- *Corresponding Author:
- Lei Zhang
State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, Zhejiang 311300, China
E-mail: starzhanglei@aliyun.com
| This article was originally published in a special issue,“Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “44-54” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Alkaline/neutral invertase is one of the key enzymes in sucrose metabolism, which irreversibly converts sucrose into fructose and glucose, the precursors of polysaccharide synthesis. Our main objective is to study the alkaline/neutral invertase gene function of Dendrobium catenatum, a rare and endangered medicinal plant, and provide evidence for revealing the biosynthesis of polysaccharides in Dendrobium catenatum. Eight alkaline/neutral invertase genes were identified from Dendrobium catenatum genome and DcNI4 gene was found to be highly expressed in stem by transcriptomic data, which was consistent with the high polysaccharide content in stem of Dendrobium catenatum. DcNI4 was confirmed to localize in cytoplasm. The expression level of DcNI4 in stem was significantly higher than that in roots and leaves at 6 mo, this difference might be due to the accumulation of polysaccharides and the energy consumption of plant growth. In Arabidopsis thaliana, DcNI4-overexpressing types had higher DcNI4 activity, increased polysaccharide content and promoted the root development. Our results provide new insights into the biological function of DcNI4 and lay a foundation for analysing the synthesis pathway of polysaccharides in Dendrobium catenatum.
Keywords
Dendrobium catenatum, DcNI4, Arabidopsis thaliana, polysaccharides
Dendrobium catenatum (D. catenatum) is a well-known traditional Chinese medicinal plant that has been utilized as food and drug materials for thousands of years. It has the classical functions of nourishing yin, clearing heat, promoting fluid and invigorating stomach[1], as well as diverse modern pharmacological activities, such as anti-fatigue, immunomodulatory, hypoglycemic, hypolipidemic, hepatoprotective and anti-cancer[2]. The stem is generally believed as the main medicinal part of D. catenatum, which contains a variety of chemical components, including polysaccharides, alkaloids, amino acids, phenanthrene and bibenzyl, etc. The polysaccharide fraction was a homogenous polysaccharide and most of these D. catenatum Polysaccharides (DCPs) are classified as 1,4-linked glucomannan or mannoglucan, and their beta (β)-configurations are acetylated to varying degrees and locations, with or without branching. As the major active compound to play the medicinal effects, the content of which has PMID: 35677385; PMID: 37092860 is also an important index to evaluate the plant quality of D. catenatum. Nowadays, wild D. catenatum has been officially listed as one of the key protected endangered species in China due to the environmental impacts[3]. However, polysaccharides are complicated and often exist as mixtures, it is difficult to qualitatively separate each molecule and determine their structures. The current research on polysaccharides is far less than that of flavonoids or alkaloids. Therefore, polysaccharides will be the promising focus in the future research.
Sucrose is not only a photosynthesis-derived compound in most plants, acting as a major molecule in carbon transport, but also plays a significant role in the functional biology and the response to environmental stresses[4]. In addition, sucrose is hydrolyzed to provide energy and carbon sources for plant growth. The hydrolyzed products are important metabolic signals to regulate genes expression and plant development[5,6]. Recent studies have found that sucrose contributes to protein storage, anthocyanin accumulation, flower induction due to the effect exerted by sucrose on gene expression at the transcriptional level[7]. When plants face stress, sucrose can regulate cell metabolism and participate in the stress response. Sucrose has an important role in the regulation of cell division and differentiation. The hydrolysis and synthesis of sucrose is a key issue in plant carbon metabolism and it has been extensively studied [8]. The contents of polysaccharides and alcohol-soluble extracts in D. catenatum from rock epiphytic plants were the highest and the root system was developed[9], which shows a close correlation between root development and the accumulation of secondary metabolites in D. catenatum.
Sucrose invertase irreversibly convert sucrose into fructose and glucose, the precursors of polysaccharide synthesis were blocked. Invertases can be classified into acidic and Alkaline/Neutral Invertase (NI) according to their optimal pH. Acidic Invertases (AI) are β- fructosidases with an optimum pH of 4.5-5.0, located in the cell wall and vacuole. Apart from sucrose, AI also break down raffinose and hydrothreose[10,11]. While NI are part of a family of glucosidases with an optimum pH of 6.5-8.0, which are mainly located in the cytoplasm or organelles[12,13]. NI require a high degree of substrate specificity, which appears to be the only substrate for this class of invertases[14,15]. Since NIs are not glycosylated, they are unstable and have low enzymatic activity compared to AI, which has led to a lack of understanding of their function. NI gene families have been identified in many species, such as Arabidopsis[16], pepper[17], populus[18], tea plant[19], rice[20,21], grape and other plants[22]. Furthermore, the NI gene family of plants can be divided into two distinct phylogenetic groups, called alpha (α) and β. Neutral/alkaline convertases of α-group are mainly located in the cytosol, whereas neutral/alkaline convertases of β-group are mainly located in the cytoplasmic matrix. In this study, 8 NI genes were identified from the genome of D. catenatum and the DcNI4 gene was discovered to be highly expressed in the stem by transcriptome data, which was consistent with the high polysaccharide content in the stem of D. catenatum[23]. We analyzed the enzymatic properties, the gene expression profile and the protein sublocalization of DcNI4. By ectopic expression of DcNI4 in Arabidopsis thaliana (A. thaliana) we also analyzed the polysaccharide content which was found higher than in control plants. These results provided a basis for revealing the biosynthetic process of D. catenatum and breeding varieties with high polysaccharide content and good quality.
Materials and Methods
Plant materials:
D. catenatum cultivar “Jingpin No. 1” (Breed No. Zhe R-SV-DO-015-2014) used in this study was obtained from the tissue culture room of the State Key Laboratory of Subtropical Silviculture in Zhejiang Province, China. Those D. catenatum were cultured under a 16 h/d light photoperiod at 25°. Fresh roots, stems and leaves from the plants were collected and frozen in liquid nitrogen for subsequent Ribonucleic Acid (RNA) isolation. A. thaliana cultivar ecotype Columbia-0 (Col-0) was used for stable genetic transformation. Seeds were sterilized with 75 % alcohol for 10 min and then sterilized with 95 % alcohol for 10 min, alcohol-treated seeds were thoroughly dried with sterile filter paper in a fume hood. The sterilized seeds were sown on Murashige and Skoog (MS) medium and vernalized at 4° for 2 d in the dark. Transferred to an artificial climate chamber and cultured under a 16 h/d light photoperiod at 25°. At 10 d after germination, seedlings grown uniformly were transferred to soil for further growth.
