- *Corresponding Author:
- Liu Bingbing
Department of Anesthesiology, Kailuan General Hospital, Tangshan, Hebei Province 063000, China
E-mail: liubingbingsci@163.com
| Date of Received | 07 June 2021 |
| Date of Revision | 14 August 2022 |
| Date of Acceptance | 18 May 2023 |
| Indian J Pharm Sci 2023;85(3):721-730 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Glioma is the most frequent primary intracranial tumor worldwide. Dexmedetomidine is an efficient anesthetic used in surgery and has an impact on glioma. Yet, the mechanism of dexmedetomidine on the biological characteristics of glioma cells is unknown. Circ_BRAF, microRNA-381-3p and thrombospondin 2 was detected via quantitative reverse transcription-polymer chine reaction. Proliferation detection was conducted using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and colony formation assay. Transwell assay was used to determine cell migratory and invasive capacities. The protein expression was determined using Western blot. Binding between microRNA- 381-3p and circ_BRAF or thrombospondin 2 was validated using dual-luciferase reporter. Role between dexmedetomidine and circ_BRAF on glioma growth was analyzed using the xenograft tumor model in mice. Dexmedetomidine inhibited circ_BRAF expression in glioma cells in a dose-dependent manner. Dexmedetomidine induced glioma cell proliferation, migration and invasion was neutralized via circ_ BRAF up regulation. Meanwhile, circ_BRAF could facilitate thrombospondin 2 expression via binding with miR-381-3p. Besides, dexmedetomidine was revealed to promote glioma tumourogenesis in vivo via circ_BRAF/ microRNA-381-3p/thrombospondin 2 pathways. Dexmedetomidine exposure expedited glioma development via circ_BRAF/microRNA-381-3p/thrombospondin 2 axis, providing a theoretical basis for glioma therapy.
Keywords
Glioma, dexmedetomidine, circ_BRAF, microRNA-381-3p, thrombospondin 2
Glioma is a commonest malignant intracranial neoplasm that mainly occurs in Central Nervous System (CNS) and remains largely incurable[1,2]. Glioma has highly proliferative and invasive capacities with high drug-resistance and incidence rate[3]. At present, despite numerous medical studies have been conducted on glioma etiology, the survival rate remain extremely low. Hence, the development of effective targeted therapies for glioma is essential.
Dexmedetomidine (DEX) is a highly selective agonist of the alpha (α) 2-adrenoceptor with sedative, anxiolytic, sympatholytic and analgesic properties, which has been widely applied for sedation during surgical operations[4,5]. DEX has been attested to facilitate breast cancer cell malignant behaviors[6,7]. Moreover, DEX at 10 nm could aggrandize the proliferation of both glioma U251 and U87MG cells[8]. Nevertheless, the underlying mechanism of DEX on glioma tumourogenesis is needed to be explained.
Circular RNAs (circRNAs) represent a newly described type of non-coding RNAs (ncRNAs) possessing high stabilization and closed-loop structures[9,10]. Deregulated circRNAs can regulate the proliferative and invasive abilities of glioma[11,12]. For example, circ-SMAD7 could promote glioma cell proliferation and metastasis by up regulating Proliferating Cell Nuclear Antigen (PCNA)[13]. CircMMP1 promoted the progression of glioma via microRNA (miR)-433/High-Mobility Group Box (HMGB) 3 axis[14]. Moreover, circ_BRAF (hsa_circ_0082755) was down regulated in glioma tissues[15]. However, studies about the role of circ_BRAF on glioma are far from enough.
miRNAs are small ncRNAs, Ribonucleic Acid (RNA) lack protein-coding abilities like circRNAs and can modulate cancer malignant progression by targeting the 3’Untranlasted Regions (3’UTRs) of messenger RNA (mRNAs) to regulate gene expression[16,17]. MiRNAs could function as latent diagnostic markers in glioma[18,19]. For instance, miRNA-9 was able to distinctly enhance glioma cell proliferation[20]. MiR-136-3p could block glioma tumourogenesis by sponging KLF7[21]. Meanwhile, miR-381 silence might impede glioma cell malignant progression via binding to LRRC4[22]. While the targeted relationship between circ_BRAF and miR-381-3p in glioma tumor genesis has been unexplored.
Herein, the impacts of DEX on glioma proliferation, metastasis and circ_BRAF expression were first revealed. Available bioinformatics software exhibited binding between miR-381-3p and circ_BRAF or THBS2. This current study disclosed a novel circRNA/miRNA/mRNA pathway based on circ_ BRAF in glioma and provided potential therapeutic target for glioma treatment.
Materials and Methods
Cell culture and DEX treatment:
Under a humidified 5 % Carbon dioxide (CO2) environment at 37°, human glioma cell lines A172 and U251 (Bena, Beijing, China) were maintained in Dulbecco's Modified Eagle Medium (DMEM) plus 10 % Fetal Bovine Serum (FBS) (Gibco, Carlsbad, California, United States of America (USA)).
To investigate optimum concentration of DEX treatment on the malignant behaviors of glioma cells, different final concentrations of DEX (5 nm, 15 nm, 30 nm and 45 nm) (Santa Cruz Laboratories, Dallas, Texas, USA) were applied to treat A172 and U251 cells for different times, with 0 nm DEX served as control group. Finally, 30 nm DEX was chosen for subsequent experiments.
Cell transfection:
Briefly, circ_BRAF overexpression vector was acquired via cloning circ_BRAF fragment into pCD-ciR (vector). In terms of miR-381-3p overexpression/knockdown, its mimics/inhibitor (miR-381-3p/anti-miR-381-3p) was introduced into A172 and U251 cells, (miR-NC/anti-miRNC) as their corresponding control. For the knockdown of Thrombospondin 2 (THBS2), si- THBS2 was transfected into A172 and U251 cells, with non-target siRNA (si-NC) as the negative control. Using Lipofectamine™ 3000 (Invitrogen, Paisley Scotland, UK), A172 and U251 cells were transfected with above oligonucleotides and plasmids GenePharma (Shanghai, China).
For stable overexpression of circ_BRAF, lentivirus carrying circ_BRAF vector (lenti-circ_BRAF) or corresponding non-target Negative Control (lenti-NC) was stably transfected into A172 cells in the presence of polybrene (8 μg/ml; Millipore, Billerica, Massachusetts, USA) by Lipofectamine™ 3000 (Invitrogen).
Cell proliferation detection:
Briefly, 1×104 A172 and U251 cells with different transfection following by exposing to different concentrations of DEX (0, 5, 15, 30 and 45 nm) for 24 h were planted into 96-well plates, respectively. Then, 3-(4,5-Dimethylthiazol-2-yl)-2,5 Diphenyl Tetrazolium Bromide (MTT) (Sigma-Aldrich, St. Louis, Missouri, USA) was introduced. After removing supernatant, 150 μl dimethyl sulfoxide (Sigma-Aldrich) was added.
Colony formation experiment:
After treatment, A172 and U251 cells were routinely cultivated 2 w. Generated colonies were interacted using crystal violet (Sangon, Shanghai, China) for staining, followed by count using a microscope.
