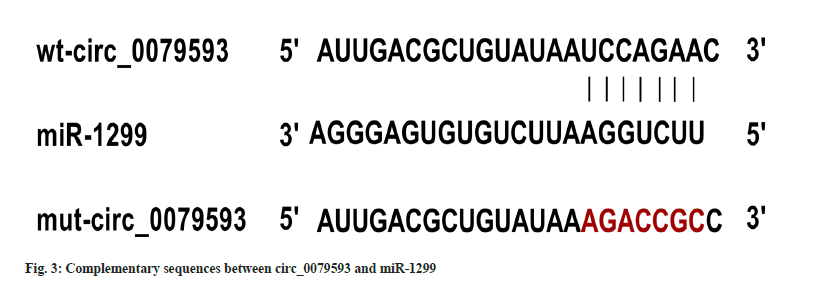- *Corresponding Author:
- Ren Liu
Department of Nursing, Chongqing Nursing Vocational College, Chongqing 404100, China
E-mail: m15923026913_1@163.com
| Date of Received | 13 March 2023 |
| Date of Revision | 05 October 2023 |
| Date of Acceptance | 10 February 2024 |
| Indian J Pharm Sci 2024;86(1):202-210 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the effects of croton alkaloids on malignant behaviors of breast cancer cells T47D and the possible mechanism. Different doses of croton alkaloids were exposed to T47D cells. T47D cells were transfected with small interfering-circular_0079593 and small interfering-negative control by liposome transfection, respectively. The plasmid cloning deoxyribonucleic acid-circular_0079593 transfected T47D cells were performed with 150 μg/ml croton alkaloids treatment. Circular_0079593 and microRNA-1299 were determined via qualitative reverse transcription polymerase chain reaction in breast cancer samples and cells. 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide assay and colony formation assay for proliferation, scratch assay for migration and transwell assay for invasion were performed. Circular_0079593 and microRNA-1299 target relationship was examined using dual-luciferase reporter assay. Protein analysis was implemented through Western blot. Croton alkaloids increased cell proliferation inhibition rate and E-cadherin protein level (p<0.05) while reduced colony formation number and invasive cells (p<0.05), and croton alkaloids suppressed scratch healing rate and N-cadherin protein level (p<0.05), and these effects were dose-dependent. Circular_0079593 up-regulation and microRNA-1299 down-regulation were detected in breast cancer tissues contrasted to adjacent samples (p<0.05). Croton alkaloids induced circular_0079593 expression up-regulation (p<0.05) and microRNA-1299 level inhibition (p<0.05) in a dose-dependent manner. Circular_0079593 could targeted regulate the expression of microRNA-1299. Transfection of small interfering-circular_0079593 elevated microRNA-1299 level, and suppressed cell malignant behaviors. Circular_0079593 overexpression could weaken the regulation of croton alkaloids in T47D cell behaviors. Croton alkaloids could repress breast cancer cell proliferation, colony formation, migration and invasion via mediating circ_0079593/microRNA-1299 axis.
Keywords
Breast cancer, croton alkaloids, circ_0079593, microRNA-1299, cell proliferation, migration, invasion
Breast Cancer (BC) is a commonly diagnosed tumor with high occurrence rate in female, and the age distribution is younger in recent years[1]. With the advances of therapeutic technologies, survival prognosis has signally improved in BC patients[2]. However, some patients still have high recurrence and metastasis to result in poor treatment effect[3,4]. Hence, it is of great importance to discover safe and effective medicines in BC treatment. Some active ingredients isolated from plants appear anti-tumor function and can be used for BC treatment[5,6]. Croton Alkaloids (CA) is extracted from croton in the Euphorbia family, and exerts inhibiting effect on cell development in colorectal cancer and ovarian cancer[7,8]. Nevertheless, the function of CA in BC has not been fully studied.
Circular RNA (circRNA) is a closed non-coding RNA (ncRNA) molecule with tissue specificity and stability, and possesses the characteristic as microRNA (miR)-sponge[9,10]. circRNA exhibits aberrant expression in tumors and vital molecular function in accelerating or suppressing biological processes of tumor cells[11]. circ_0038632 was identified as a promoter of BC through interacting with miR-520a-3p[12]. circNCOR1 affected radiotherapy efficacy in BC cells via miR-638 binding[13]. Yang et al.[14] have shown that circ_0079593 overexpression promoted cell metastasis and proliferation of glioma. Level inhibition of miR-1299 was discovered in BC and miR-1299 up-regulation reduced BC cell growth[15]. However, the relation of circ_0079593/ miR-1299 in BC has not been clarified.