Seeds of tobacco (Nicotiana benthamiana (N. benthamiana)) were sown on soil, 1 w after germination, the seedlings were transferred into the new pots and cultivated under the same conditions as D. catenatum. The leaves of tobacco can be used for Agrobacterium tumefaciens (A. tumefaciens)transiently expression after 5 w.
Plant treatments:
Drought treatment was carried out according to the previously reported method[24]. The D. catenatum were cultured at 28°/22° d/night temperature, at 50 %-70 % Relative Humidity (RH) and a 12/12 h photoperiod, a light intensity of 100 μmol/m2/s and watering every 2 d at 15:30. D. catenatum plants used in experiments were 8 mo old and 12 cm tall. The plants were irrigated on the 1st and 8th d. Half an hour after the light was turned on and off on the 2nd d (DRY1, DRY2), 7th d (DRY3, DRY4) and 9th d (DRY6, DRY7) d and half an hour after the light was turned off on the 8th d (DRY5), the fourth leaf that matured at the top was collected. Each time point contained three biological replications. D. catenatum plants were treated at low temperature and samples were collected. Each treatment (0°) and control group (20°) was replicated 3 times and the incubation time was 20 h. All collected samples were immediately frozen in liquid nitrogen and stored at -80°.
Screening of DcNIs and cloning of DcNI4:
Transcriptome data were used to analyze tissue expression levels of DcNI genes and their expression changes in response to stress to screen key genes.
The full-length complementary Deoxyribonucleic Acid (cDNA) of the DcNI4 gene was then cloned from D. catenatum total RNA by reverse transcribed RNA using Polymerase Chain Reaction (PCR) using primers like Forward: CCGCAAGCTTGTCGAATGGAAGGGACCTCATGCATCAG and Reverse: TTC GAGCTCCGTCGAGCATGTCCAGGAAGTAGATCTCTTG designed for the DcNI4 gene.
Characterization of DcNI4 in D. catenatum:
The DcNI4 sequence was analyzed with various online tools. ProtParam, Softberry, SignalP-6.0 and TMHMM2.0 were used to predict the physicochemical properties, subcellular localization, protein signal peptide and protein transmembrane region of the protein respectively. Protein phosphorylation sites and protein secondary and tertiary structures were predicted using online tools like NetPhos-3.1, Self-Optimized Prediction Method with Alignment (SOPMA) and SWISS-MODEL respectively. We downloaded the D. catenatum genome from the National Center for Biotechnology Information (NCBI) database (GenBank: JSDN00000000.2), obtained the nucleotide sequence 2000 base pair (bp) upstream of the start codon through TBtools (an integrative toolkit developed for interactive analyses of big biological data) and submitted it to the PlantCare website to predict cis acting elements or motifs[25].
Phylogenetic analysis:
Multiple sequence alignment of protein sequences of 39 NIs genes from A. thaliana, D. catenatum, Capsicum annuum (C. annuum), Morchella esculenta (M. esculenta), Phalaenopsis equestris (P. equestris) and Vitis vinifera (V. vinifera)were analysed using ClustalW sequence alignment tool. The phylogenetic tree with 1000 bootstrap replicates was constructed via the Neighbor-Joining (NJ) method in MEGA X software. Bootstrap analysis was used to estimate the reliability of the tree.
Vector construction:
For the DcNI4 overexpression, the ?Open Reading Frame (ORF) of DcNI4 was amplified from D. catenatum genomic DNA and inserted into the plant expression vector pCAMBIA1300 (Cambia, Canberra, Australia) included a Green Fluorescent Protein (GFP) tag with the 35S promoter of Cauliflower mosaic virus (CaMV) using Sal? by the reaction of homologous recombination.
Transiently expression in N. benthamiana leaves by A. tumefaciens:
The plasmids pCAMBIA1300-DcNI4-GFP and pCAMBIA1300-GFP were transformed into A. tumefaciens strain GV3101 and transiently transferred into leaf epidermal cells of 5 w old N. benthamiana. Agroinfiltration method was used to transiently expression in N. benthamiana leaves by A. tumefaciens[26]. The transformed A. tumefaciens cells were resuspended in 2-(N-Morpholino)ethanesulfonic Acid (MES) liquid medium buffer with 150 μM acetosyringone at Optical Density (OD600)=0.4-0.6 and incubated at 25° for at least 2 h before being infiltrated into the abaxial side of N. benthamiana leaves. Tobacco leaves were injected with A. tumefaciens cells containing pCAMBIA1300-GFP as a control group. After incubation at 23° for 72 h of dark treatment, GFP signals were observed with a ZEISS 710 confocal laser scanning microscope (ZEISS Microsystems, Germany). Three biological repeats were performed to verify these results.
Plant transformation and phenotype analysis:
Wild type A. thaliana was cultivated until florescence and cut off the top inflorescence immediately. The mature pods and fully opened flowers were cut off when blooms were at their peak, and used A. tumefaciens to infect A. thaliana. Sprayed flowers and buds with Agrobacterium resuspended and activated for more than 2 h. Arabidopsis plants were placed in the dark for 24 h. A week after the completion of the first infection, the second infection was conducted in the same way. After screening positive plants to T3 generation, homozygous transgenic Arabidopsis was obtained. We extracted RNA from Arabidopsis leaves and set three biological replicates for each line. Three line plants were randomly selected to observe the root tips of A. thaliana seedlings with DcNI4 overexpression by fluorescence microscopy. We observed whether there was any significant difference in appearance of phenotype between transgenic Arabidopsis and wild type.