Transwell assay:
Cell migration and invasion was illustrated employing Transwell chamber (Corning Inc., Corning, New York, USA) pre-coated without and with Matrigel, respectively. Cells with FBSfree medium were added into upper unit. 10 % FBS was supplied into the lower unit. After 24 h of cell culture, cells in lower surface were dyed using crystal violet (Sangon) after washed with phosphate buffered saline (Phosphate Buffer Solution (PBS), BD Biosciences, San Jose, California, USA) and immobilized using methanol (Sangon). Ultimately, the metastasis ability was assessed using an inverted microscope.
Quantitative Reverse Transcription-Polymerase Chine Reaction (qRT-PCR):
Trizol reagent (Beyotime, Shanghai, China) was utilized and the complementary Deoxyribonucleic Acid (cDNA) was synthesized via Prime Script RT Reagent kit (Takara, Dalian, China). Subsequently, specific primers (Sangon) and iQ SYBR Green Super mix (Bio-Rad) was employed qRT-PCR. The 2-ΔΔCt method computed gene expression. GAPDH and U6 were internal parameters, for circ_BRAF/ BRAF/THBS2 and miR-381-3p. Primers were presented in Table 1.
Actinomycin D treatment:
Extracted RNA from cells was incubated with 2 μg/ml Actinomycin D (Solarbio, Beijing, China) to block transcription for 0, 8, 16 and 24 h. Finally, qRT-PCR was conducted for the examination of circ_BRAF expression and linear BRAF mRNA expression.
Western blot:
Protein was isolated with Radioimmunoprecipitation (RIPA) lysis buffer (Beyotime). After being electrophoresed on 10 % Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) gel, samples were blotted onto polyvinylidene fluoride membranes, which were kept with the primary antibodies (Abcam, Cambridge, UK) as follows; Ki67 (ab243878; 1:1000), Snail (ab216347; 1:1000), E-cadherin (ab133597; 1:2000), THBS2 (ab84469; 1 μg/ml) and Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) (ab181602; 1:10 000). After that, samples were incubated with second antibody (ab6721, 1:3000), followed by analysis using Rapid Step Enhanced Chemiluminescence (ECL) reagent.
Dual-luciferase reporter assay:
Generally, circ_BRAF/THBS2-WT and circ_ BRAF/THBS2-MUT were acquired by amplifying and synthesizing the sequences of Wide-Type (WT) circ_BRAF or THBS2 3’UTR possessing miR-381-3p binding sites and Mutant (MUT) sequence lacking miR-381-3p binding sites into pmirGLO vector (Promega, Madison, Wisconsin, USA). Then, tumor cells were transfected with these constructs with miR-381-3p mimic or miRNC, followed by analysis using dual-luciferase reporter system (Promega).
Xenograft tumor model:
First of all, 24 male mice (Vital River Laboratory, Beijing, China) were introduced. Then, A172 cells (2×106/0.2 ml PBS) introduced with lenticirc_ BRAF or lenti-NC were suspended with PBS and then injected subcutaneously into the left flank of nude mice (about 6 w old, 25 g) to establish xenograft models. DEX (25 μg/kg) was then administrated mice every 2 h for three times immediately after transfection. These nude mice were arbitrarily divided into 4 groups (n=6 mice/ group); control group, DEX group, DEX+lenti- NC group and DEX+lenti-circ_BRAF group. The tumor volume was continuously monitored every 5 d with the formula of (length×width2)/2. At 30 d post inoculation, removed tumors were weighed and collected for abundances of circ_BRAF, miR- 381-3p and THBS2. The current animal experiment got authorization by Animal Ethical Committee of Kailuan General Hospital.
Statistical analysis:
Data was presented as means±standard deviations with three duplicates. We analyzed all statistics involved according to GraphPad Prism. Two-tailed Student’s t-test or Analysis of Variance (ANOVA) followed by Tukey’s test was applied for difference analysis. Statistical significance was set at p<0.05.
Results and Discussion
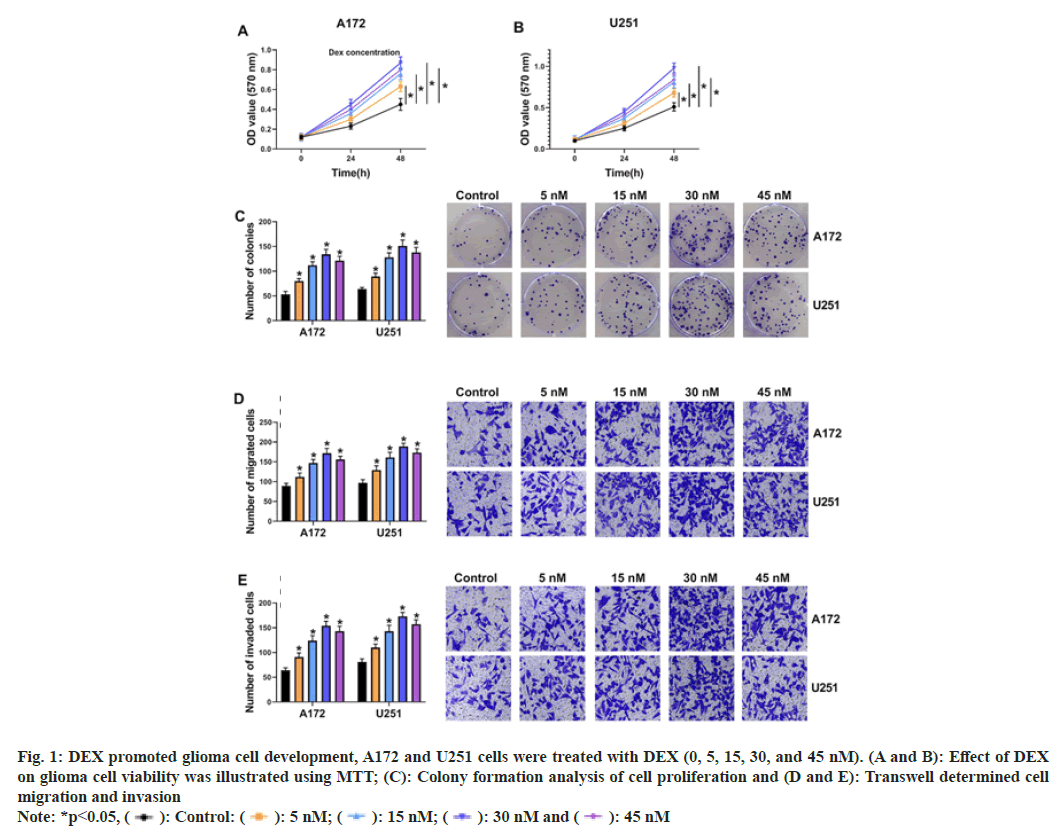
To explore the influence of DEX on cell progression, glioma cells were exposed to DEX (0, 5, 15, 30 and 45 nm). MTT and cell colony formation assay displayed that DEX treatment (5, 15 and 30 nm) facilitated cell proliferation in a concentration-dependent manner in A172 and U251 cells at 48 h after DEX treatment, but cell proliferation was decreased again after 45 nm DEX treatment relative to 30 nm DEX treatment (fig. 1A-fig. 1C). Transwell assay illustrated that cell migratory and invasive abilities were gradually accelerated after DEX treatment (5 nm, 15 nm and 30 nm), while the effects were alleviated after 45 nm DEX treatment than 30 nm DEX treatment (fig. 1D and fig. 1E). Therefore, 30 nm DEX was chosen for further experiments. These data demonstrated that DEX could promote glioma progression.