This study concentrated on the influences of CA on proliferation and motility of BC cells. Moreover, circ_0079593 was hypothesized to have a target relation with miR-1299. The regulatory mechanism of CA with circ_0079593/miR-1299 was elucidated.
Materials and Methods
Materials and reagents:
BC and adjacent tissues (n=31 of each group) were collected after surgical resection in our hospital from May 2019 to July 2020, then immediately preserved at -80°. The age of BC patients was 50- 68 (56.35±4.11) y old. Patients or close relatives have written informed consent forms, and this study was following the Declaration of Helsinki of the world medical association. CA (purity ≥99 %) was purchased from Google Biological Chemical Co., Ltd. (Wuhan, China). Human BC cell line T47D was purchased from Kebai Biology (Nanjing, China). Dulbecco's Modified Eagle Medium (DMEM) and Fetal Bovine Serum (FBS) were purchased from Gibco (United States of America (USA)). Trizol reagent, Lipofectamine2000 and circ_0079593 overexpression vector (plasmid cloning Deoxyribonucleic Acid (pcDNA)- circ_0079593) were provided by Invitrogen (USA). The complementary DNA (cDNA) synthesis and quantitative Reverse Transcription- Polymerase Chain Reaction (qRT-PCR) kits were provided by Thermo Fisher (USA). The oligonucleotides contained miR-1299 mimic, mimic NC, si-circ_0079593 and si-NC (Ribobio, Guangzhou, China). 3-(4,5-Dimethylthiazol-2-yl)- 2,5-Diphenyltetrazolium Bromide (MTT) reagent and matrigel were bought from Solarbio (Beijing). Transwell chamber was purchased from Corning (USA). Rabbit antibody for E-cadherin, N-cadherin and goat anti-Rabbit secondary antibody were obtained from CST (USA).
Grouping:
After cell culture (3×103 cells/well) in 96-well plates overnight, T47D cells were disposed to increasing concentrations of CA for 24 h[16], which was recorded as CA 50 μg/ml group, CA 100 μg/ ml group and CA 150 μg/ml group, respectively. T47D cell line with normal culture was recorded as control group. Lipofectamine2000 was used for si-NC and si-circ_0079593 transfection according to the instruction book, which was denoted as sicirc_ 0079593 group and si-NC group. T47D cells with transfection of pcDNA-circ_0079593 were disposed with 150 μg/ml CA for 24 h, which was labeled as CA+pcDNA-circ_0079593 group.
RT-qPCR for circ_0079593 and miR-1299 quantification:
Total Ribonucleic Acid (RNA) was isolated employing Trizol reagent, and concentration detection was performed under an ultraviolet spectrophotometer. The reverse transcription into cDNA was administrated through cDNA synthesis kit, followed by PCR amplification with cDNA as the template. The reaction system was as follows; 10 μl SYBR Green Master Mix, 1 μl cDNA, 0.8 μl forward and reverse primer, and system was supplemented by double distilled Water (ddH2O) to 20 μl. Reaction procedures included Predenaturation (95°, 2 min), denaturation (95°, 30 s), annealing (60°, 30 s), and extension (72°, 30 s) with 40 cycles. The relative expression was examined through ABI StepOnePlus fluorescence qPCR instrument.
MTT assay for cell proliferation:
The 96-well plates were inoculated with T47D cells with 3×103 cells/well, then 20 μl/well MTT incubation was administrated at 37° for 4 h. Then the supernatant was discarded after cell centrifugation (3000 r/min, 5 min), followed by lucifugal incubation of 150 μl/well Dimethyl Sulfoxide (DMSO) for 5 min. OD value was examined via microplate reader and proliferation inhibition rate was calculated as follows:
(Optical Density (OD)control group-ODexperimental group)/ (ODcontrol group-ODblank group)×100 %
Colony formation assay for cell growth:
500 T47D cells of each well in 6-well plates were cultivated for 2 w. Subsequently, culture medium needed to be changed every 2 d. The fastened (4 % paraformaldehyde, 30 min) and dyed (1 % crystal violet, 20 min) colonies were washed in Phosphate Buffer Solution (PBS) and counted.
Scratch assay for cell migration:
1×104 T47D cells/well in 6-well plates were cultured to 100 % confluence, followed by producing two straight scratches using a sterile pipette tip. PBS was added for cell washing and cell culture was performed in serum-free DMEM for 24 h. Following detection of scratch width, scratch healing rate was calculated.
