- *Corresponding Author:
- Z. Kai
Department of Blood Transfusion, Tianjin Hospital, Tianjin 300211, China
E-mail: 13752590428@139.com
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “101-107” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of this study is to investigate the methylation and expression of kallikrein 12 gene in triple-negative breast cancer cells. We evaluated the methylation status of the promoter region of kallikrein 12 gene in breast cancer using bisulfite sequencing polymerase chain reaction to compare the expression of kallikrein 12 gene in MDA-MB-231 and breast cancer cells and examined the expression of kallikrein 12 messenger ribonucleic acid by real-time quantitative polymerase chain reaction. In addition, we detected the expression of kallikrein 12 protein using Western blotting. The elevated methylation of the kallikrein 12 promoter in triple-negative cancer cells led to a reduction in its protein and messenger ribonucleic acid levels. But, after the treatment with 5-aza-2'-deoxycytidine, the protein and messenger ribonucleic acid expression increased. The use of 5-aza-2'- deoxycytidine can significantly reduce the level of kallikrein 12 promoter methylation in the MDA-MB-231 cells. These findings suggest that the hypermethylation may be responsible for the reduced gene expression in these cancer cells. In this study, we introduced a new concept and methodology for the treatment and diagnosis of triple-negative breast cancer.
Keywords
Kallikrein 12 gene, methylation, 5-aza-2'-deoxycytidine, triple negative breast cancer
Around a half of all women worldwide are generally diagnosed with breast cancer in their lifetime. It is also the 2nd leading cause of death among women. In United States, around 41 individuals among 100 000 individuals are diagnosed for this disease. Due to the complexity of breast cancer, treatment options are often difficult to choose. Recent studies have shown that certain types of receptors, such as the Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal Growth Factor Receptor 2 (HER2), play a significant role in the development of this disease[1]. Currently, breast cancer is categorized into different types based on the levels of receptors known as ER, PR, and HER2[2]. Triple-negative breast cancer is a type of cancer that occurs when all of these receptors are negative. It is commonly seen among women of Hispanic or African descent.
Currently, there are significant challenges in the field of breast cancer research regarding the treatment of triple negative disease. The Kallikrein (KLK) family related peptidases of human tissue comprises KLK1 to KLK15, which are serine endopeptidases with trypsin and chymotrypsin-like activity[2]. This family is a group of serine endopeptidases with trypsin and chymotrypsin-like activity. The members of KLKs family have multiple biological functions, including blood coagulation, complement activation, embryonic development, and wound healing. KLK12 is a synaptic serine endopeptidase involved in multiple biological activities, such as epidermal proliferation, differentiation, and keratogenesis[3]. Recently, it has been discovered that KLK is closely associated with the incidence and progression of tumors. An increasing number of studies have demonstrated the abnormality of KLK in numerous types of tumors, including ovarian cancer[4], cervical cancer[5], and lung cancer[6].
It is increasingly evident that KLK12 serves as a biological marker for tumor diagnosis and prognosis[7]. A later study noted that the development of triple-negative cancer was linked to the hypermethylation of promoter regions of the KLK12. This finding contradicts the conventional wisdom regarding the link between structural changes and the development of cancer[2].
In this study, we utilized 5-Aza-2'-Deoxycytidine (5-Aza-2' Deoxyc) in the treatment of breast cancer cells, especially for the cell lines, breast cancer cells (MCF-7) and MDA-MB-231. Our objective was to analyze the effect of 5-Aza-2' Deoxyc on the re-expression and demethylation of the KLK12. We also explored the link between the promoter's methylation level and the cell's gene expression.
Materials and Methods
Cell culture and treatment:
The study was conducted between two groups, the control group and the experimental group which included the triple-negative and ER1-positive breast cancer cells. Different cell culture protocols were utilized, including the use of a Dulbecco's Modified Eagle Medium (DMEM) and the demethylation of 5-Aza-2' Deoxyc. Different cell cultures were then used, and the demethylated drug was administered at 2.5 μmol/l. The medium was changed daily for 6 d by adding fresh drug in the medium. The cells were harvested, and total nucleic acids namely, Deoxyribonucleic Acid (DNA), Ribonucleic Acid (RNA) and proteins were extracted for follow-up experiments.