Fig. 1: DEX promoted glioma cell development, A172 and U251 cells were treated with DEX (0, 5, 15, 30, and 45 nM). (A and B): Effect of DEX
on glioma cell viability was illustrated using MTT; (C): Colony formation analysis of cell proliferation and (D and E): Transwell determined cell
migration and invasion
Note: *p<0.05, ( ): Control: (
): Control: ( ): 5 nM; (
): 5 nM; ( ): 15 nM; (
): 15 nM; ( ): 30 nM and (
): 30 nM and ( ): 45 nM
): 45 nM
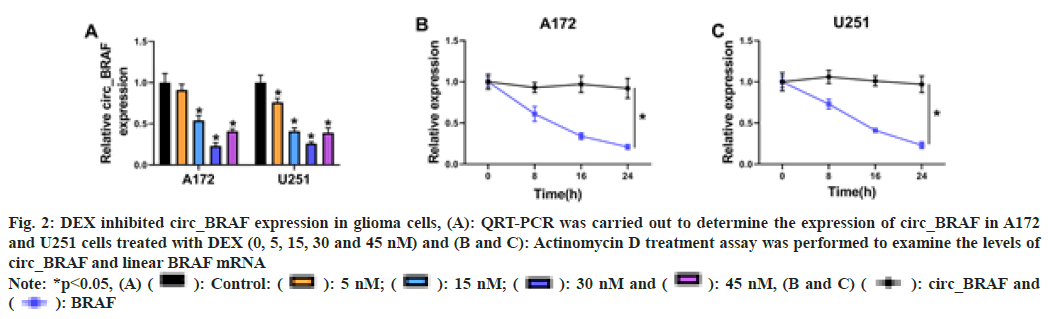
In order to illustrate the impact of DEX exposure on circ_BRAF expression in A172 and U251 cells, qRT-PCR assay was performed. The results showed that circ_BRAF expression was dramatically down regulated by DEX exposure (5 nm, 15 nm and 30 nm) in a concentrationdependent manner, but it was again elevated after 45 nm DEX treatment than 30 nm DEX treatment (fig. 2A). Then, Actinomycin D treatment showed half-life of linear BRAF was lower than circ_BRAF (fig. 2B and fig. 2C). These results suggested that circ_BRAF was a circular RNA and the facilitating role of DEX in glioma progression might be related with circ_ BRAF expression.
Fig. 2: DEX inhibited circ_BRAF expression in glioma cells, (A): QRT-PCR was carried out to determine the expression of circ_BRAF in A172
and U251 cells treated with DEX (0, 5, 15, 30 and 45 nM) and (B and C): Actinomycin D treatment assay was performed to examine the levels of
circ_BRAF and linear BRAF mRNA
Note: *p<0.05, (A) ( ): Control: (
): Control: ( ): 5 nM; (
): 5 nM; ( ): 15 nM; (
): 15 nM; ( ): 30 nM and (
): 30 nM and ( ): 45 nM, (B and C) (
): 45 nM, (B and C) ( ): circ_BRAF and
(
): circ_BRAF and
( ): BRAF
): BRAF
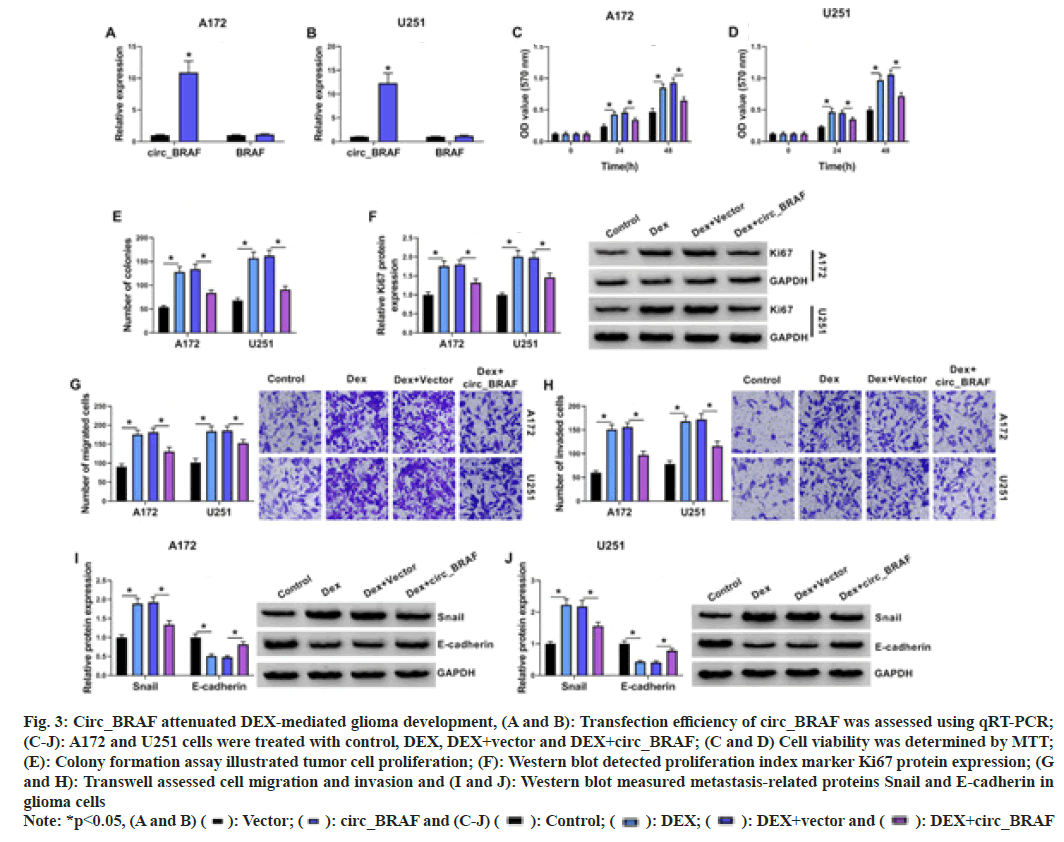
First of all, qRT-PCR results elucidated that circ_BRAF, rather than BRAF, was dramatically raised by circ_BRAF introduction in A172 and U251 cells, (fig. 3A and fig. 3B), hinting the high transfection efficiency of circ_BRAF. Subsequently, the effects of circ_BRAF overexpression on glioma cell development after DEX exposure were exposed. As shown in fig. 3C-fig. 3E, DEX treatment enhanced A172 and U251 cell proliferation, which was partially attenuated via circ_BRAF overexpression. Meanwhile, Ki67 level was raised via DEX addition, while circ_BRAF overexpression hindered the phenomenon in tumor cells (fig. 3F). With the employment of Transwell assay, the promotional effect of DEX on migration and invasion abilities of A172 and U251 cells were alleviated by circ_BRAF overexpression (fig. 3G and fig. 3H). Additionally, Western blot results unveiled that DEX exposure accelerated Snail and repressed E-cadherin protein expression in glioma cells; however, this impact was impeded via circ_BRAF (fig. 3I and fig. 3J). Doubtlessly, these findings illustrated that circ_ BRAF participated in DEX-mediated glioma cell progression.