Scratch healing rate=(scratch width0 h-scratch width24 h)/scratch width0 h×100 %
Transwell assay for invasion examination:
The top chamber was coated with the diluted matrigel in pre-cooled medium for 4 h. Respectively, the top and bottom chambers were pipetted with 200 μl T47D cells (2.5×105 cells/ ml) and 600 μl DMEM+10 % FBS medium. Transwell chamber was incubated for 24 h, then the transmembrane cells were administrated with fixation (paraformaldehyde, 20 min) and staining (crystal violet, 10 min). Eventually, invasive cells were numbered.
Dual-luciferase reporter assay for circ_0079593 and miR-1299 target detection:
Circular RNA interactive was employed for predicting circ_0079593 and miR-1299 binding region. The complementary sequence was mutated by gene mutation technique. The recombinant vectors wt-circ_0079593 and mut-circ_0079593 were obtained by cloning the complementary sequence and mutant sequence into pmirGLO vector, respectively. miR-1299 mimic or miRNC and wt-circ_0079593 or mut-circ_0079593 were co-transfected into T47D cells by liposome transfection. 48 h later, cells were gathered for relative luciferase detection.
Western blot for protein analysis:
Radioimmunoprecipitation Assay (RIPA) lysis solution was utilized to extract total proteins, and concentration was examined via Bicinchoninic Acid (BCA) assay method. Protein sample was isolated through Sodium Dodecyl Sulphate- Polyacrylamide Gel Electrophoresis (SDS-PAGE), followed by transferring membranes and blocking for 2 h. The primary antibody for E-cadherin (1:800), N-cadherin (1:800) or Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) (1:1000) was incubated for 24 h at 4°. After incubation with the secondary antibody (1:3000) at 25° for 1 h, gray values of bands were analyzed by ImageJ software after Enhanced Chemiluminescence (ECL) detection.
Statistical analysis:
Data were indicated as x±s in accordance with normal distribution, and Statistical Package for the Social Sciences (SPSS) 21.0 was utilized for data analysis. Independent sample t test was employed for comparison between two groups, and comparison among multiple groups was implemented via one-way Analysis of Variance (ANOVA). Statistically, the significant difference was represented as p<0.05.
Results and Discussion
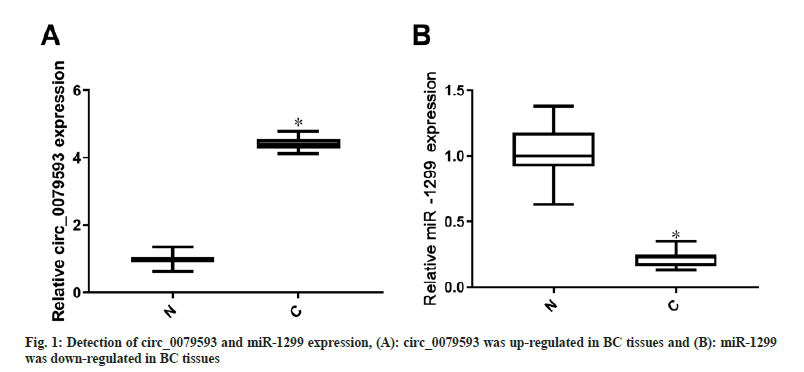
Circ_0079593 expression was elevated (p<0.05) but miR-1299 level was lessened (p<0.05) in BC samples contrasted to adjacent samples, as shown in fig. 1.
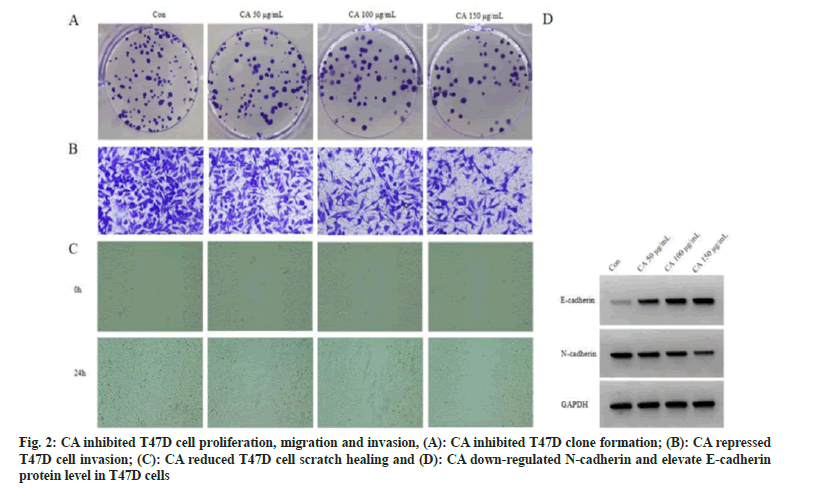
Relative to control group, cell proliferation inhibition rate was gradually increased in three CA groups (p<0.05). Cell colonies, invaded cells and scratch healing rate of three CA treatment groups were reduced in a dose-independent way (p<0.05). As the results of CA treatment, E-cadherin was enhanced (p<0.05) while N-cadherin protein level was lessened (p<0.05). Difference among different dose groups was statistically significant (p<0.05), as displayed in fig. 2 and Table 1.