The total polysaccharide content determination:
The aerial parts of A. thaliana were collected and firstly pre-frozen at -80° for 12 h, then quickly placed into a freeze-dryer for 3 d. The analytical balance was used to weigh 0.3 g powder after grinding and dissolved it into 25 ml double distilled water, weighed and recorded. The sample was then boiled in a 100° water bath for 2 h. After cooling, distilled water was added up to original weight at room temperature. We collected the supernatant after centrifuging for 60 min, at 6000 rpm. 20 ml of absolute ethanol was added to 5 ml of supernatant and kept at 4° overnight for precipitation. Then centrifuged at 6000 rpm for 1 h and discarded the supernatant. We added 20 ml of 80 % ethanol to resuspend, centrifuged at 6000 rpm for 30 min, discarded the supernatant and repeated operation once. We dissolved the precipitate in distilled water and made up to 50 ml to analyze. The experiment referred to the "Chinese Pharmacopoeia" Dendrobium candidum content determination method to extract total polysaccharides. The content of polysaccharide was determined by phenol sulfuric acid method. With 1 ml of water as a blank, the absorbance was measured at 488 nm and the experiment was performed in parallel three times. The total polysaccharide content was calculated by anhydrous glucose.
Relative expression analysis by real-time quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR):
Fresh roots, stems and leaves from the D. candidum plants were collected and frozen in liquid nitrogen for subsequent RNA isolation by MiniBEST plant RNA extraction kit (TaKaRa) according to the manufacturer’s instructions. The isolated total RNA was reverse-transcribed into cDNA using the PrimeScriptTM RT reagent kit with genomic DNA (gDNA) Eraser (TaKaRa). qRT-PCR assays using TB GreenTM Premix Ex TaqTM ? Kit (TaKaRa) according to the manufacturer’s instructions. The relative transcript levels of the samples estimated by the Cycle Threshold (CT) method were compared to the mean of three independent PCRs. The actin gene of D. candidum was used to estimate relative messenger RNA (mRNA) levels. The primers of qRT-PCR are listed in Table 1. The process for amplification was 30 s at 95°, followed by 40 cycles of 95° for 5 s and 20 s at 60° for primer annealing and elongation. Then the dissociation stage was 40 s at 95°, 45 s at 60° and 20 s at 95°. According to the following formula of 2−ΔΔCT, we calculated relative gene expression[27], ΔΔCT=(CT, Target–CT, Actin) Time x– (CT, Target–CT, Actin) Time 0.
| Primer | Sequence |
|---|---|
| DcNI4-RT-F | TGGCGGACGAGGACGACG |
| DcNI4-RT-R | CCCAAGCCTCCGCAATCGTA |
| Actin-RT-F | TTGTGTTGGATTCTGGTGATGGTGT |
| Actin-RT-R | TTTCCCGTTCTGCTGTTGTTGTGAA |
| DcNI4-F1 | GGGTACCACAGTCGAATGGAAGGGACCTCATGCATCAG |
| DcNI4-R1 | AGGTTGTCATGTCGAGCATGTCCAGGAAGTAGATCTCTTG |
| DcNI4-F2 | GACGACAAGGCCATGGAAGGGACCTCATGCATCAG |
| DcNI4-R2 | AGGTGGTGGTGCTCGAGGCATGTCCAGGAAGTAGATCTCTTG |
Note: RT: Reverse Transcription; F: Forward primer and R: Reverse primer
Table 1: Primers for DCNI4 Gene Cloning, Vector Construction and qPCR
Statistical analysis:
Seven independent transgenic lines were identified from regenerated plants. Line 1, line 10, line 11 were selected for subsequent analysis. The Col-0 lines were used as controls. All data are expressed as mean±Standard Deviation (SD) of 3 independent biological replicates, each with 3 technical replicates. Significant differences between treatments using Student’s t-test were analyzed using one-way Analysis of Variance (ANOVA).
Results and Discussion
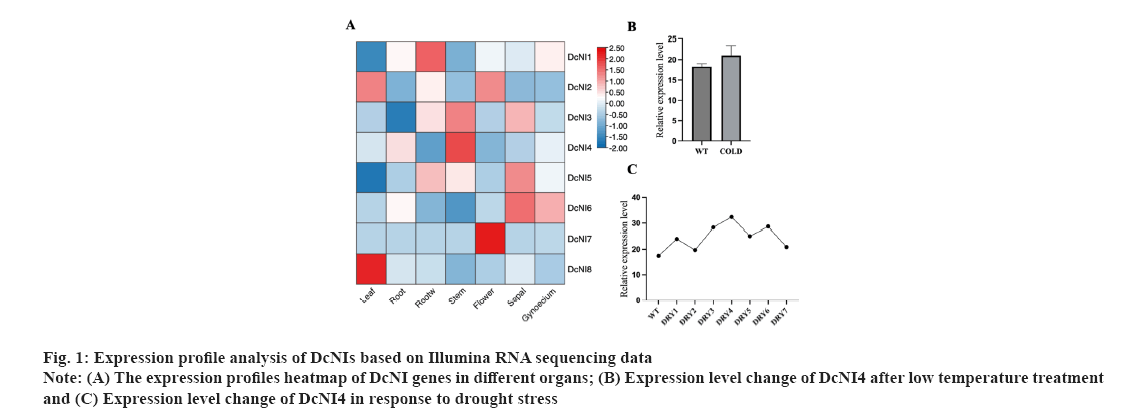
Screening of DcNIs and cloning of DcNI4 were shown in fig. 1A-fig. 1C. A total of 8NI genes were blasted and screened from the genome of D. catenatum based on the sequences of NI genes that have been reported in A. thaliana. They were named as DcNI1 to DcNI8. Based on the transcriptome data of D. catenatum obtained earlier by our research group, a heat map of the expression levels of the eight identified NI genes in roots, stems, leaves, flowers and other organs of D. catenatum was drawn. A colour bar indicates expression differences in different organs (fig. 1A). Expression level change of DcNI4 after low temperature treatment was shown in fig. 1B where control: 20°, 20 h and cold: 0°, 20 h. Each group had three repetitions. Under cold conditions, DcNI4 expression level did not decrease due to the plant blocking sucrose catabolism to resist low temperature (fig. 1B). The results showed that DcNIs were expressed in various organs of D. catenatum and highest in the stem, which conformed to the characteristics of polysaccharide distribution. By analyzing the expression differences of DcNI genes under drought and cold stress, we found that they showed different changes and speculated that their functions were also different. Under drought conditions, the expression level of DcNI4 gene was high and it still maintain a high level after rehydration. DRY1/DRY2 indicates the 2nd d treatment at 06:30 and 18:30; DRY3/DRY4 indicates the 7th d treatment at 06:30 and 18:30; DRY5 indicates the 8th d treatment at 18:30 after watering the seedlings; DRY6/DRY7 indicates the 9th d treatment at 06:30 and 18:30 (fig. 1C). We speculated that DcNI4 might be involved in regulating the resistance to low temperature and drought stress. Coupled with its significant high expression in the stem, we took it as the research object and the full-length cDNA sequence was cloned.