Fig. 3: Circ_BRAF attenuated DEX-mediated glioma development, (A and B): Transfection efficiency of circ_BRAF was assessed using qRT-PCR;
(C-J): A172 and U251 cells were treated with control, DEX, DEX+vector and DEX+circ_BRAF; (C and D) Cell viability was determined by MTT;
(E): Colony formation assay illustrated tumor cell proliferation; (F): Western blot detected proliferation index marker Ki67 protein expression; (G
and H): Transwell assessed cell migration and invasion and (I and J): Western blot measured metastasis-related proteins Snail and E-cadherin in
glioma cells
Note: *p<0.05, (A and B) ( ): Vector; (
): Vector; ( ): circ_BRAF and (C-J) (
): circ_BRAF and (C-J) ( ): Control; (
): Control; ( ): DEX; (
): DEX; ( ): DEX+vector and (
): DEX+vector and ( ): DEX+circ_BRAF
): DEX+circ_BRAF
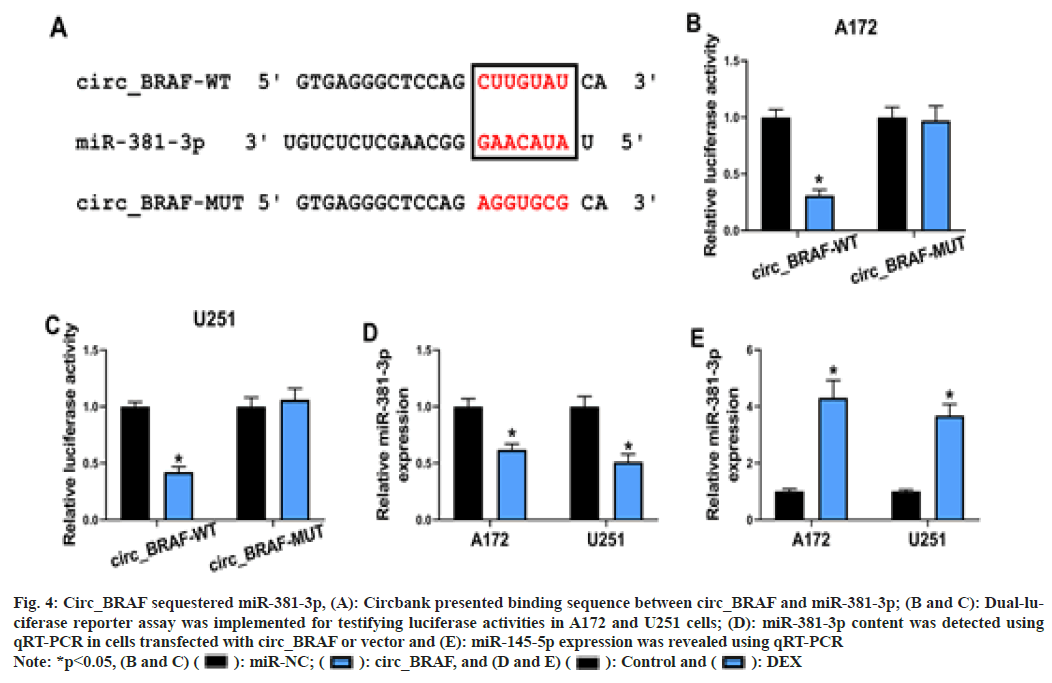
Based on Circbank online database, there are binding sequence between circ_BRAF and miR- 381-3p (fig. 4A). Meanwhile, the luciferase activity was dramatically repressed in circ_ BRAF-WT group in cells with miR-381-3p relative to miR-NC, whereas the luciferase activity had no obvious change in circ_BRAFMUT group (fig. 4B and fig. 4C), based on the dual-luciferase reporter assay. Subsequently, compared with transfection of vector, miR- 381-3p was down regulated in glioma cell lines by circ_BRAF elevation (fig. 4D). Later, the impact of DEX on miR-381-3p expression was also examined. The result attested that DEX observably accelerated miR-381-3p expression in glioma cells (fig. 4E). All these evidences illustrated circ_BRAF directly targeted miR- 381-3p in glioma cells.
Fig. 4: Circ_BRAF sequestered miR-381-3p, (A): Circbank presented binding sequence between circ_BRAF and miR-381-3p; (B and C): Dual-luciferase
reporter assay was implemented for testifying luciferase activities in A172 and U251 cells; (D): miR-381-3p content was detected using
qRT-PCR in cells transfected with circ_BRAF or vector and (E): miR-145-5p expression was revealed using qRT-PCR
Note: *p<0.05, (B and C) ( ): miR-NC; (
): miR-NC; ( ): circ_BRAF, and (D and E) (
): circ_BRAF, and (D and E) ( ): Control and (
): Control and ( ): DEX
): DEX
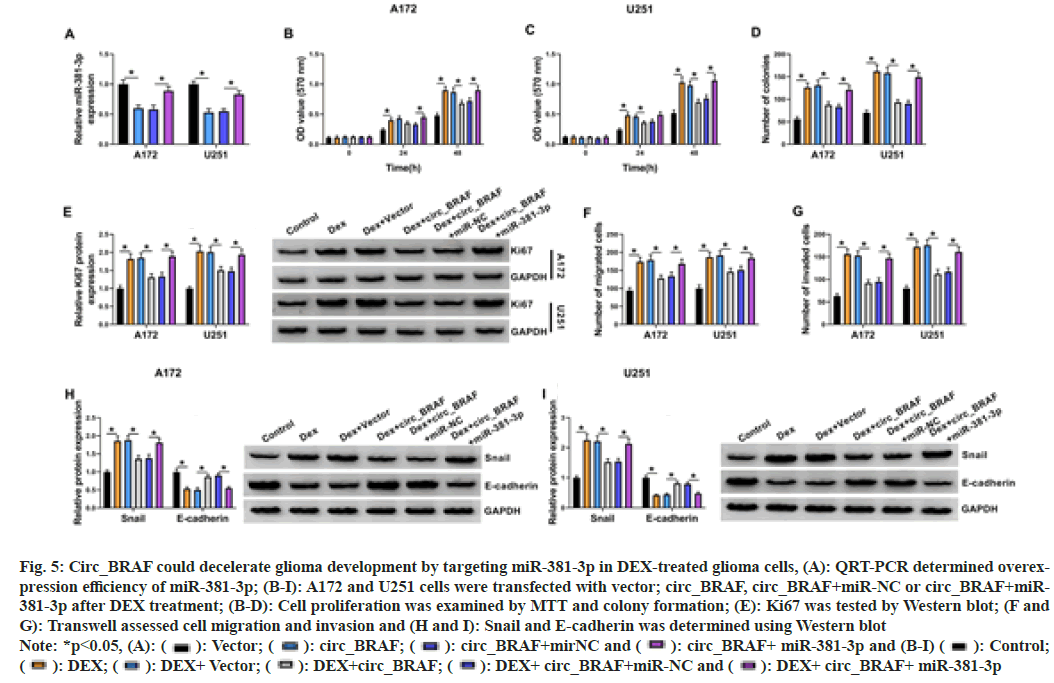
Furthermore, regulatory of circ_BRAF and miR- 381-3p on tumor development was validated. At first, miR-381-3p expression was down regulated in glioma cells via circ_BRAF transfection, while it was elevated by miR-381-3p addition (fig. 5A). Meanwhile, enhanced circ_BRAF restrained DEX-triggered A172 and U251cell proliferation promotion, which was relieved via introduction of miR-381-3p (fig. 5B-fig. 5E). In addition, circ_BRAF overexpression attenuated DEX treatment-induced cell migration and invasion, which were abolished via miR-381- 3p addition (fig. 5F-fig. 5I). Collectively, these data demonstrated that circ_BRAF/miR-381-3p modulated DEX-mediated glioma development.