| Group | Inhibition rate (%) | Colony formation (number) | Scratch healing rate (%) | Invaded cells (number) | E-cadherin | N-cadherin |
|---|---|---|---|---|---|---|
| Control | 0.00±0.00 | 116.67±3.30 | 70.67±3.97 | 170.67±5.44 | 0.17±0.01 | 0.69±0.04 |
| CA 50 μg/ml | 14.45±0.711) | 100.67±3.091) | 60.92±2.791) | 148.00±5.101) | 0.33±0.021) | 0.55±0.031) |
| CA 100 μg/ml | 38.08±1.251)2) | 78.33±2.051)2) | 44.86±1.431)2) | 109.67±3.861)2) | 0.56±0.041)2) | 0.38±0.021)2) |
| CA 150 μg/ml | 55.19±2.771)2)3) | 54.00±0.821)2)3) | 32.56±0.731)2)3) | 74.33±1.251)2)3) | 0.80±0.041)2)3) | 0.21±0.021)2)3) |
| F | 746.868 | 352.491 | 131.192 | 300.583 | 244.865 | 157.424 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Contrasted to control group, 1)p<0.05; contrasted to CA 50 μg/ml group, 2)p<0.05 and contrasted to CA 100 μg/ml group, 3)p<0.05
Table 1: CA Inhibited T47d cell growth and motility (X±S, N=9)
Contrasted to control group, circ_0079593 level was reduced (p<0.05) and miR-1299 was upregulated (p<0.05) in three CA treatment groups. The statistical difference was found in different groups (p<0.05), as exhibited in Table 2.
| Group | circ_0079593 | miR-1299 |
|---|---|---|
| Control | 1.00±0.00 | 1.00±0.00 |
| CA 50 μg/ml | 0.88±0.021) | 1.53±0.061) |
| CA 100 μg/ml | 0.57±0.041)2) | 2.47±0.081)2) |
| CA 150 μg/ml | 0.29±0.031)2)3) | 3.67±0.101)2)3) |
| F | 422.759 | 823.695 |
| p | 0.000 | 0.000 |
Note: Relative to control group, 1)p<0.05; relative to CA 50 μg/ml group, 2)p<0.05 and relative to CA 100 μg/ml group, 3)p<0.05
Table 2: CA down-regulated Circ_0079593 and up-regulated miR-1299 (X±S, N=9)
Circ_0079593 and miR-1299 binding region by circular RNA interactive was revealed in fig. 3. In cell experiment with co-transfection of wt-circ_0079593 vector, the relative luciferase activity of miR-1299 group was suppressed by comparison to miR-NC transfection (p<0.05). No statistical difference of luciferase activity was observed in miR-1299 and miR-NC groups with co-transfection of mut-circ_0079593, as shown in Table 3.