DNA and RNA extraction:
The DNA extraction method was carried out using phenol-chloroform extraction, and the RNA extraction procedure was performed according to the instructions of the kit from Shanghai Bioengineering Technology Service Co.
Real-Time quantitative Polymerase Chain Reaction (RT-PCR):
Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) was used as an internal reference. The primer sequences of the GAPDH and the target gene KLK12 were purchased from Suzhou Jinweizhi Biotechnology Co. Ltd. The reaction was performed using Platinum Quantitative PCR SuperMix-UDG Kit (Invitrogen, USA) with a reaction system of 20 μl under the conditions of 95° 30 s for pre-denaturation, 95° 30 s for denaturation, 60° 60 s for annealing and 72° 60 s for extension, 40 cycles in total.
DNA sodium bisulfite was modified using the EpiTect plus DNA Bisulfite Kit (QIAGEN, Inc., USA). 2 μg DNA was dissolved in 50 μl deionised water and denatured by adding 3 mol/l Sodium Hydroxide (NaOH), 30 μl 10 mmol/l hydroquinone and 520 μl 3.6 mol/l pH 5.0 sodium bisulfite at 50°, every 10 min for 18 h. After purification using the Wizard®DNA Clean-up System, it was precipitated with absolute ethanol and re-dissolved in 50 μl deionised water.
KLK12 promoter CpG island detection:
3 main start site sequences were used to search sites for common confirmation namely, Reference Sequence (RefSeq): National Center for Biotechnology Information (NCBI) Database (http://www.ncbi.nlm.nih.gov/RefSeq/); New Energy and Industrial Technology Development Organization (NEDO) (http://www.kazusa.or.jp/NEDO/) and Kazusa DNA Research Institute (Kazusa DNA Res Inst) (http://www.kazusa.or.jp/en/human/). In the sequence of the KLK12 gene, the ATG locus and the proximal promoter of the KLK12 gene were determined. The number of CpG was counted according to the sequence.
Bisulfite Sequencing PCR (BSP) method:
The BSP method followed the protocol established by Dupuy et al. [8]. The region from -600 bp to +100 bp in the KLK12 promoter was amplified using ZymoTaqTM DNA polymerase (Zymo Research, USA) with sodium bisulfite modified DNA as the template. The PCR products were observed using agarose gel electrophoresis and gel imaging system, and the target fragments were recovered using QIAquick gel extraction kit (QIAGEN). pGEM®-T Easy Vector System (Promega Corporation, USA) was used to ligate the recovered DNA fragments to the T-vector in a molar ratio of 8:1. After the ligated product was transfected into Escherichia coli, it was grown on a solid medium containing ampicillin at a temperature of 37°. 10 colonies were randomly selected from each plate and inoculated into 1 l Luria Broth (LB) liquid medium containing 100 μg ampicillin at 230 rpm for 16 h at 37°. Plasmid DNA was extracted using Plasmid MiniKit (QIAGEN) and methylation of the KLK12 promoter was analyzed by sequencing.
Western blotting:
The total proteins from cells were collected and concentration was determined using the Bicinchoninic Acid (BCA) method. 50 μg protein was subjected to Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE).
The membranes were transferred on semidry transfer apparatus, blocked with 5 % nonfat dry milk powder at 37° for 1.5 h. The corresponding primary antibodies (1:1000 dilutions) were added; the membrane was then incubated overnight at 4°, following washed extensively with Phosphate Buffered Saline (PBS) solution containing 0.1 % tween, 23 times for 10 min each. The horseradish peroxidase labeled secondary antibodies (1:10 000 dilution) were incubated at room temperature for 1 h, the membrane was washed extensively, then developed by adding Enhanced Chemiluminescence (ECL) developer, and photographed by ultrasensitive multifunctional imager (Carl Zeiss meditec, Hilden, Germany). The KLK12 antibodies were obtained from Jackson Laboratory.
