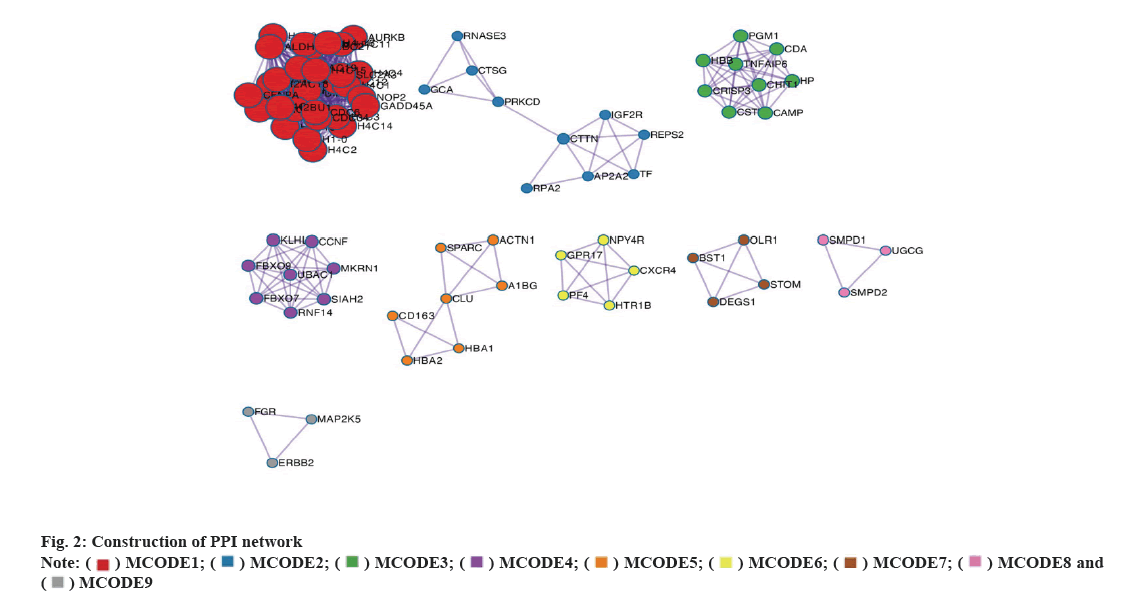
Construction of PPI network was explained here. A STRING database was used to construct a PPI network for the 896 DEGs and the gene interactions with a confidence score≥0.4 were selected for visualization using Cytoscape software. The MCODE plug-ins were run to identify significant clustering modules (fig. 2). Within these, there are 9 prominent MCODEs. The primary up-regulated genes (Signal Recognition Particle 72 (SRP72) and B-Cell Lymphoma-2 Associated Transcription Factor 1 (BCLAF1)) were localized in MCODE1, while the primary down- regulated genes (Cathelicidin Antimicrobial Peptide (CAMP) and Defensin Alpha 1B (DEFA1B)) were localized in MCODE3. MCODE1 and MCODE3 were also the leading MCODEs for AI-sensitive breast cancer patients (with the highest nodal grade). Thus, the DEGs including SRP72, BCLAF1, CAMP and DEFA1B could be considered as the key target genes for AI treatment in breast cancer patients. As shown in fig. 2, in the network, each point represents a node. The more line segments connected to the node, the higher the degree of the node. The size of the node directly reflects the degree; the thicker the connection between the nodes, the stronger the interaction between the two nodes.