Fig. 5: Circ_BRAF could decelerate glioma development by targeting miR-381-3p in DEX-treated glioma cells, (A): QRT-PCR determined overexpression
efficiency of miR-381-3p; (B-I): A172 and U251 cells were transfected with vector; circ_BRAF, circ_BRAF+miR-NC or circ_BRAF+miR-
381-3p after DEX treatment; (B-D): Cell proliferation was examined by MTT and colony formation; (E): Ki67 was tested by Western blot; (F and
G): Transwell assessed cell migration and invasion and (H and I): Snail and E-cadherin was determined using Western blot
Note: *p<0.05, (A): ( ): Vector; (
): Vector; ( ): circ_BRAF; (
): circ_BRAF; ( ): circ_BRAF+mirNC and (
): circ_BRAF+mirNC and ( ): circ_BRAF+ miR-381-3p and (B-I) (
): circ_BRAF+ miR-381-3p and (B-I) ( ): Control;
(
): Control;
( ): DEX; (
): DEX; ( ): DEX+ Vector; (
): DEX+ Vector; ( ): DEX+circ_BRAF; (
): DEX+circ_BRAF; ( ): DEX+ circ_BRAF+miR-NC and (
): DEX+ circ_BRAF+miR-NC and ( ): DEX+ circ_BRAF+ miR-381-3p
): DEX+ circ_BRAF+ miR-381-3p
Since the identification of circ_BRAF/miR-381- 3p axis in glioma cells, downstream functional target of miR-381-3p was further predicted and validated. Star base prediction software manifested that THBS2 3’UTR harbored the potential pairing sequence of miR-381-3p (fig. 6A). Dual-luciferase reporter results confirmed combination between THBS2-WT and miR-381- 3p (fig. 6B and fig. 6C). Then, the overexpression or knockdown efficiency of miR-381-3p was certified (fig. 6D). Simultaneously, THBS2 mRNA and protein level was restricted through miR-381-3p mimic and facilitated via miR-381- 3p inhibition (fig. 6E and fig. 6F). Besides, DEX could inhibit THBS2 content in glioma cell lines (fig. 6G and fig. 6H). These analyses explained the targeted relation between miR-381-3p and THBS2.
Fig. 6: MiR-381-3p targeted THBS2 in glioma, (A): Binding sites between miR-381-3p and THBS2 were shown; (B and C): Binding was attested
using dual-luciferase reporter assay; (D): Overexpression or knockdown efficiency of miR-381-3p was verified; (E and F): Effect of miR-381-3p
overexpression or knockdown on THBS2 content was tested using qRT-PCR or Western blot and (G and H): THBS2 content in tumor cells after
DEX treatment were examined using qRT-PCR or Western blot
Note: *p<0.05, (B and C) ( ): miR-NC; (
): miR-NC; (  ): miR-381-3p; (D-F) (
): miR-381-3p; (D-F) ( ): miR-NC; (
): miR-NC; (  ): miR-381-3p; (
): miR-381-3p; ( ): Anti-miR-NC and (
): Anti-miR-NC and ( ): AntimiR-
381-3p and (G and H) (
): AntimiR-
381-3p and (G and H) ( ): Control and (
): Control and ( ): DEX
): DEX
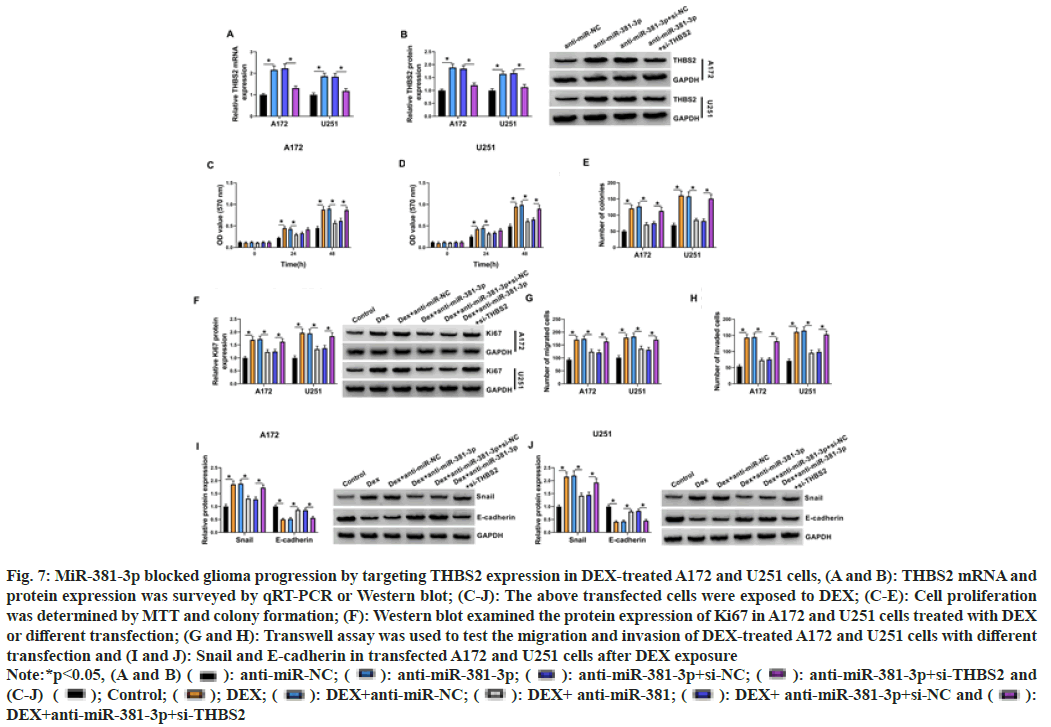
THBS2 content were reinforced by anti-miR- 381-3p, where the enhancements were attenuated in anti-miR-381-3p+si-THBS2 group (fig. 7A and fig. 7B), indicating the high transfection efficiency of anti-miR-381-3p and si-THBS2 in A172 and U251 cells. Importantly, DEX-induced cell proliferation (fig. 7C-fig. 7F) and metastasis (fig. 7G-fig. 7J) promotion were partly assuaged by anti-miR-381-3p transfection in A172 and U251 cells; however, these effects were all further abated by the addition of si-THBS2 (fig. 7C-fig. 7J). These outcomes indicated that the action of miR-381-3p in DEX-treated glioma cells was achieved by the negative modulation of THBS2.