Statistical analysis:
The statistical software Statistical Package of Social Sciences (SPSS) 22.0 was used to carry out the Chi-square (χ2) test on the experimental data, and p<0.05 was considered statistically significant.
Results and Discussion
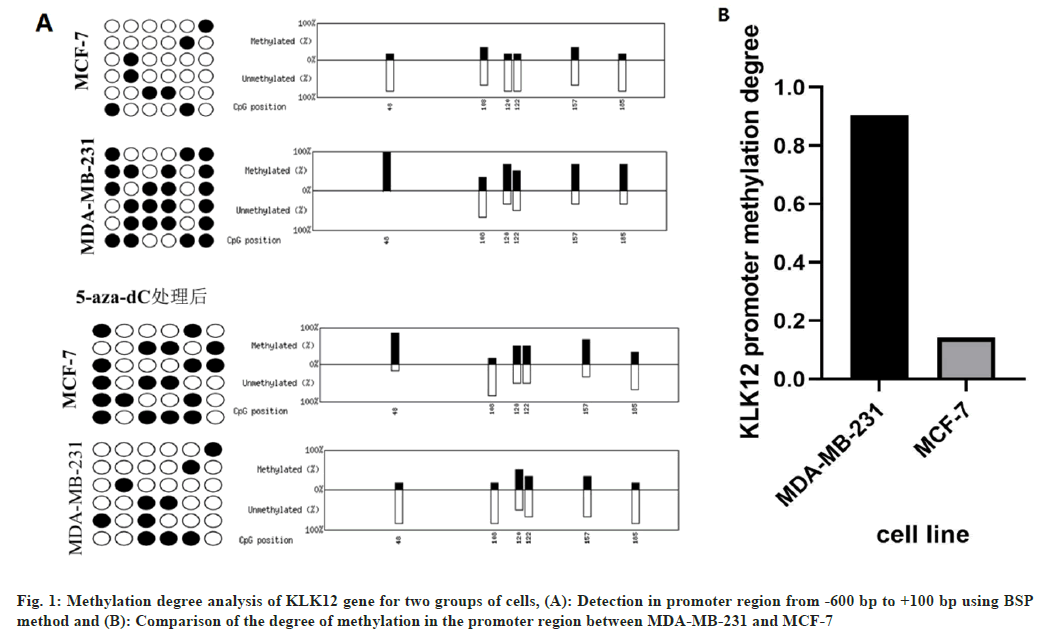
The methylation degree analysis for triple negative breast cancer cells and the control group was carried out. Bioinformatic analysis revealed that the region from -600 bp to +100 bp in the proximal promoter of the KLK12 gene was rich in CpG islands. BSP method was used to detect the methylation of KLK12 promoter from -600 bp to +100 bp.
The results of the study revealed that the level of KLK12 promoter methylation was 90.4 % in the MDA-MB-231 cells and 14.3 % in the control group's MCF-7. The difference between the two groups has a significantly lower level of methylation (p=0.0014) (fig. 1A and fig. 1B). After 1 w of treatment with 5-Aza-2' Deoxyc, the promoter of the gene KLK12 was unmethylated in the MCF-7 cell lines, while its methylated counterpart was found in the MDA-MB231 cell lines, as shown in fig. 1.