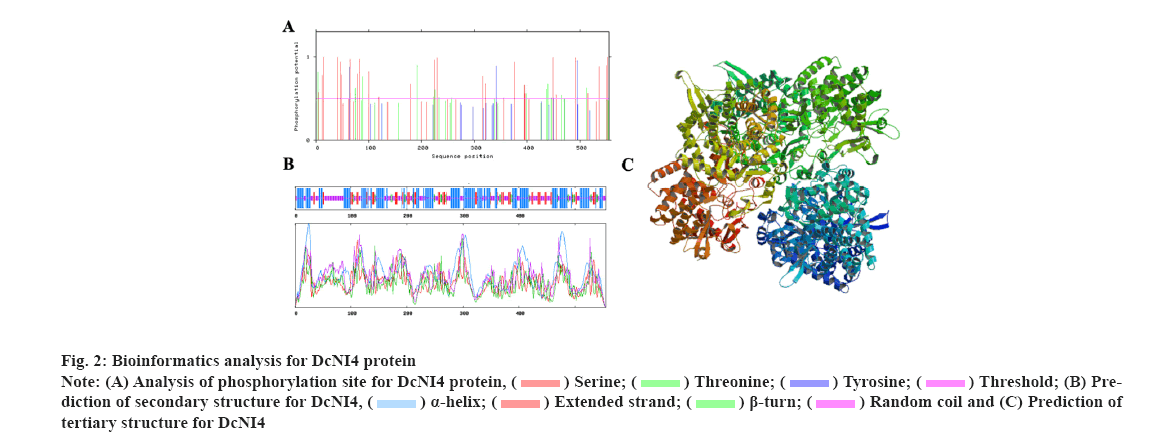
Characterization of DcNI4 in was explained here. To investigate the DcNI4 gene function, a 1668 bp segment of the DcNI4 ORF region was isolated from cDNA. The full-length cDNA of DcNI4 encodes a polypeptide of 556 amino acid residues with a calculated mass of 63.23 kDa and the Isoelectric point (pI) of 6.02. The instability index and Grand Average of Hydropathy (GRAVY) were computed to be 48.24 and −0.121 respectively, with a fat coefficient of 87.91. These findings indicated that DcNI4 was an unstable and acidic hydrophobic protein. The online tool SignalP-6.0 predicted that DcNI4 had no signal peptide and was a non-secreted protein, which speculated that DcNI4 might play a function in the cytoplasm. According to the results predicted by TMHMM2.0, DcNI4 protein had no transmembrane helical region and belonged to the extracellular protein. Phosphorylation site prediction results showed that the protein contains 44 phosphorylation sites including 26 serine Ser sites, 14 threonine Thr sites and 4 tyrosine Tyr sites (fig. 2A). The secondary structure prediction showed that the alpha (α)-helix (42.34 %), random coils (36.94 %), extended chain (14.59 %) and beta (β)-sheet accounting for the least 6.20 % were scattered throughout the protein (fig. 2B). The tertiary structure of DcNI4 protein predicted by SWISS-MODEL showed that the protein had 80.84 % sequence similarity with A. thaliana NI Cytosolic Invertase 1 (AtCINV1) and homologous modeling used the enzyme protein (6ttj.1) A chain as the template, the modeling range was 87-538 amino acids, the Global Model Quality Estimate (GMQE) value was 0.81 and the three Dimensional (3D) modeling quality was good (fig. 2C).
Cis-acting elements can regulate gene expression and do not encode any protein parts. Analysis of the cis-acting elements in the DcNI4 promoter sequence by the PlantCARE database indicated that the promoter contains not only the basic promoter elements such as the TATA-box and CAAT-box, but also some cis-acting elements related to stress responses, such as Abscisic Acid Responsive Elements (ABRE), Anaerobic Responsive Elements (AREs), drought responsive elements like MYB TF Binding Site involved in drought inducibility (MBS), MeJA-responsive and photoresponsive elements (G-box) and Low-Temperature Responsive elements (LTR).
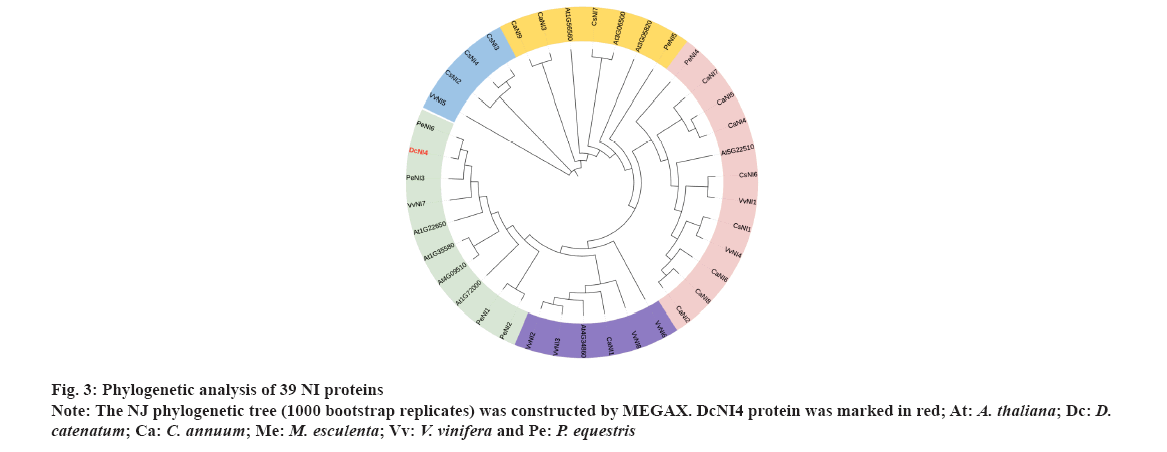
Phylogenetic analysis NI proteins were shown in fig. 3. To determine the evolutionary relationships among DcNI4 protein and NIs from other species, we constructed a NJ phylogenetic tree, sequences of 39 NI family members from 6 species (A. thaliana, D. catenatum, C. annuum, M. esculenta, V. vinifera and P. equestris) by using MEGAX software. These protein sequences were divided into 5 groups. The DcNI4 and NIs from A. thaliana, V. vinifera and P. equestris were clustered into a large branch with high homology. AT1G35580 had been reported to inhibit lateral root development by involving in osmotic stress-induced in Arabidopsis[28]. In addition, AT1G35580 and AT4G09510 had been found important in plant growth and fitness in Arabidopsis after the removal of apical dominance[29]. Therefore, we speculated that DcNI4 could affect sucrose metabolism and polysaccharide synthesis, and played an important role in plant development and stress resistance.