Fig. 7: MiR-381-3p blocked glioma progression by targeting THBS2 expression in DEX-treated A172 and U251 cells, (A and B): THBS2 mRNA and
protein expression was surveyed by qRT-PCR or Western blot; (C-J): The above transfected cells were exposed to DEX; (C-E): Cell proliferation
was determined by MTT and colony formation; (F): Western blot examined the protein expression of Ki67 in A172 and U251 cells treated with DEX
or different transfection; (G and H): Transwell assay was used to test the migration and invasion of DEX-treated A172 and U251 cells with different
transfection and (I and J): Snail and E-cadherin in transfected A172 and U251 cells after DEX exposure
Note:*p<0.05, (A and B) ( ): anti-miR-NC; (
): anti-miR-NC; ( ): anti-miR-381-3p; (
): anti-miR-381-3p; ( ): anti-miR-381-3p+si-NC; (
): anti-miR-381-3p+si-NC; (  ): anti-miR-381-3p+si-THBS2 and
(C-J) (
): anti-miR-381-3p+si-THBS2 and
(C-J) ( ); Control; (
); Control; ( ); DEX; (
); DEX; ( ): DEX+anti-miR-NC; (
): DEX+anti-miR-NC; (  ): DEX+ anti-miR-381; (
): DEX+ anti-miR-381; ( ): DEX+ anti-miR-381-3p+si-NC and (
): DEX+ anti-miR-381-3p+si-NC and (  ):
DEX+anti-miR-381-3p+si-THBS2
):
DEX+anti-miR-381-3p+si-THBS2
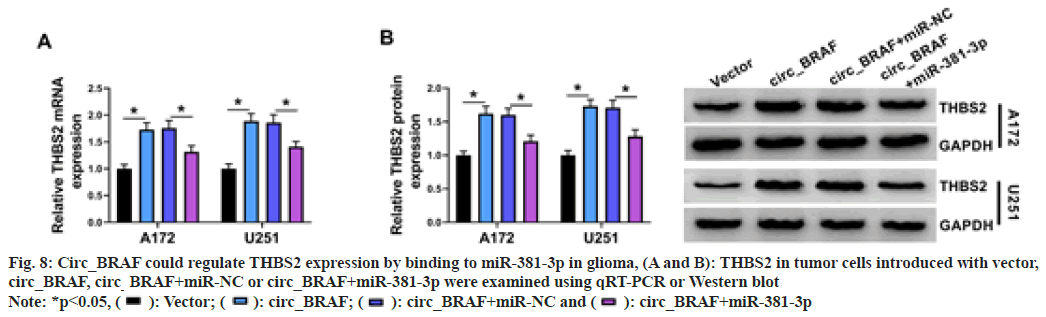
Next, the circ_BRAF/miR-381-3p/THBS2 was examined, tumor cells were introduced with circ_BRAF, circ_BRAF+miR-381-3p, or corresponding controls. Results presented THBS2 content was strengthened via circ_ BRAF overexpression, while the influence was counteracted via miR-381-3p introduction (fig. 8A and fig. 8B). Overall, circ_BRAF interacted with miR-381-3p to increase THBS2 expression.
Fig. 8: Circ_BRAF could regulate THBS2 expression by binding to miR-381-3p in glioma, (A and B): THBS2 in tumor cells introduced with vector,
circ_BRAF, circ_BRAF+miR-NC or circ_BRAF+miR-381-3p were examined using qRT-PCR or Western blot
Note: *p<0.05, ( ): Vector; (
): Vector; (  ): circ_BRAF; (
): circ_BRAF; ( ): circ_BRAF+miR-NC and (
): circ_BRAF+miR-NC and ( ): circ_BRAF+miR-381-3p
): circ_BRAF+miR-381-3p
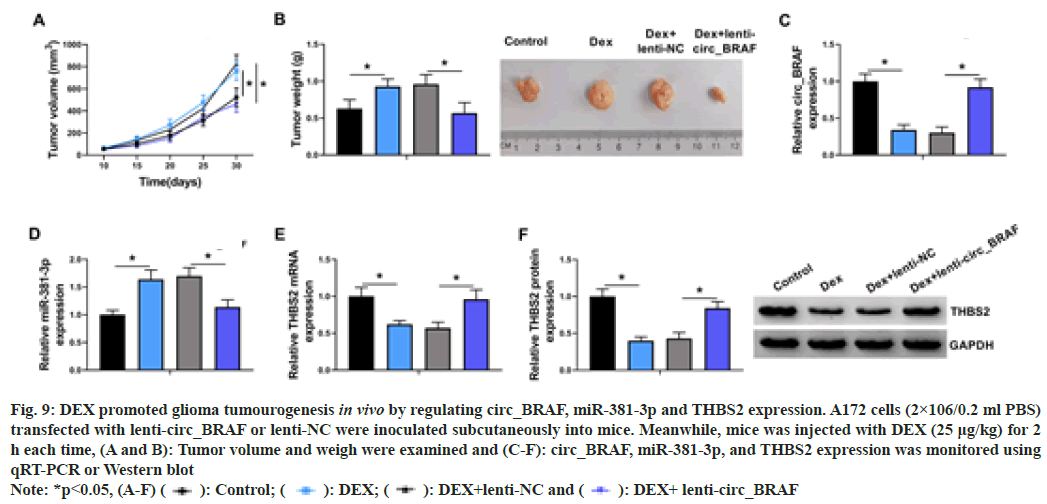
In the animal experiment, DEX exposure could improve tumor volume and weight, while lentivirus-mediated circ_BRAF overexpression (lenti-circ_BRAF) hindered this impact (fig. 9A and fig. 9B). Subsequently, qRT-PCR results revealed that DEX treatment dramatically down regulated circ_BRAF and up regulated miR- 381-3p expression, which were overturned via circ_BRAF introduction (fig. 9C and fig. 9D). Similarly, THBS2 expression was down regulated by DEX treatment and circ_BRAF overexpression partially neutralized the impacts (fig. 9E and fig. 9F). Together, in vivo, DEX contributed to glioma growth through the dependence of circ_BRAF/miR-381-3p/THBS2 axis.