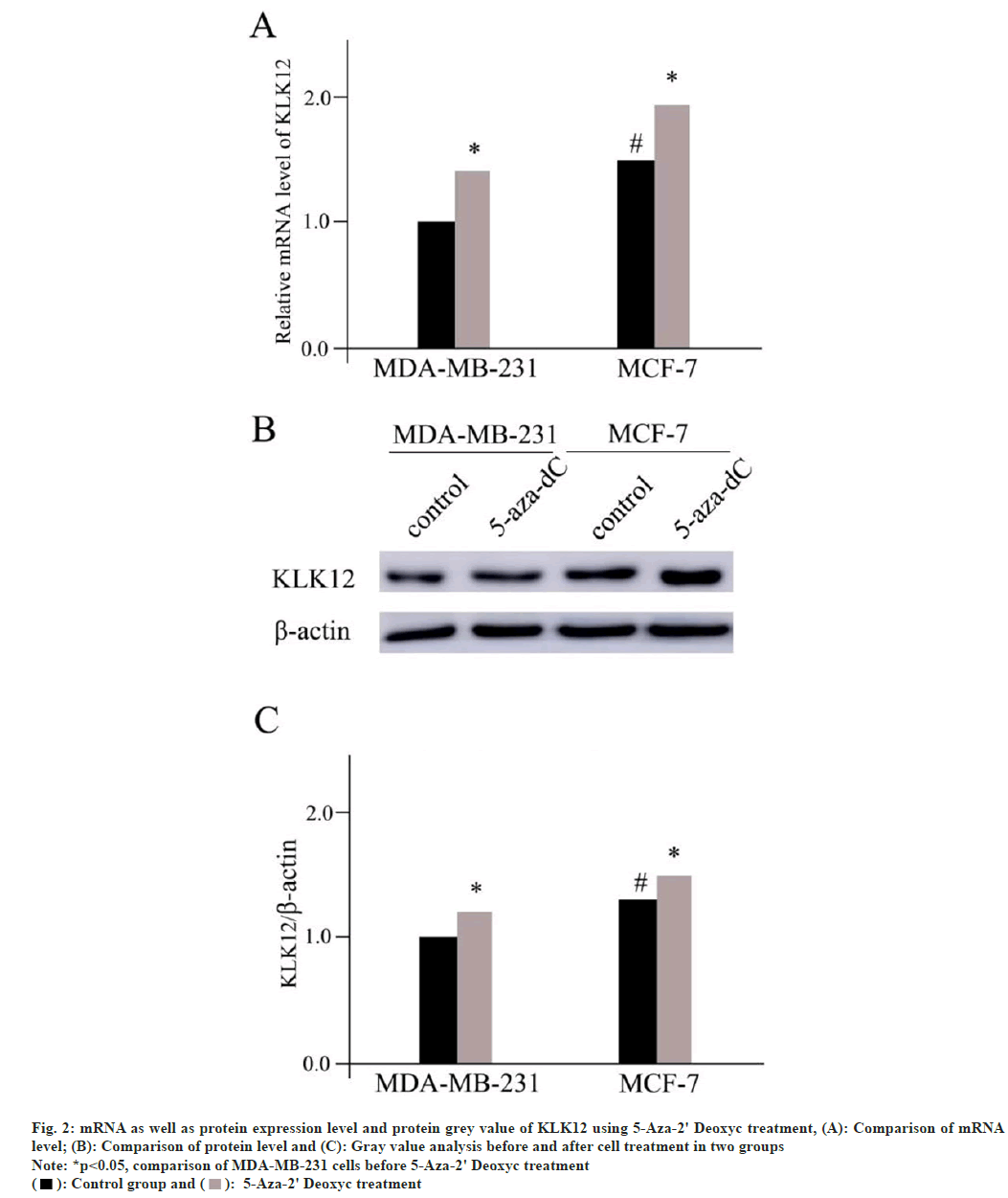
Messenger RNA (mRNA) as well as protein expression level and protein grey value of KLK12 using 5-Aza-2' Deoxyc treatment has been studied. The levels of protein and mRNA expression in the two different cell lines improved after 1 w of treatment with 5-Aza-2' Deoxyc demethylated drug (MDA-MB-231, p=0.015 and MCF-7, p=0.017). The difference between the control group and the MDA-MB-231 was significant. The levels of the protein and mRNA were still lower in the latter (mRNA, p=0.021 and protein p=0.028) (fig. 2).