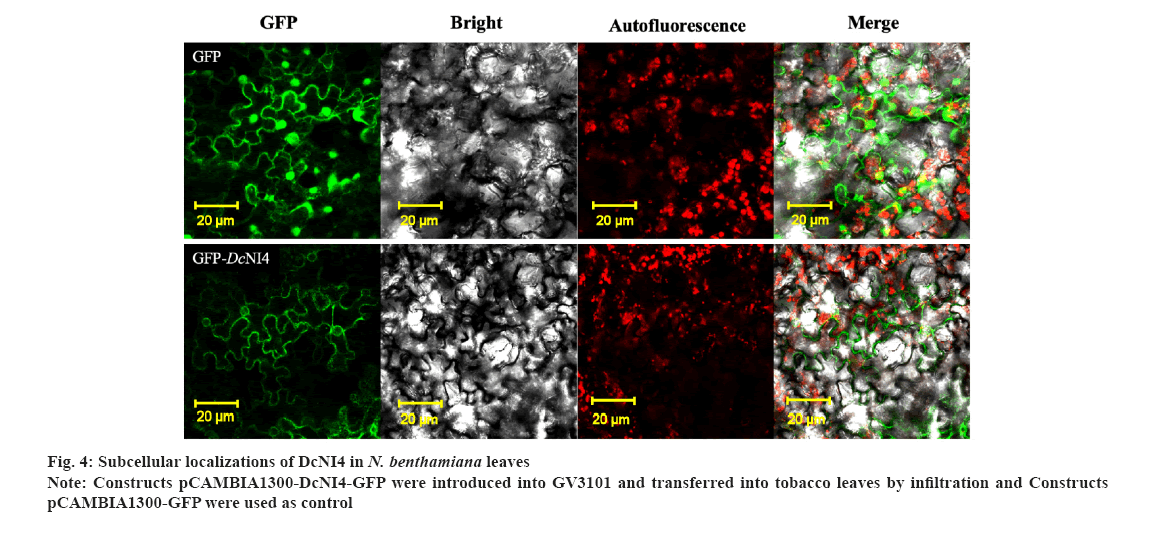
Subcellular localization of DcNI4 was shown in fig. 4. The online prediction software Softberry (http://www.softberry.com/berry.phtml) showed that DcNI4 was localized in the cytoplasm. The constructed vector pCAMBIA1300-DcNI4-GFP and pCAMBIA1300-GFP plasmids were transformed into A. tumefaciens GV3101 and injected into the tobacco leaves.
The results showed that GFP green fluorescence was observed in whole cells in pCAMBIA1300-GFP-transformed leaves, DcNI4-GFP fluorescence signals were detected in the cytoplasm as shown in fig. 4.
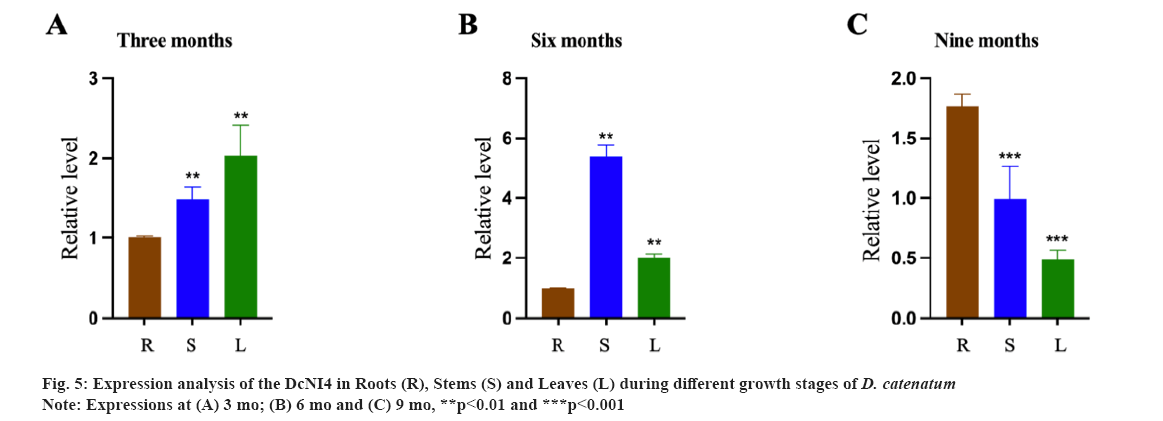
Expression patterns of DcNI4 in D. catenatum were shown in fig. 5. DcNI genes are important for plant growth and development. In order to preliminarily elucidate the function of DcNI4 in different growth stagesof D. catenatum, we used qRT-PCR to examine the expression of DcNI4 in root, stem and leaf at 3 mo, 6 mo and 9 mo. qRT-PCR results showed that DcNI4 was expressed in the whole plant and the expression level of DcNI4 varied in different parts at different growth stages. In 6 mo, the expression of DcNI4 in the stems was the highest, which was significantly higher than that in the roots and leaves at the same period. It was speculated that the stem cells of D. catenatum were rapidly dividing and expanding during the vigorous growth period, so they needed more sugar to provide energy. DcNI can break down sucrose irreversibly and provide precursors for the synthesis of polysaccharides. The high expression of the DcNI4 may be related to the massive accumulation of DCPs in the stem as shown in fig. 5A-fig. 5C.