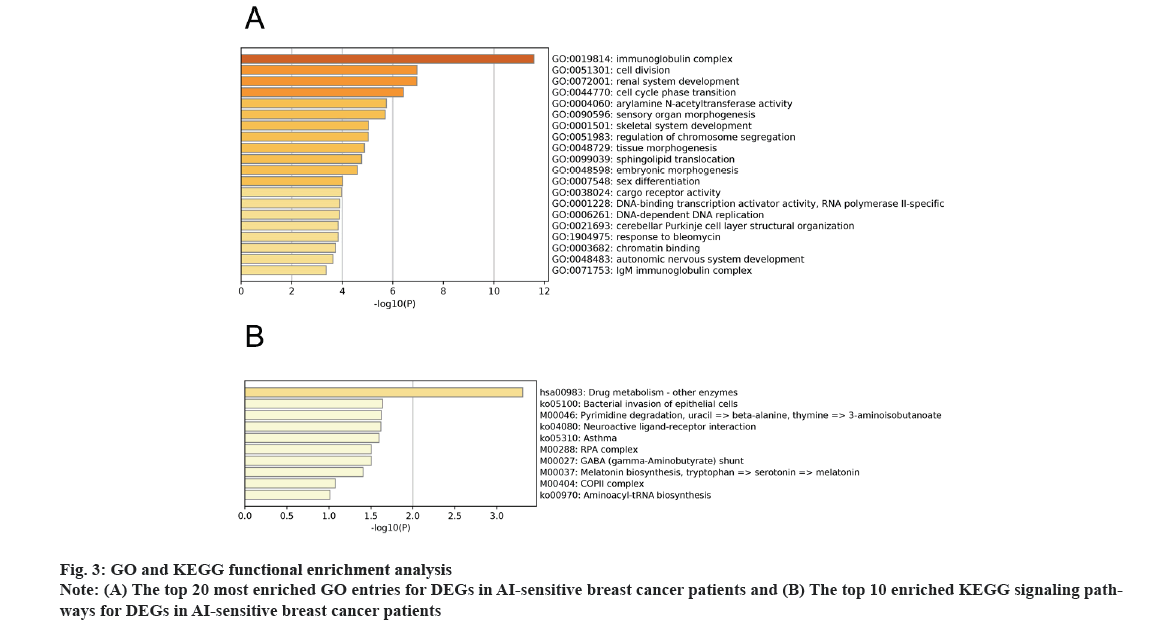
GO and KEGG functional enrichment analysis was explained here. For the 896 DEGs, we performed both GO and KEGG analysis for AI treatment in breast cancer patients. The GO enrichment analysis included BPs and CCs as well as MFs. As shown in fig. 3A, DEGs were enriched in processes involving immunoglobulin complex, cell division and others. At the same time, the KEGG analysis reflected the enriched signalling pathways. As shown in fig. 3B, drug metabolism and other signalling mechanisms were enriched in AI treatment of breast cancer patients.
Like other types of cancer, breast cancer is a truly complex disease and is the second leading cause of cancer-related death in women worldwide[17-19]. Accumulating evidence has suggested that breast cancer is closely associated with a variety of internal (genetic) as well as external (environmental) factors. Due to the characteristics of breast cancer, different therapies have emerged in the era of breast cancer in recent years[20]. For example, Antibody- Drug Conjugation systems (ADCs), nanoparticles (albumin, metal, lipid, polymer and micelle-based nanoparticles) and Breast Cancer Stem Cells (BCSCs)-based therapies as well as AI treatment in this study. However, even with the breakthrough of these novel therapies, resistance to treatment has severely hampered the overall effect.
There are several AIs available in clinical practice, including exemestane, formestane, anastrozole, letrozole, fadrozole and vorozole, etc. Currently, AI is recommended in clinical practice for postmenopausal women. In fact, postmenopausal women are at high risk of developing breast cancer due to estrogen production in peripheral tissues of the body other than the ovaries[21]. However, aromatase has been identified in breast tissue, leading to local estrogen accumulation that can be effectively suppressed by AIs. A previous study by the Early Breast Cancer Trialists Collaborative Group (EBCTCG) compared the clinical outcomes between AI and a common anticancer drug (tamoxifen)[22].
They claimed that for women with early-stage ER- positive breast cancer, adjuvant tamoxifen reduces 15 y risk of breast cancer mortality by one third. While AIs were more effective than tamoxifen in postmenopausal women, they are ineffective in premenopausal women when used without ovarian suppression. In addition, using an AI instead of tamoxifen in premenopausal women with ovarian suppression could significantly reduce the risk of breast cancer recurrence. However, the most promising challenge for AIs in breast cancer therapy is resistance and toxicity, especially for the steroid class of drugs. All of this could be due to the fact that the molecular mechanism behind AIs is unclear.