Fig. 2: mRNA as well as protein expression level and protein grey value of KLK12 using 5-Aza-2' Deoxyc treatment, (A): Comparison of mRNA
level; (B): Comparison of protein level and (C): Gray value analysis before and after cell treatment in two groups
Note: *p<0.05, comparison of MDA-MB-231 cells before 5-Aza-2' Deoxyc treatment  5-Aza-2' Deoxyc treatment
5-Aza-2' Deoxyc treatment
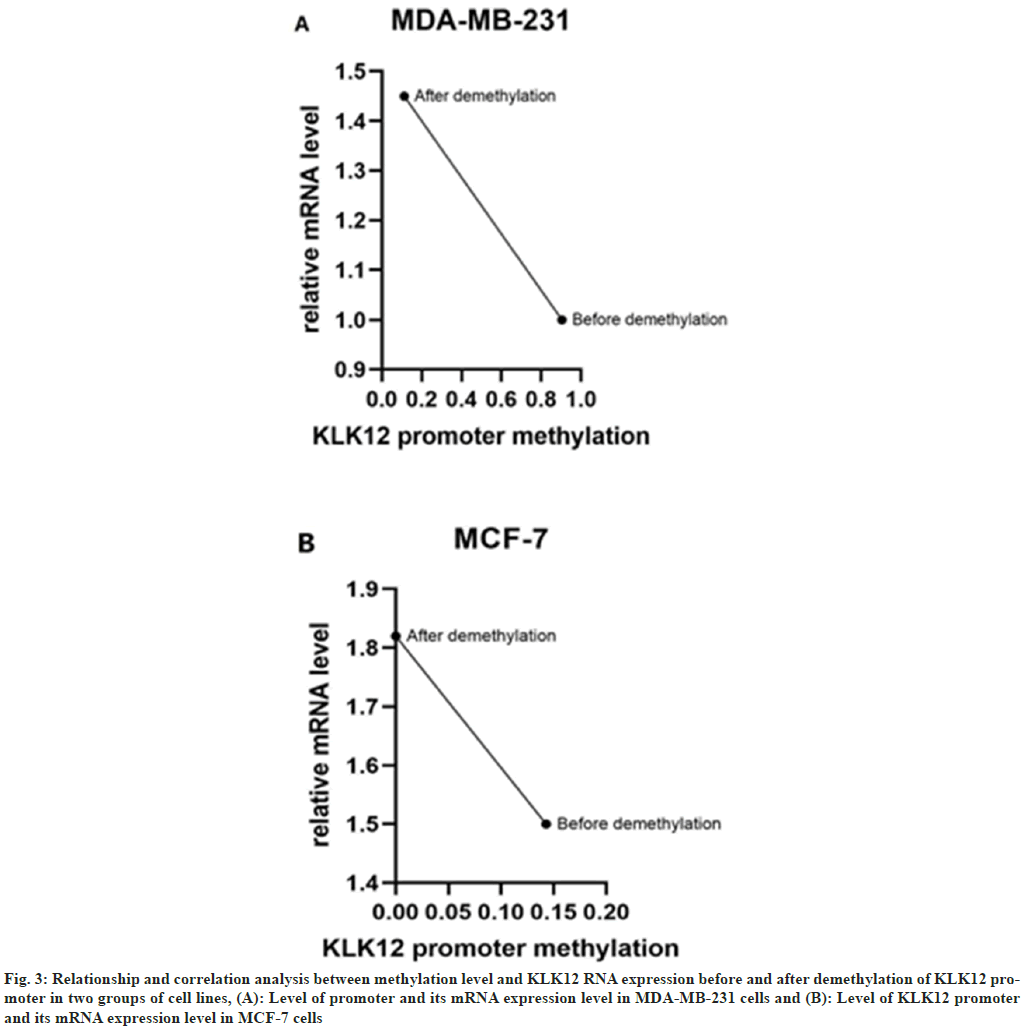
The relationship between the methylation level of KLK12 promoter before and after demethylation and the expression of KLK12 RNA in two groups of cell lines was studied. In triple-negative breast cancer cells MDA-MB-231, KLK12 expression was lower in those with hypermethylation of the KLK12 gene promoter, while in ER-positive cells MCF-7, KLK12 expression was higher in those with hypomethylation of the KLK12 gene promoter. The expression level of KLK12 was inversely proportional to the methylation level of the KLK12 promoter (fig. 3).