Fig. 9: DEX promoted glioma tumourogenesis in vivo by regulating circ_BRAF, miR-381-3p and THBS2 expression. A172 cells (2×106/0.2 ml PBS)
transfected with lenti-circ_BRAF or lenti-NC were inoculated subcutaneously into mice. Meanwhile, mice was injected with DEX (25 μg/kg) for 2
h each time, (A and B): Tumor volume and weigh were examined and (C-F): circ_BRAF, miR-381-3p, and THBS2 expression was monitored using
qRT-PCR or Western blot
Note: *p<0.05, (A-F) ( ): Control; (
): Control; ( ): DEX; (
): DEX; ( ): DEX+lenti-NC and (
): DEX+lenti-NC and (  ): DEX+ lenti-circ_BRAF
): DEX+ lenti-circ_BRAF
Glioma is intracranial tumor with high mortality and morbidity[23]. Therefore, it is of great importance to seek effective therapies for glioma. There has been plentiful research presenting that circRNAs might exert crucial roles in carcinogenesis of glioma[11]. Herein, the function of circ_BRAF/miR-381-3p/THBS2 axis on DEX-induced glioma cells was investigated.
DEX is an effective anesthetic employed in surgical operation and has been testified to own special impact on neuro protection of brain injury[5]. Meanwhile, DEX played a crucial role on brain protection in glioma patients bearing craniotomy[24]. Nevertheless, the protective effect may be disadvantageous for tumor removing during surgery. DEX facilitated cell proliferation and migration and up regulated anti-apoptotic proteins in lung carcinoma and neuro glioma cells[25]. In addition, DEX had obvious influence on glioma cell proliferation in the presence or absence of cisplatin[8]. Whereas, the underlying mechanisms of DEX in glioma growth remain elusive. In our view, the impacts of DEX on glioma cell development were described. Herein, DEX was also illustrated to facilitate glioma cell proliferation, migration and invasion and hinder circ_BRAF content. More importantly, the underneath regulatory mechanism between circ_ BRAF and DEX exposure on glioma development were expounded in subsequent experiments.
Convincing evidence disclosed numerous circRNAs can serve as novel biomarkers and diagnostic or therapeutic targets for glioma[26,27]. Meanwhile, the abundance of circ_BRAF was digressive in glioma tissues, especially in the tissues of patients with high pathological grade[15]. Similarly, in our study, circ_BRAF was down regulated in DEX-treated glioma cell line. After circ_BRAF overexpression in A172 and U251 cells, we found DEX-triggered cell proliferation, migration and invasion facilitating were all alleviated. These results suggested that circ_BRAF was indeed able to block the growth and metastasis of DEX-induced glioma cells.
Plenty of circRNAs are being found involved in tumor progression through serving as natural sponges for miRNAs[28,29]. Moreover, the circRNA-miRNA-mRNA interaction network has been reported to participate in the tumourogenesis and progression of glioma[30]. For example, circ0082374 could facilitate glioma cell movability by the action on miR-326/SIRT1 signal axis[31]. While circFANCL stimulated the development of glioma by regulating miR-337- 3p/HMGB1 pathway[32]. However, the action mechanism of circ_BRAF/miRNA/mRNA axis in glioma has not been clarified. Here, circ_BRAF directly targeted miR-381-3p. Study has shown that DEX could promote the proliferative and transferred capacities of Pheochromocytoma 12 (PC12) cells by inhibiting Leucine-Rich Repeats Containing 4 (LRRC4) via enhancing miR-381 expression[33]. Similarly, in this study, DEX markedly facilitated miR-381-3p content. Also, miR-381-3p overexpression promoted glioma progression to restore the inhibition effects on cell progression induced by circ_BRAF in DEXtreated glioma cells. In other words, circ_BRAF restrained glioma development through binding with miR-381-3p.
Many researches have demonstrated that miR-381- 3p is involved in cancer progression and serves as an anticancer or oncogenic miRNA through targeting different mRNAs in multifarious cancers, including cervical cancer[34], prostate cancer[35] and renal cell carcinoma[36]. Meanwhile, miR-381 has also shown to participate in multiple biological processes of glioma via regulating gene expression. For instance, miR- 381 could act as a possible therapeutic target for glioma remedy and its deletion hindered glioma growth by sponging LRRC4[22]. Tang et al.[37] also certified the involvement of the interaction between miR-381 and LRRC4 in the pathogenic mechanism of glioma. Here, binding between miR-381-3p and THBS2 was revealed in glioma cells. Meanwhile, DEX reduced THBS2 level in A172 and U251 cells. Therefore, we speculated that there was a correlation between DEX/circ_ BRAF/miR-381-3p axis and THBS2.
The effects of THBS2 in the advancement of various cancers have been illustrated by functioning as the targets of miRNAs. For example, THBS2 was decreased in cervical cancer tissues and diminished tumor metastasis[38]. Circ_0020123 promoted NSCLC cell growth through up regulating THBS2[39]. THBS2 was also reported to be a crucial gene that modulated by CEBPB in glioma cells[40]. Meanwhile, THBS2 was a target of miR-9 and was related to miR-9-induced malignant behaviors in glioma cells[20]. Herein, THBS2 knockdown reversed the inhibiting action of miR-381-3p silence on glioma development under DEX exposure cells. Furthermore, THBS2 was positively modulated by circ_BRAF via sponging miR-381-3p. Moreover, DEX promoted glioma growth through reducing circ_BRAF and THBS2 expression and augmenting miR-381-3p level in vivo. Therefore, these findings together elaborated that DEX could accelerate the carcinogenesis of glioma via circ_BRAF/miR-381-3p/THBS2 axis.
In conclusion, DEX treatment down regulated circ_BRAF and THBS2, while up regulated miR- 381-3p expression in A172 and U251 cells. DEX exposure suppressed cell proliferation, migration and invasion in glioma cells. Furthermore, circ_ BRAF overexpression mitigated the facilitating influence of DEX on glioma progression by miR-381-3p/THBS2 axis, providing a new sight for glioma therapy.
Author Contributions:
Lisong Cai and Zheng Yajing have contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Ferris SP, Hofmann JW, Solomon DA, Perry A. Characterization of gliomas: From morphology to molecules. Virchows Arch 2017;471(2):257-69.
[Crossref] [Google Scholar] [PubMed]
- Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol 2017;14(7):434-52.
[Crossref] [Google Scholar] [PubMed]
- Cahill D, Turcan S. Origin of gliomas. Semin Neurol 2018;38(1):5-10.
[Crossref] [Google Scholar] [PubMed]
- Constantin JM, Momon A, Mantz J, Payen JF, de Jonghe B, Perbet S, et al. Efficacy and safety of sedation with dexmedetomidine in critical care patients: A meta-analysis of randomized controlled trials. Anaesth Crit Care Pain Med 2016;35(1):7-15.