| Group | wt-circ_0079593 | mut-circ_0079593 |
|---|---|---|
| miR-NC | 0.96±0.08 | 1.01±0.09 |
| miR-1299 | 0.31±0.021) | 0.95±0.08 |
| t | 13.653 | 0.863 |
| p | 0.000 | 0.437 |
Note: Contrasted to miR-NC group, 1)p<0.05
Table 3: Detection of relative luciferase activity (X±S, N=9)
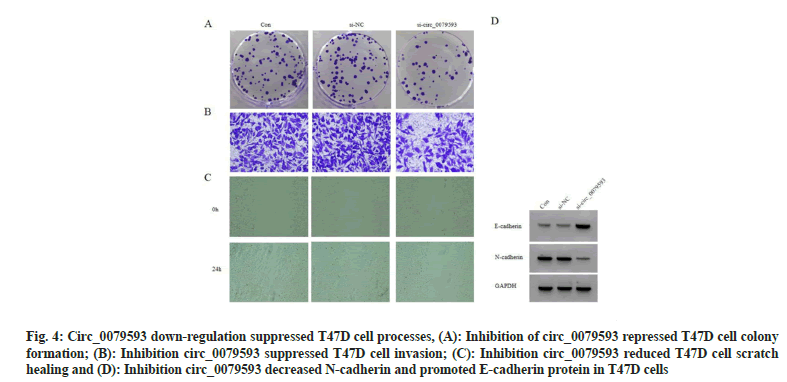
In contrast with control group and si-NC group, miR-1299 level in si-circ_0079593 group was increased (p<0.05). Knocking down circ_0079593 increased cell proliferation inhibition rate (p<0.05), while colony formation, invasive cells and scratch healing rate were suppressed (p<0.05). circ_0079593 inhibition resulted in E-cadherin protein up-regulation (p<0.05) and N-cadherin protein reduction (p<0.05), as exhibited in fig. 4 and Table 4.
Fig. 4: Circ_0079593 down-regulation suppressed T47D cell processes, (A): Inhibition of circ_0079593 repressed T47D cell colony formation; (B): Inhibition circ_0079593 suppressed T47D cell invasion; (C): Inhibition circ_0079593 reduced T47D cell scratch healing and (D): Inhibition circ_0079593 decreased N-cadherin and promoted E-cadherin protein in T47D cells
| Group | circ_0079593 | miR-1299 | Inhibition rate (%) | Colony formation (number) | Scratch healing rate (%) | Invaded cells (number) | E-cadherin | N-cadherin |
|---|---|---|---|---|---|---|---|---|
| Control | 1.00±0.00 | 1.00±0.00 | 0.00±0.00 | 116.33±4.50 | 71.30±3.38 | 170.00±6.16 | 0.17±0.01 | 0.69±0.05 |
| si-NC | 0.99±0.03 | 1.02±0.02 | 0.01±0.01 | 118.00±4.32 | 70.77±4.23 | 172.00±6.48 | 0.16±0.02 | 0.68±0.05 |
| si-circ_0079593 | 0.18±0.021)2) | 4.33±0.061)2) | 63.96±1.271)2) | 45.33±0.471)2) | 25.41±0.531)2) | 63.33±1.251)2) | 0.93±0.041)2) | 0.13±0.011)2) |
| F | 1533 | 25437 | 7607.39 | 395.752 | 211.011 | 426.848 | 836.143 | 181.235 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Contrasted to control group, 1)p<0.05 and relative to si-NC group, 2)p<0.05
Table 4: Knockdown of Circ_0079593 inhibited T47d cell processes (X±S, N=9)
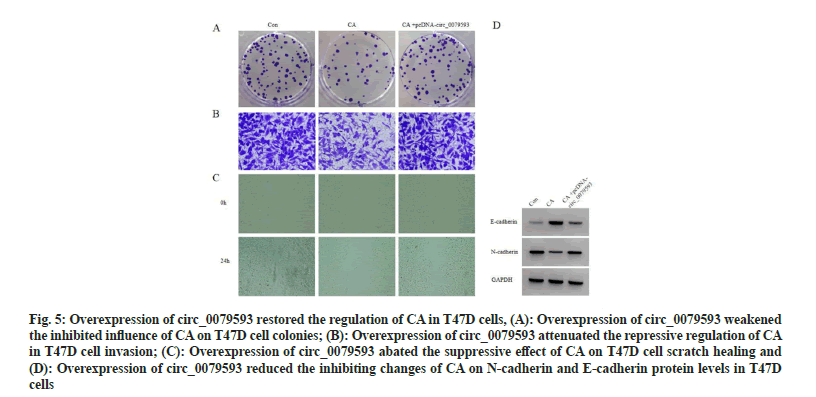
Compared with CA group, miR-1299 level down-regulation was detected in CA+pcDNAcirc_ 0079593 group (p<0.05). Overexpression of circ_0079593 evoked reduction of cell proliferation inhibition rate (p<0.05), as well as the promotion of cell colonies, invaded cells and scratch healing rate (p<0.05). E-cadherin protein level was inhibited (p<0.05) while N-cadherin was elevated (p<0.05) with circ_0079593 upregulation, as exhibited in fig. 5 and Table 5.