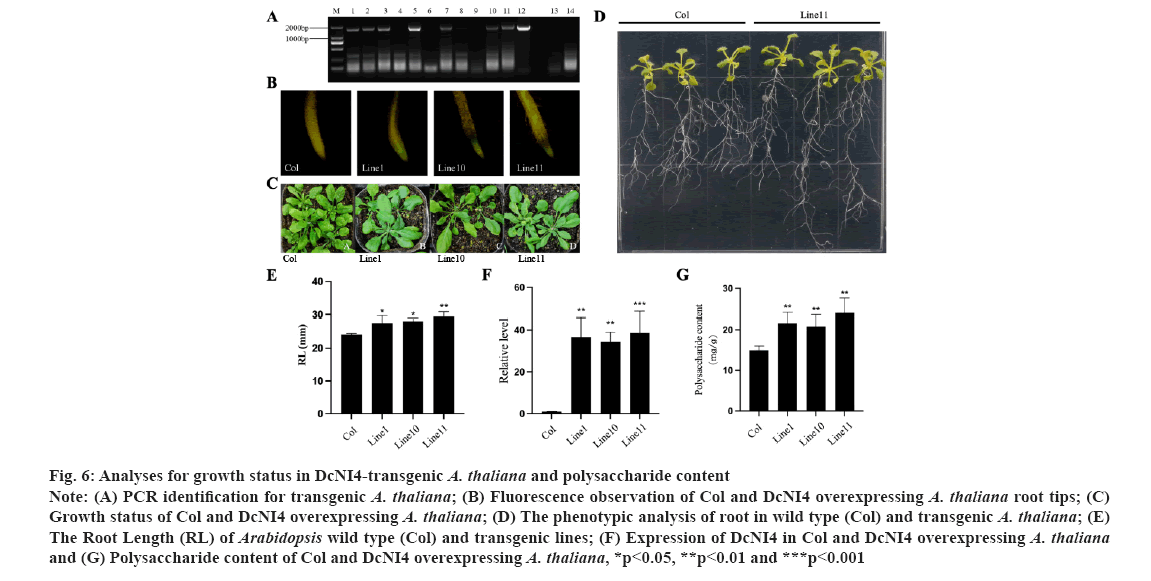
Overexpression of DcNI4 in Arabidopsis increased polysaccharide biosynthesis. To analyze the function of DcNI4 gene in regulating the polysaccharide synthesis, the DcNI4 overexpression vector was introduced into Arabidopsis plants and seven transgenic lines were obtained by PCR and sequencing. Here M indicates DNA marker, 1-11 indicates PCR product from DcNI4 transgenic A. thaliana, 12 indicates PCR product from recombinant plasmids, 13-14 indicates PCR product from other gene transgenic and Col A. thaliana (fig. 6A). Three transgenic Arabidopsis lines 1, 10 and 11 were randomly selected, and the root tips of the seedlings were observed under a fluorescent microscope. Compared with the wild type, there was obvious green fluorescence, named line 1, line 10 and line 11 (fig. 6B). The DcNI4 homozygous lines, line 1, line 10, line 11 and wild type Arabidopsis were cultured under the same conditions (fig. 6C). The leaf morphologies of DcNI4 transgenic Arabidopsis were not significantly different from those of wild type. Many studies had shown that NI gene family played an important role in root growth and development[28]. Statistical analysis showed that there was a significant difference in root length between the 2 w old transgenic and wild type Arabidopsis, and the root length of transgenic plants increased by 21.2 % (fig. 6D and fig. 6E). In addition, use gene-specific primers and qRT-PCR to detect the expression of DcNI4 in transgenic lines. In transgenic Arabidopsis lines, the expression levels of DcNI4 were significantly higher than those in wild type (fig. 6F). The polysaccharide contents of three overexpression transgenic Arabidopsis were 24.86 mg/g, 22.46 mg/g and 24.92 mg/g. Compared with the wild type (18.92 mg/g), it was increased by 31.36 %, 18.70 % and 31.70 % respectively (fig. 6G). These results indicated that DcNI4 gene could increase polysaccharide content in transgenic Arabidopsis.
Fig 6: Analyses for growth status in DcNI4-transgenic A. thaliana and polysaccharide content
Note: (A) PCR identification for transgenic A. thaliana; (B) Fluorescence observation of Col and DcNI4 overexpressing A. thaliana root tips; (C) Growth status of Col and DcNI4 overexpressing A. thaliana; (D) The phenotypic analysis of root in wild type (Col) and transgenic A. thaliana; (E) The Root Length (RL) of Arabidopsis wild type (Col) and transgenic lines; (F) Expression of DcNI4 in Col and DcNI4 overexpressing A. thaliana and (G) Polysaccharide content of Col and DcNI4 overexpressing A. thaliana, *p<0.05, **p<0.01 and ***p<0.001
NI can hydrolyze sucrose to fructose and glucose irreversibly. It is highly substrate specific and sucrose appears to be the only substrate[14,15,30]. Polysaccharides are the main effective ingredient in D. candidum, the catabolism of sucrose supplies the required precursors for polysaccharide synthesis. Here, we identified eight members ofDcNI family in D. candidum using genome-wide analysis to study the mechanism by which DcNI regulated polysaccharide synthesis. Transcriptome data showed that DcNI4 was expressed in multiple tissues of D. candidum, with the highest expression level in the stems (fig. 1A), which was consistent with the high content of polysaccharides in the stems[31]. In plants kingdom, sucrose is an important energy substance and the catabolism of sucrose produces different signal transduction, which plays an important role in plant biotic and abiotic stress[32]. Under drought and low temperature conditions, the expression of DcNI4 did not decrease due to the reduction of sucrose catabolism, but increased to a certain extent (fig. 1B and fig. 1C). We speculated that DcNI4 might regulate the resistance to low temperature and drought stress.