[Crossref] [Google Scholar] [PubMed]
- Keating GM. Dexmedetomidine: A review of its use for sedation in the intensive care setting. Drugs 2015;75(10):1119-30.
[Crossref] [Google Scholar] [PubMed]
- Chi M, Shi X, Huo X, Wu X, Zhang P, Wang G. Dexmedetomidine promotes breast cancer cell migration through Rab11-mediated secretion of exosomal TMPRSS2. Ann Transl Med 2020;8(8):531.
- Xia M, Ji NN, Duan ML, Tong JH, Xu JG, Zhang YM, et al. Dexmedetomidine regulates the malignancy of breast cancer cells by activating α2-adrenoceptor/ERK signaling pathway. Eur Rev Med Pharmacol Sci 2016;20(16):3500-6.
[Google Scholar] [PubMed]
- Yang H, Chen Y, Yan H, Wu H. Effects of dexmedetomidine on glioma cells in the presence or absence of cisplatin. J Cell Biochem 2020;121(1):723-34.
[Crossref] [Google Scholar] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LV, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genetics 2019;20(11):675-91.
[Crossref] [Google Scholar] [PubMed]
- Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365(2):141-8.
[Crossref] [Google Scholar] [PubMed]
- Hao Z, Hu S, Liu Z, Song W, Zhao Y, Li M. Circular RNAs: Functions and prospects in glioma. J Mol Neurosci 2019;67(1):72-81.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Zhao K, Huang N, Zhang N. Circular RNAs and human glioma. Cancer Biol Med 2019;16(1):11-23.
- Zuo CY, Qian W, Huang CJ, Lu J. Circular RNA circ-SMAD7 promoted glioma cell proliferation and metastasis by up regulating PCNA. Eur Rev Med Pharmacol Sci 2020;24(14):7542.
[Crossref] [Google Scholar] [PubMed]
- Yin K, Liu X. CircMMP1 promotes the progression of glioma through miR-433/HMGB3 axis in vitro and in vivo. IUBMB Life 2020;72(11):2508-24.
[Crossref] [Google Scholar] [PubMed]
- Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang H, et al. Differential expression of circular RNAs in glioblastoma multiforme and its correlation with prognosis. Transl Oncol 2017;10(2):271-9.
[Crossref] [Google Scholar] [PubMed]
- Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 2019;11(1):1-24.
[Crossref] [Google Scholar] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141(4):1202-7.
[Crossref] [Google Scholar] [PubMed]
- Zhong Z, Zhou F, Wang D, Wu M, Zhou W, Zou Y, et al. Expression of KLF9 in pancreatic cancer and its effects on the invasion, migration, apoptosis, cell cycle distribution and proliferation of pancreatic cancer cell lines. Oncol Rep 2018;40(6):3852-60.
[Crossref] [Google Scholar] [PubMed]
- Tong XD, Liu TQ, Wang GB, Zhang CL, Liu HX. MicroRNA-570 promotes lung carcinoma proliferation through targeting tumor suppressor KLF9. Int J Clin Exp Pathol 2015;8(3):2829-34.
[Google Scholar] [PubMed]
- Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y, et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res 2019;38(1):99.
- Xu Y. MicroRNA-136-3p inhibits glioma tumorigenesis in vitro and in vivo by targeting KLF7. World J Surg Oncol 2020;18(1):169.
- Tang H, Wang Z, Liu Q, Liu X, Wu M, Li G. Disturbing miR-182 and-381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PloS One 2014;9(1):e84146.
[Crossref] [Google Scholar] [PubMed]
- Davis ME. Epidemiology and overview of gliomas. Semin Oncol Nurs 2018;34(5):420-9.
[Crossref] [Google Scholar] [PubMed]
- Luo X, Zheng X, Huang H. Protective effects of dexmedetomidine on brain function of glioma patients undergoing craniotomy resection and its underlying mechanism. Clin Neurol Neurosurg 2016;146:105-8.
[Crossref] [Google Scholar] [PubMed]
- Wang C, Datoo T, Zhao H, Wu L, Date A, Jiang C, et al. Midazolam and dexmedetomidine affect neuroglioma and lung carcinoma cell biology in vitro and in vivo. Anesthesiology 2018;129(5):1000-14.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Lin X, Geng X, Shi L, Li Q, Liu F, et al. Advances in circular RNAs and their role in glioma. Int J Oncol 2020;57(1):67-79.
[Crossref] [Google Scholar] [PubMed]
- Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer 2020;19(1):34.
[Crossref] [Google Scholar] [PubMed]
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 2017;14(5):514-21.
[Crossref] [Google Scholar] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495(7441):384-8.
[Crossref] [Google Scholar] [PubMed]
- Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu J, Maling G, et al. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol 2018;137(3):493-502.
[Crossref] [Google Scholar] [PubMed]
- Wang B, Li B, Si T. Knockdown of circ0082374 inhibits cell viability, migration, invasion and glycolysis in glioma cells by miR-326/SIRT1. Brain Res 2020;1748:147108.
[Crossref] [Google Scholar] [PubMed]
- Tao W, Jia Z, Mengshi W, Wei L, Feng L. Circular RNA circFANCL motivates the glioma progression via the action on the miR-337-3p/HMGB1 signal axis. Minerva Med; 2020.
[Crossref] [Google Scholar] [PubMed]
- Xue Y, Xu T, Jiang W. Dexmedetomidine protects PC12 cells from ropivacaine injury through miR-381/LRRC4/SDF-1/CXCR4 signaling pathway. Regen Ther 2020;14:322-9.
[Crossref] [Google Scholar] [PubMed]
- Shang A, Zhou C, Bian G, Chen W, Lu W, Wang W, et al. miR-381-3p restrains cervical cancer progression by down regulating FGF7. J Cell Biochem 2019;120(1):778-89.
[Crossref] [Google Scholar] [PubMed]
- Hu J, Wu X, Yang C, Rashid K, Ma C, Hu M, et al. Anticancer effect of icaritin on prostate cancer via regulating miR-381-3p and its target gene UBE2C. Cancer Med 2019;8(18):7833-45.
[Crossref] [Google Scholar] [PubMed]
- Zhao C, Zhou Y, Ran Q, Yao Y, Zhang H, Ju J, et al. MicroRNA-381-3p functions as a dual suppressor of apoptosis and necroptosis and promotes proliferation of renal cancer cells. Front Cell Dev Biol 2020;8:290.
[Crossref] [Google Scholar] [PubMed]
- Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res 2011;1390:21-32.
[Crossref] [Google Scholar] [PubMed]
- Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen XJ, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis 2017;8(12):3220.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Zhao L, Wang Y. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by sponging miR-590-5p to regulate THBS2. Cancer Cell Int 2020;20(1):387.
[Crossref] [Google Scholar] [PubMed]
- Du C, Pan P, Jiang Y, Zhang Q, Bao J, Liu C. Microarray data analysis to identify crucial genes regulated by CEBPB in human SNB19 glioma cells. World J Surg Oncol 2016;14(1):258.
[Crossref] [Google Scholar] [PubMed]