Fig. 5: Overexpression of circ_0079593 restored the regulation of CA in T47D cells, (A): Overexpression of circ_0079593 weakened the inhibited influence of CA on T47D cell colonies; (B): Overexpression of circ_0079593 attenuated the repressive regulation of CA in T47D cell invasion; (C): Overexpression of circ_0079593 abated the suppressive effect of CA on T47D cell scratch healing and (D): Overexpression of circ_0079593 reduced the inhibiting changes of CA on N-cadherin and E-cadherin protein levels in T47D cells
| Group | circ_0079593 | miR-1299 | Inhibition rate (%) | Colony formation (number) | Scratch healing rate (%) | Invaded cells (number) | E-cadherin | N-cadherin |
|---|---|---|---|---|---|---|---|---|
| Control | 1.00±0.00 | 1.00±0.00 | 0.00±0.00 | 117.00±4.55 | 71.21±4.17 | 170.00±6.16 | 0.17±0.02 | 0.70±0.05 |
| CA | 0.29±0.031) | 3.68±0.101) | 55.78±3.211) | 54.67±0.471) | 32.80±1.041) | 74.67±2.491) | 0.79±0.051) | 0.21±0.021) |
| CA +pcDNA-circ_0079593 | 0.92±0.052) | 1.27±0.062) | 8.04±0.372) | 106.33±3.682) | 65.10±2.892) | 163.33±4.112) | 0.24±0.022) | 0.62±0.042) |
| F | 400.324 | 1440.82 | 783.712 | 290.185 | 142.936 | 417.599 | 314.455 | 138.2 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Contrasted with control group, 1)p< 0.05 and relative to CA group, 2)p<0.05
Table 5: Overexpression of Circ_0079593 restored the regulation of Ca in T47d cells (X±S, N=9)
Active ingredients extracted from plants can play anti-cancer roles in BC through multiple targets and pathways. For example, curcumin repressed migration and induced apoptosis in colorectal cancer cells by modulating circHN1[17]. Astragaloside IV restrained cell growth and metastasis in BC via up-regulating long noncoding RNA (lncRNA) Thyrotropin Releasing Hormone Degrading Enzyme-Antisense RNA 1 (TRHDEAS1)[ 18]. circRNA with dysregulation in BC can induce the negative regulation of miRNA by serving as a miRNA sponge to participate in BC development[19,20]. However, whether circRNA can act as a potential target in BC treatment with traditional Chinese medicine or active ingredients of plants needs further research.
CA was reported to reduce cell growth and induce cell apoptosis of lung cancer[21]. Cell proliferation and apoptosis-related proteins were regulated by CA in gastric cancer[22]. Also, CA treatment resulted in liver cancer cell growth inhibition and apoptosis acceleration[23]. It is unknown about the effects of CA on BC cell malignant behaviors. The results during this study attested that proliferation inhibition rate was elevated and colony formation number was decreased with the concentration increase of CA, suggesting that CA could inhibit BC cell proliferation. Epithelial- Mesenchymal Transformation (EMT) is an essential biological process with transformation from polar epithelial cells to mesenchymal cells under certain conditions, consequently leading to cell migration and invasion[24]. E-cadherin (a biomarker for epithelial cells) and N-cadherin (a biomarker for mesenchymal cells) dysregulation can evoke EMT occurrence to partake in cell metastasis[25]. Herein, scratch healing rate and invaded cells were hindered by increasing doses of CA. CA induced E-cadherin up-regulation and N-cadherin down-regulation, which certified that CA suppressed EMT process and BC cell motility. In a dose-dependent pattern, CA was implicated in the regulation of BC cellular behaviors.
The published studies indicated that circ_0079593 functioned as a pivotal molecule in cancer regulation via miRNA sponging effect. circ_0079593 exhibited high expression in melanoma, and promoted proliferation as well as invasion[26]. The oncogenic function of circ_0079593 was associated with miR-516b/ GRM3 or miR-433/EGFR axis[27,28]. circ_0079593 was discovered to facilitate glioma cell motility and angiogenesis through sponging miR-324-5p to elevate XBP1 level[29]. In the current study, aberrant up-regulation of circ_0079593 in BC samples hinted that circ_0079593 might have vital regulation in the pathological processes of BC. The further evidences affirmed that circ_0079593 interacted with miR-1299 and negatively regulated miR-1299 level. Anti-tumor role of miR-1299 has been elucidated in human cancer research. By acting as a target of circ_0006404, miR-1299 repressed cell viability and metastasis in prostate cancer[30]. circ_0058608 contributed to lung cancer cell malignant progression and chemo resistance via absorbing miR-1299[31]. Also, miR-1299 was down-regulated in BC and up-regulated miR-1299 expression inhibited BC progression[32]. Also, circ_0001925 expedited cell processes in BC through miR-1299 sponging mechanism to mediate YY1[33]. Consistently, our expression detection suggested that miR-1299 was down-regulated in BC tissues. CA could reduce circ_0079593 level and enhance miR-1299 expression in BC. Moreover, circ_0079593 expression inhibition impeded BC cell progression and up-regulated miR-1299. Overexpression of circ_0079593 attenuated the cancer-inhibitory influences of CA on BC cells and sequestered miR-1299 level. It was suggested that CA suppressed cell malignant behaviors through down-regulating the expression of circ_0079593 to induce miR-1299 up-regulation.