The characteristic analysis of DcNI4 protein showed that it was an acidic hydrophobic proteins and non-transmembrane protein. The instability index greater than 40 indicated that the protein was less stable in vitro. In other species, the function of NIs was poorly understood due to their instability and low enzyme activity[13]. Signal peptide analysis indicated that DcNI4 was a non-secreted protein and it was related to its biological function in the cytoplasm (fig. 2). The phylogenetic tree indicated that DcNI4 may be homologous to the NIs in A. thaliana, V. vinifera and P. equestris (fig. 3). The sequence of DcNI4 was highly similar to AtCINV1 and the similarity was 80.84 %. AtCINV1 has been reported to regulate lateral root growth in response to stress[28].
NI was divided into α-group and β-group according to the evolutionary relationships and β-group NI had been reportedly located in the cytoplasm[33]. The online software Softberry was used in this study to predict the possible localization of DcNI4 in the cytoplasm. Subsequently, DcNI4-GFP was transiently expressed in N. benthamiana leaves and DcNI4 was located in the cytoplasm (fig. 4). This was consistent with the fact that it belongs to β-group and was predicted to have no signal peptide.
The expression patterns indicated that DcNI4 exhibited different expression levels in roots, stems and leaves at different growth stages in D. candidum. At 6 mo, the expression of DcNI4 in stems was significantly higher than that in roots and leaves (fig. 5B). This might be related to the large amount of sucrose decomposed to provide the energy for growth and the large accumulation of active ingredient polysaccharides in stem, indicating that DcNI4 might be a key gene in this process.
Sucrose regulates plant growth and development through the catabolism by NI to produce glucose and fructose[34]. Mutations in OsCYT-INV1 cause sucrose accumulation and hexose reduction, leading to root growth defects in Oryza sativa[35]. The homozygous mutants of LjINV1 showed a severe reduction in the growth of roots in Lotus japonicus[36]. An NI gene of M. esculenta Neutral Invertase (MeNINV1) could enhance sucrose catabolism and promote vegetative growth[37]. The gene expression and enzymatic activity of CaNINV5 showed that it might be the main NINV gene for sucrose hydrolysis during pepper fruit development[17]. By analyzing root phenotypes of 2 w old transgenic Arabidopsis plants overexpression DcNI4, we found the increase of DcNI4 expression promoted root development and increased root length (fig. 6D and fig. 6E). It had been reported that hexose facilitated cell division and expansion, whereas sucrose facilitated differentiation and maturation[8]. The increase of primary root length in transgenic Arabidopsis might be related to sucrose hydrolysis. These studies fully demonstrated that NI regulated plant growth and development by affecting sucrose metabolism.
The determination of total polysaccharide content showed that the polysaccharide content of transgenic Arabidopsis was significantly higher than wild type Arabidopsis (fig. 6G). The result suggested that DcNI4 could promote polysaccharide accumulation by involving in sucrose hydrolysis. In follow-up research, if the genetic transformation system could be established, the function of DcNI4 can be further verified in D. candidum. These results will provide a selectable target and reliable basis for cultivating D. candidum varieties with high medicinal ingredient contents through metabolic engineering.
In potato, the expression of the invertase gene had been found significantly upregulated under abiotic (heat, drought and salt) stress conditions[38]. Sucrose metabolism, particularly the catabolism of sucrose by invertase, played a central role in plant responses to low-temperature stress[39]. The MBS and LTR in the promoter of DcNI4 indicated that its function in abiotic stress resistance remained to be further explored.
In this study, we discovered a NI DcNI4 that was involved in the synthesis pathway of polysaccharidesin D. catenatum. DcNI4 gene was found to be highly expressed in stem, which was consistent with the high polysaccharide content in stem of D. catenatum. In A. thaliana, overexpression of DcNI4 could promote the accumulation of polysaccharides, thereby affecting the root development. Under adverse conditions, sucrose catabolism occurs but DcNI4 expression increased.
Funding:
This work was funded by the National Natural Science Foundation of China (No. 82170274 and 82225047) and the National Key Research and Development Program of China (No. 2022YFC3501703).
Author’s contributions:
Luyao Yu, Kai Hao and Zhanpin Zhu contributed equally to this work. Kai Hao, Luyao Yu and Lei Zhang conceived and designed the entire research plans; Kai Hao, Luyao Yu and Chen Liu performed most of the experiments and Kai Hao, Luyao Yu, Zhanpin Zhu, Ruiyang Yao and Lei Zhang wrote the manuscript.
Conflict of interests:
The authors declared no conflict of interest.
References
- Si JP, Zhang Y, Luo YB, Liu JJ, Liu ZJ. Herbal textual research on relationship between Chinese medicine "Shihu" (Dendrobium spp.) and "Tiepi Shihu" (D. catenatum). Zhongguo Zhong Yao Za Zhi 2017;42(10):2001-5.
[Crossref] [Google scholar] [PubMed]
- Chen WH, Wu JJ, Li XF, Lu JM, Wu W, Sun YQ, et al. Isolation, structural properties, bioactivities of polysaccharides from Dendrobium officinale Kimura et. Migo: A review. Int J Biol Macromol 2021;184:1000-13.
[Crossref] [Google scholar] [PubMed]
- Cheng J, Dang PP, Zhao Z, Yuan LC, Zhou ZH, Wolf D, et al. An assessment of the Chinese medicinal Dendrobium industry: Supply, demand and sustainability. J Ethnopharmacol 2019;229:81-8.
[Crossref] [Google scholar] [PubMed]
- Rao J, McClements DJ. Food-grade microemulsions, nanoemulsions and emulsions: Fabrication from sucrose monopalmitate & lemon oil. Food Hydrocoll 2011;25(6):1413-23.
[Crossref] [Google scholar] [PubMed]
- Koch KE, Wu Y, Xu J. Sugar and metabolic regulation of genes for sucrose metabolism: Potential influence of maize sucrose synthase and soluble invertase responses on carbon partitioning and sugar sensing. J Exp Bot 1996;47:1179-85.
[Crossref] [Google scholar] [PubMed]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell 2002;14(1):185-205.
[Crossref] [Google scholar] [PubMed]
- Yoon J, Cho LH, Tun W, Jeon JS, An G. Sucrose signaling in higher plants. Plant Sci 2021;302:110703.