Fig. 3: Relationship and correlation analysis between methylation level and KLK12 RNA expression before and after demethylation of KLK12 promoter in two groups of cell lines, (A): Level of promoter and its mRNA expression level in MDA-MB-231 cells and (B): Level of KLK12 promoter and its mRNA expression level in MCF-7 cells
The prognosis and clinical management of patients with breast cancer vary depending on the type of cancer. Currently, the molecular subtypes of this disease are used in the diagnosis and treatment of patients (2013 St. Gallen International Breast Cancer Conference). These are, luminal A (ER/PR positive, HER2 negative tumors, Antigen Kiel 67 (Ki-67) positive <20 % and the percentage is the result of immunohistochemical staining of patient samples); luminal B (ER/PR positive <20 %, HER2 negative tumors and Ki-67 positive >20 %), HER2 positive b2 (ER/PR positive, HER2 negative and overexpressing, Ki-67 positive >20 %); HER2 overexpressing (ER-negative, PR-positive, HER2-overexpressing); triple-negative (ER-negative, PR-negative, and HER2-negative), and other specific subtypes[9]. The clinical definition of the disease referred to as triple-negative cancer was 1st established during the 2000s. It is a group of breast cancer cells that are not considered to be cancerous. According to the data collected by the American Cancer Society (ACS), this type of cancer mainly occurs in women under 40 y old[9]. A study conducted on the pathological characteristics of this type of cancer revealed that it had poor prognosis. It also had characteristics that allow it to spread easily and locally.
About 46 % of triple-negative breast cancer patients have distant metastasis of tumor cells, usually involving the brain and internal organs. Median survival was only 13.3 mo and the recurrence rate was 25 %[10]. The median time to recurrence was (35-67) mo for non-triple negative patients and (19-40) mo for triple negative patients.
Breast cancer patients with triple-negative status have a 75 % likelihood of dying within 3 y following their operation[10]. The lack of targeted drugs has limited the available treatment options for this type of cancer. Patients with this condition are not sensitive to molecularly targeted or endocrine therapy. The use of postoperative chemoradiotherapy has been known to have poor results for patients with triple-negative breast cancer. One of the most common treatment options for this type of cancer is the use of the drug, bevacizumab. However, it has not been shown to prolong the survival of these patients. Studying the molecular markers and the pathogenesis of this disease could lead to the development of new effective and personalized treatment strategies.
In recent years, epigenetic research has attracted much attention from cancer researchers, particularly the methylation of key target genes. Methylation is a process that can lead to the silencing of certain genes. It plays a significant role in the development of cancer cells. In a study, the researchers focused on the methylation status of several target genes in triple negative breast cancer. They were able to use drugs that can treat these cancer cells.
It is widely involved in physiological and pathological processes[11]. In cancer-related diseases, some differentially expressed members of the KLK family have become markers for screening and diagnosis of hormone-dependent cancers. For example, KLK2 and KLK3 (commonly referred to as Prostate-Specific Antigen, (PSA)) are used for prostate and breast cancer, KLK6 and KLK2, KLK10 for ovarian cancer, KLK11 for ovarian and prostate cancer, and KLK13 for breast and ovarian cancer[12-14].