In the study, we compared 7 AI-sensitive (S1-S7) and AI-resistant (R1-R7) breast cancer patients in detail. Compared with AI-resistant patients, 896 genes showed significantly different expression. With further PPI analysis, we found that the DEGs such as SRP72, BCLAF1, CAMP and DEFA1B could be considered as central target genes for AI treatment in breast cancer patients. SRP72 stands for stress response (heat shock) protein (SRP) 72. A previous study by Prevo et al. suggested SRP72 as a novel gene involved in radioresistance for tumour progression[23]. Deletion of SRP72 resulted in significant radiosensitization of immortal Human cell line (HeLa, cervical), Pancreatic Adenocarcinoma cell line (PSN-1, pancreatic), Urinary Bladder Carcinoma cell line (T24, bladder), BT-549 (breast) and MCF7 (breast) tumour lines as measured by colony formation assays. The identification of SRP72s that underpin tumour radioresistance could help us to better understand the development of targeted radiosensitizers or help to personalize radiotherapy treatment. BCLAF1 has been shown to manipulate Programmed Cell Death Ligand 1 (PD-L1) in response to Ionizing Radiation (IR)[24]. Knockdown of BCLAF1 greatly reduced PD-L1 expression by promoting PD-L1 ubiquitination. Previously, using a similar method, a study focused on the expression profile of dysregulated proteins in sublethally heat- treated breast cancer cells[25]. The overall expression of BCLAF1 was inhibited and the proliferation and invasion ability of breast cancer cells were activated in the sublethal heat-treated group. The CAMP is a classical G-protein dependent signaling pathway. A large body of evidence has shown that hormone- bound steroid receptors in breast cancer cells activate complex signaling networks that include the Mitogen-Activated Protein Kinase (MAPK) and cyclic Adenosine Monophosphate (cAMP) pathways. Collectively, these pathways lead to breast cancer progression and resistance to hormone therapy[26].
Breast cancer is a common cancer with a rapidly increasing prevalence and its aetiology is still poorly understood. AIs are the recommended therapy for breast cancer with high frequency of drug resistance. In this study, we systematically investigated the differentially expressed mRNAs for AI-treated breast cancer patients. The DEGs were approved and the associated genetic network was established. Overall, several novel genes were initiated and the innovative signaling cascade was established.
Conflict of Interest
The authors declared no conflict of interest.
References
- Hutchinson L. Challenges, controversies, breakthroughs. Nat Rev Clin Oncol 2010;7(12):669-70.
[Crossref] [Google scholar] [PubMed]
- Woolston C. Breast cancer. Nature 2015;527(7578):S101.
[Crossref] [Google scholar] [PubMed]
- Veronesi U, Boyle P, Goldhirsch A, Orecchia R, Viale G. Breast cancer. Lancet 2005;365(9472):1727-41.
[Crossref] [Google scholar] [PubMed]
- Li D, Zhao W, Zhang X, Lv H, Li C, Sun L. NEFM DNA methylation correlates with immune infiltration and survival in breast cancer. Clin Epigenetics 2021;13(1):112.
[Crossref] [Google scholar] [PubMed]
- Rajana N, Mounika A, Chary PS, Bhavana V, Urati A, Khatri D, et al. Multifunctional hybrid nanoparticles in diagnosis and therapy of breast cancer. J Control Release 2022;352:1024-47.
[Crossref] [Google scholar] [PubMed]
- Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LA, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst 2014;106(5).
[Crossref] [Google scholar] [PubMed]
- Subramanian A, Salhab M, Mokbel K. Oestrogen producing enzymes and mammary carcinogenesis: A review. Breast Cancer Res Treat 2008;111:191-202.