In conclusion, CA exhibited anti-tumor regulation in BC cell proliferation, migration and invasion. Additionally, CA function was achieved by circ_0079593/miR-1299 sponging mechanism. circ_0079593/miR-1299 axis could participate in BC development, as a possible therapeutic target for CA in BC. However, this research remains some limitations. The downstream targets for circ_0079593/miR-1299 still need further exploration.
Authors contribution:
Li Yanjiang and Pei Liu have contributed equally
to this study.
Funding:
This work was supported by Chongqing Research Institution Performance Incentive Guidance Special Project (cstc2020jxjl130005).
Conflict of interests:
The authors declared no conflict of interests.
References
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71(1):7-33.
[Crossref] [Google Scholar] [PubMed]
- Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72(5):409-36.
[Crossref] [Google Scholar] [PubMed]
- Jin J, Qiu S, Wang P, Liang X, Huang F, Wu H, et al. Cardamonin inhibits breast cancer growth by repressing HIF-1α-dependent metabolic reprogramming. J Exp Clin Cancer Res 2019;38(1):377.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Zhao W, Wang W, Lin S, Yang L. Puerarin suppresses LPS-induced breast cancer cell migration, invasion and adhesion by blockage NF-κB and Erk pathway. Biomed Pharmacother 2017;92:429-36.
[Crossref] [Google Scholar] [PubMed]
- Wang WD, Shang Y, Li Y, Chen SZ. Honokiol inhibits breast cancer cell metastasis by blocking EMT through modulation of snail/slug protein translation. Acta Pharmacol Sin 2019;40(9):1219-27.
[Crossref] [Google Scholar] [PubMed]
- Jiao L, Wang S, Zheng Y, Wang N, Yang B, Wang D, et al. Betulinic acid suppresses breast cancer aerobic glycolysis via caveolin-1/NF-κB/c-Myc pathway. Biochem Pharmacol 2019;161:149-62.
[Crossref] [Google Scholar] [PubMed]
- Zhao X, Chen J, Cai P. Effects of croton alkaloids on the proliferation and apoptosis of colorectal cancer SW620 cells by regulating the expression of miR-4317 in vitro and its action mechanism. Chin Gen Pract 2010;13(21):2345-8.
- Zhao XY, Chen J, Cai PS. Effect of croton alkaloid on cell cycle, apoptosis and PCNA gene expression in human ovarian cell. Chin Gen Pract 2010;13:2345-8.
- Wang H, Gao X, Yu S, Wang W, Liu G, Jiang X, et al. Circular RNAs regulate parental gene expression: A new direction for molecular oncology research. Front Oncol 2022;12:947775.
[Crossref] [Google Scholar] [PubMed]
- Yehui L, Zhihong L, Fang T, Zixuan Z, Mengyuan Z, Zhifang Y, et al. Bibliometric analysis of global research on circular RNA: Current status and future directions. Mol Biotechnol 2023.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Shan G. circRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 2021;505:49-57.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Zheng Y, Yu L, Liu M, Zhang Y. Identification of circ_0038632 as a promoter of breast cancer through miR-520a-3p-dependent modulation of CDCA3. Ann Clin Lab Sci 2023;53(4):562-72.
- He ZY, Zhuo RG, Yang SP, Zhou P, Xu JY, Zhou J, et al. circNCOR1 regulates breast cancer radiotherapy efficacy by regulating CDK2 via hsa-miR-638 binding. Cell Signal 2023;109:110787.
[Crossref] [Google Scholar] [PubMed]
- Yang Z, Li C, Fan XY, Liu LJ. Circular RNA circ_0079593 promotes glioma development through regulating KPNA2 expression by sponging miR-499a-5p. Eur Rev Med Pharmacol Sci 2020;24(3):1288-301.