Previous studies have shown that KLK12 is a tumor suppressor gene with low expression in breast cancer, and its splice KLK12sv1/2 is upregulated in breast cancer patients, but KLK12sv3 is downregulated. KLK12 appears to play an opposing role in breast cancer progression through the expression of its alternative splicing variants, with KLK12sv1/2 and KLK12sv3 splicing being good prognostic markers in breast cancer patients[2]. Gong et al.[15] and his colleagues claimed that diverse expression accompanied either by increased or decreased enzymatic activity of KLK12 has been linked to several diseases including triple-negative breast cancer. Moreover, positive but low KLK12 mRNA levels were detected in about half of the triple-negative breast cancer patients (54 out of 116; 47 %), the other samples were negative for KLK12 mRNA expression. Meanwhile, using univariate Cox analysis, positive KLK12 mRNA expression was significantly associated with shortened Disease-Free Survival (DFS), Hazard Ratio (HR)=2.12, 95 % Confidence Interval (CI)=1.19-3.78, (p=0.010); Overall Survival (OS), HR=1.91, 95 % CI=1.04-3.50 and (p=0.037), so that it could been shown that the positive KLK12 expression is closely associated with shortened DFS and OS, suggesting that KLK12 plays a tumor-supporting role in triple-negative breast cancer. Another study by Sato et al. measured the expression profile of 15 KLK genes in breast carcinomas using microarray data[16]. They focused on immunolocalized KLK12 in 140 breast carcinomas and evaluated its clinical significance, showing KLK12 was the most prominent member associated with low-grade malignancy in breast carcinomas. Immunohistochemical KLK12 status was positively associated with ER and PR status, while it was inversely associated with stage, pathological T factor, lymph node metastasis, and distant metastasis. Prognostic analysis demonstrated that KLK12 was a favorable prognostic factor for both DFS and breast cancer-specific survival of the patients. However, the role and function of KLK12 promoter methylation in hormone-dependent breast cancer has not been reported.
In this study, the 5-Aza-2' Deoxyc was utilized as demethylation drug. The agent was widely used for cancer study based on its demethylation functions. Previously, in Esophageal Squamous Cancer Cell (ESCC), 5-Aza-2' Deoxyc treatment significantly increased the expression level of Maternally Expressed Gene 3 (MEG3). While, ectopic expression of MEG3 in ESCC cells inhibited cell proliferation, promoted apoptosis, and suppressed metastasis[17]. Except for the demethylation functions, 5-Aza-2' Deoxyc was also applied in canonical Wingless-related integration site (Wnt) signaling cassette and resulted in suppression of the Wnt/catenin Beta (β) signaling pathway and rescue of the Cadmium (Cd) induced cell radioresistance[18].
The aim of this study was to investigate the role of KLK12 promoter methylation in the diagnosis and treatment of triple negative breast cancer. Although the findings of the study showed that the level of methylation in the breast cancer cells was lower, it did not determine the exact role of this process in the development of this type of cancer. The researchers noted that future studies should focus on the use of silenced or hypermethylated cell lines to analyze the role of this process in the development of this disease.
In this study, the researchers discovered that the methylation level of the gene KLK12 in the MCF-7 cell line was lower than that of the cancer cells that are known to develop triple-negative status. It could be also found that the level of methylation in the promoter of the gene was significantly lower in the cancer cells that are known to grow. All the promising findings here provide beneficial hints for the future clinical study of breast cancer.
Author’s contributions:
Jixian Wu contributed to the study design, data acquisition, data analysis and initial drafting. Aimei He contributed to the study design, revisions of the initial draft and experimental supervision.
Conflict of interests:
The authors declared no conflict of interest.
References
- Radosevic-Robin N, Selenica P, Zhu Y, Won HH, Berger MF, Ferrando L, et al. Recurrence biomarkers of triple negative breast cancer treated with neoadjuvant chemotherapy and anti EGFR antibiotics. NPJ Break Cancer 2021;7(1):124.
[Crossref] [Google Scholar] [PubMed]
- Papachristopoulou G, Tsapralis N, Michaelidou K, Ardavanis-Loukeris G, Griniatsos I, Scorilas A, et al. Human Kallikrein related peptidase 12 (KLK12) splice variants discriminate benigns from cancer break students. Clin Biochem 2018;58:78-85.
[Crossref] [Google Scholar] [PubMed]
- Hua Q, Li T, Liu Y, Shen X, Zhu X, Xu P. Upregulation of KLK8 predicts poor prognosis in pancreative cancer. Front Oncol 2021;11:624837.