[Crossref] [Google scholar] [PubMed]
- Santen RJ, Brodie H, Simpson ER, Siiteri PK, Brodie A. History of aromatase: Saga of an important biological mediator and therapeutic target. Endocr Rev 2009;30(4):343-75.
[Crossref] [Google scholar] [PubMed]
- Zhao H, Zhou L, Shangguan AJ, Bulun SE. Aromatase expression and regulation in breast and endometrial cancer. J Mol Endocrinol 2016;57(1):19-33.
[Crossref] [Google scholar] [PubMed]
- Brueggemeier RW, Hackett JC, Diaz-Cruz ES. Aromatase inhibitors in the treatment of breast cancer. Endocr Rev 2005;26(3):331-45.
[Crossref] [Google scholar] [PubMed]
- Burstein HJ, Lacchetti C, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: American society of clinical oncology clinical practice guideline update on ovarian suppression. J Clin Oncol 2016;34(14):1689-701.
[Crossref] [Google scholar] [PubMed]
- Chen S, Masri S, Wang X, Phung S, Yuan YC, Wu X. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol 2006;102(1):232-40.
[Crossref] [Google scholar] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139-40.
[Crossref] [Google scholar] [PubMed]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012;16(5):284-7.
[Crossref] [Google scholar] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(1):607-13.
[Crossref] [Google scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google scholar] [PubMed]
- Jafari SH, Saadatpour Z, Salmaninejad A, Momeni F, Mokhtari M, Nahand JS, et al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J Cell Physiol 2018;233(7):5200-13.
[Crossref] [Google scholar] [PubMed]
- Thorat MA, Balasubramanian R. Breast cancer prevention in high-risk women. Best Pract Res Clin Obstet Gynaecol 2020;65:18-31.
[Crossref] [Google scholar] [PubMed]
- Kashyap D, Pal D, Sharma R, Garg VK, Goel N, Koundal D, et al. Global increase in breast cancer incidence: Risk factors and preventive measures. Biomed Res Int 2022;2022:1-16.
[Crossref] [Google scholar] [PubMed]
- Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, et al. Breast cancer: Biology, biomarkers and treatments. Int Immunopharmacol 2020;84:106535.
[Crossref] [Google scholar] [PubMed]
- Kharb R, Haider K, Neha K, Yar MS. Aromatase inhibitors: Role in postmenopausal breast cancer. Arch Pharm 2020;353(8):2000081.
[Crossref] [Google scholar] [PubMed]
- Bradley R, Braybrooke J, Gray R, Hills RK, Liu Z, Pan H, et al. Aromatase inhibitors versus tamoxifen in premenopausal women with oestrogen receptor-positive early-stage breast cancer treated with ovarian suppression: A patient-level meta-analysis of 7030 women from four randomised trials. Lancet Oncol 2022;23(3):382-92.
[Crossref] [Google scholar] [PubMed]
- Prevo R, Tiwana GS, Maughan TS, Buffa FM, McKenna WG, Higgins GS. Depletion of signal recognition particle 72kDa increases radiosensitivity. Cancer Biol Ther 2017;18(6):425-32.
[Crossref] [Google scholar] [PubMed]
- Ma Z, Wang H, Meng F, Han Y, Chen Y, Xiao M, Jiang H, Yu Z, Xu B. Role of BCLAF‐1 in PD‐L1 stabilization in response to ionizing irradiation. Cancer Sci 2021;112(10):4064-74.
[Crossref] [Google scholar] [PubMed]
- Xia S, Li X, Xu S, Ni X, Zhan W, Zhou W. Sublethal heat treatment promotes breast cancer metastasis and its molecular mechanism revealed by quantitative proteomic analysis. Aging 2022;14(3):1389-406.
[Crossref] [Google scholar] [PubMed]
- Castoria G, Migliaccio A, D'Amato L, di Stasio R, Ciociola A, Lombardi M, et al. Integrating signals between cAMP and MAPK pathways in breast cancer. Front Biosci 2008;13(4):1318-27.
[Crossref] [Google scholar] [PubMed]