[Crossref] [Google Scholar] [PubMed]
- Liu G, Zhang Z, Song Q, Guo Y, Bao P, Shui H. circ_0006528 contributes to paclitaxel resistance of breast cancer cells by regulating miR-1299/CDK8 axis. Onco Targets Ther 2020;13:9497-511.
[Crossref] [Google Scholar] [PubMed]
- Xu Q, Fang Y, Zhao X. The apoptosis of Hela cells induced by croton alkaloid and its mechanism. Chin J Biochem Pharm 2010;31(6):392-4.
[Crossref] [Google Scholar] [PubMed]
- Wu Z, Zhao X, Sun Y, Yu H. Curcumin suppresses colorectal cancer development with epithelial-mesenchymal transition via modulating circular RNA HN1/miR-302a-3p/PIK3R3 axis. J Physiol Pharmacol 2022;73(2):219-31.
[Google Scholar] [PubMed]
- Hu S, Zheng W, Jin L. Astragaloside IV inhibits cell proliferation and metastasis of breast cancer via promoting the long noncoding RNA TRHDE-AS1. J Nat Med 2021;75(1):156-66.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 2019;10(2):55.
[Crossref] [Google Scholar] [PubMed]
- Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis 2019;10(3):175.
[Crossref] [Google Scholar] [PubMed]
- Wx LF, Wp ZC. Influence and mechanism study of crotonic alkaloids on apoptosis of lung adenocarcinoma cell. China Med Herald 2016;13(1):17-20.
- Sl GY, Lw GY. Effects of croton alkaloids on expression of PCNA, Ki-67, Bax and Fas in human gastric cancer SGC-7901 cells. Jiangsu J Tradit Chin Med 2014(6):77-9.
- Cw CP, Lp LY. Effect of Crotonis fructus alkaloid on apoptosis of human hepatoma SMMC-7721 cells and Bcl-2/Bax expression. Chin J Exp Tradit Med Formulae 2011;17(11):199-201.
- Yoodee S, Thongboonkerd V. Epigenetic regulation of epithelial-mesenchymal transition during cancer development. Int Rev Cell Mol Biol 2023;380:1-61.
- Niu XY, Zhang ZQ, Ma PL. miRNA-221-5p promotes breast cancer progression by regulating E-cadherin expression. Eur Rev Med Pharmacol Sci 2019;23(16):6983-90.
[Crossref] [Google Scholar] [PubMed]
- Zhao F, Jia Z, Feng Y, Li Z, Feng J. Circular RNA circ_0079593 enhances malignant melanoma progression by the regulation of the miR-573/ABHD2 axis. J Dermatol Sci 2021;102(1):7-15.
[Crossref] [Google Scholar] [PubMed]
- Lu J, Li Y. circ_0079593 facilitates proliferation, metastasis, glucose metabolism and inhibits apoptosis in melanoma by regulating the miR-516b/GRM3 axis. Mol Cell Biochem 2020;475(1-2):227-37.
[Crossref] [Google Scholar] [PubMed]
- Hao YL, Wang XQ. circ-0079593 promotes proliferation and migration of melanoma cells by sponging microRNA-433 and elevating EGFR expression. Eur Rev Med Pharmacol Sci 2021;25(2):779-86.
[Crossref] [Google Scholar] [PubMed]
- Wang P, Wang T, Dong L, Xu Z, Guo S, Chang C. Circular RNA circ_0079593 facilitates glioma development via modulating miR-324-5p/XBP1 axis. Metab Brain Dis 2022;37(7):2389-403.
[Crossref] [Google Scholar] [PubMed]
- Li P, Wang Z, Li S, Wang L. circ_0006404 accelerates prostate cancer progression through regulating miR-1299/CFL2 signaling. Onco Targets and Ther 2021;14:83-95.
[Crossref] [Google Scholar] [PubMed]
- Xuan X, Wang Z, Wang Y. circ_0058608 contributes to the progression and taxol resistance of non-small cell lung cancer by sponging miR-1299 to upregulate GBP1. Anticancer Drugs 2023;34(1):103-14.
[Crossref] [Google Scholar] [PubMed]
- Liu LH, Tian QQ, Liu J, Zhou Y, Yong H. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J Cell Biochem 2019;120(8):12684-93.
[Crossref] [Google Scholar] [PubMed]
- Shen BJ, Yang YF, Zhang XX. Hsa_circ_0001925 promotes malignant progression in triple-negative breast cancer via miR-1299/YY1 axis. Thorac Cancer 2023;14(8):746-57.
[Crossref] [Google Scholar] [PubMed]