[Crossref] [Google Scholar] [PubMed]
- Borgoño CA, Kishi T, Scorilas A, Harbeck N, Dorn J, Schmalfeldt B, et al. Human kallikrein 8 protein is a feasible diagnostic marker in ovarian cancer. Clin Cancer Res 2006;12(5):1487-93.
[Crossref] [Google Scholar] [PubMed]
- Bignotti E, Bellone S, Palmieri M, de Las Casas L, Roman JJ, Pecorelli S, et al. The novel serine protein tuber associated differentially expressed gene-14 (KLK8/neuropsin/ovasin) is highly overexpressed in clinical cancer. Am J Obstet Gynecol 2004;190(1):60-6.
[Crossref] [Google Scholar] [PubMed]
- Sher YP, Chou CC, Chou RH, Wu HM, Chang WS, Chen CH, et al. Human kallikrein 8 protocol negotiations a viable clinical output in non-small cell lung cancer by suppressing tube cell invasivesses. Cancer Res 2006;66(24):11763-70.
[Crossref] [Google Scholar] [PubMed]
- Lavergne M, Guillon-Munos A, Bonda WL, Attucci S, Kryza T, Barascu A, et al. Tissue factor pathway inhibitor 2 is a potential kallikrein related protein 12 inhibitor. Biol Chem 2021;402(10):1257-68.
[Crossref] [Google Scholar] [PubMed]
- Dupuy AM, Ruffel L, Bargnoux AS, Badiou S, Cristol JP. Analytical evaluation of the performances of a new procalcitonin immunoassay. Clin Chem Lab Med 2021;60(3):77-80.
[Crossref] [Google Scholar] [PubMed]
- Jaiswal A, Murakami K, Elia A, Shibahara Y, Done SJ, Wood SA, et al. Therapeutic invasion of USP9x mediated notch signaling in triple negative bread cancer. Proc Natl Acad Sci USA 2021;118(38):1-10.
[Crossref] [Google Scholar] [PubMed]
- Guo Y, Yin J, Dai Y, Guan Y, Chen P, Chen Y, et al. A novel CpG calculation risk indicator for predicting prognosis in blade cancer. Front Cell Dev Biol 2021;9:1-13.
[Crossref] [Google Scholar] [PubMed]
- Clements J, Hooper J, Dong Y, Harvey T. The expanded human Kallikrein (KLK) gene family: Genetic organization, tissue-specific expression and potential functions. Biol Chem 2001;382(1):5-14.
[Crossref] [Google Scholar] [PubMed]
- Borgono CA, Diamandis EP. The emerging roles of human issue kallikreins in cancer. Nat Rev Cancer 2004;4(11):876-90.
[Crossref] [Google Scholar] [PubMed]
- Sotiropoulou G, Pampalakis G, Diamandis EP. Functional roles of human kallikrein related peptides. J Biol Chem 2009;284(48):32989-94.
[Crossref] [Google Scholar] [PubMed]
- Stefanini AC, da Cunha BR, Henrique T. Investigation of kallikrein related peptides in normal and pathological processes. Dis Markers 2015:1-17.
[Crossref] [Google Scholar] [PubMed]
- Gong W, Liu Y, Preis S, Geng X, Petit-Courty A, Kiechle M, et al. Prognostic value of Kallikrein-related peptidase 12 (KLK12) mRNA expression in triple-negative breast cancer patients. Mol Med 2020;26(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Sato A, Takagi K, Yoshimura A, Tsukamoto W, Yamaguchi-Tanaka M, Miki Y, et al. Kallikrein-related peptidase 12 (KLK12) in breast cancer as a favorable prognostic marker. Int J Mol Sci 2023:24(9):1-13.
[Crossref] [Google Scholar] [PubMed]
- Lv D, Sun R, Yu Q, Zhang X. The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumour Biol 2016;24:16259-67.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Zhou Z, Lin X, Liao J, Zhang Y, Xie B, et al. Environmental cadmium exposure promotes the development, progression and chemoradioresistance of esophageal squamous cell carcinoma. Front Cell Dev Biol 2022:10:792933.
[Crossref] [Google Scholar] [PubMed]