[Crossref] [Google scholar] [PubMed]
- Koch K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 2004;7(3):235-46.
[Crossref] [Google scholar] [PubMed]
- Lin YK, Zhu YQ, Si JP, Qin L, Zhu Y, Wu LS, Liu JJ. Effects of cultivation environments on Dendrobium catenatum. Zhongguo Zhong Yao Za Zhi 2017;42(16):3084-9.
[Crossref] [Google scholar] [PubMed]
- Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 2016;14(1):169-76.
[Crossref] [Google scholar] [PubMed]
- Shahryar N, Maali-Amiri R. Metabolic acclimation of tetraploid and hexaploid wheats by cold stress-induced carbohydrate accumulation. J Plant Physiol 2016;204:44-53.
[Crossref] [Google scholar] [PubMed]
- Ruan YL, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation and seed development. Plant Cell 2003;15(4):952-64.
[Crossref] [Google scholar] [PubMed]
- Roitsch T, González MC. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci 2004;9(12):606-13.
[Crossref] [Google scholar] [PubMed]
- Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinha AK. Extracellular invertase: Key metabolic enzyme and PR protein. J Exp Bot 2003;54(382):513-24.
[Crossref] [Google scholar] [PubMed]
- Vargas W, Cumino A, Salerno GL. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 2003;216(6):951-60.
[Crossref] [Google scholar] [PubMed]
- Arabidopsis genome initiative genomeanalysis@ tgr. org genomeanalysis@ gsf. de. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000;408(6814):796-815.
[Crossref] [Google scholar] [PubMed]
- Shen LB, Yao Y, He H, Qin YL, Liu ZJ, Liu WX, et al. Genome-wide identification, expression and functional analysis of the alkaline/neutral invertase gene family in pepper. Int J Mol Sci 2018;19(1):224.
[Crossref] [Google scholar] [PubMed]
- Chen Z, Gao K, Su X, Rao P, An X. Genome-wide identification of the invertase gene family in Populus. PLoS One 2015;10(9):e0138540.
[Crossref] [Google scholar] [PubMed]
- Qian W, Yue C, Wang Y, Cao H, Li N, Wang L, et al. Identification of the invertase gene family (INVs) in tea plant and their expression analysis under abiotic stress. Plant Cell Rep 2016;35:2269-83.
[Crossref] [Google scholar] [PubMed]
- Ji X, van den Ende W, van Laere A, Cheng S, Bennett J. Structure, evolution and expression of the two invertase gene families of rice. J Mol Evol 2005;60(5):615-34.
[Crossref] [Google scholar] [PubMed]
- Murayama S, Handa H. Genes for alkaline/neutral invertase in rice: Alkaline/neutral invertases are located in plant mitochondria and also in plastids. Planta 2007;225(5):1193-203.
[Crossref] [Google scholar] [PubMed]
- Nonis A, Ruperti B, Pierasco A, Canaguier A, Adam-Blondon AF, di Gaspero G, et al. Neutral invertases in grapevine and comparative analysis with Arabidopsis, poplar and rice. Planta 2008;229(1):129-42.
[Crossref] [Google scholar] [PubMed]
- Liu C, Xi H, Chen X, Zhao Y, Yao J, Si J, et al. Genome-wide identification and expression pattern of alkaline/neutral invertase gene family in Dendrobium catenatum. Biotechnol Biotechnol Equip 2021;35(1):527-37.
[Crossref] [Google scholar] [PubMed]
- Zou LH, Wan X, Deng H, Zheng BQ, Li BJ, Wang Y. RNA-seq transcriptomic profiling of crassulacean acid metabolism pathway in Dendrobium catenatum. Sci Data 2018;5(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Rombauts S, Déhais P, van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 1999;27(1):295-6.
[Crossref] [Google scholar] [PubMed]
- Zhang TQ, Lian H, Tang H, Dolezal K, Zhou CM, Yu S, et al. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 2015;27(2):349-60.
[Crossref] [Google scholar] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001;25(4):402-8.
[Crossref] [Google scholar] [PubMed]
- Qi X, Wu Z, Li J, Mo X, Wu S, Chu J, et al. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol Biol 2007;64:575-87.
[Crossref] [Google scholar] [PubMed]
- Siddappaji MH, Scholes DR, Krishnankutty SM, Calla B, Clough SJ, Zielinski RE, et al. The role of invertases in plant compensatory responses to simulated herbivory. BMC Plant Biol 2015;15(1):1-2.
[Crossref] [Google scholar] [PubMed]
- Sturm A. Invertases: Primary structures, functions and roles in plant development and sucrose partitioning. Plant Physiol 1999;121(1):1-8.
[Crossref] [Google scholar] [PubMed]
- Gao F, Cao XF, Si JP, Chen ZY, Duan CL. Characterization of the alkaline/neutral invertase gene in Dendrobium officinale and its relationship with polysaccharide accumulation. Genet Mol Res 2016;15(2):1-8.
[Crossref] [Google scholar] [PubMed]
- Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS. Sugar input, metabolism and signaling mediated by invertase: Roles in development, yield potential and response to drought and heat. Mol Plant 2010;3(6):942-55.
[Crossref] [Google scholar] [PubMed]
- Rende U, Wang W, Gandla ML, Jönsson LJ, Niittylä T. Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol 2017;214(2):796-807.
[Crossref] [Google scholar] [PubMed]
- Ruan YL. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol 2014;65:33-67.
[Crossref] [Google scholar] [PubMed]
- Jia L, Zhang B, Mao C, Li J, Wu Y, Wu P, et al. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.). Planta 2008;228(1):51-9.
[Crossref] [Google scholar] [PubMed]
- Welham T, Pike J, Horst I, Flemetakis E, Katinakis P, Kaneko T, et al. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J Exp Bot 2009;60(12):3353-65.
[Crossref] [Google scholar] [PubMed]
- Wang YJ, Zhen XH, Zhou YJ, Wang YL, Hou JY, Wang X, et al. MeNINV1: An alkaline/neutral invertase gene of Manihot esculenta, enhanced sucrose catabolism and promoted plant vegetative growth in transgenic Arabidopsis. Plants 2022;11(7):946.
[Crossref] [Google scholar] [PubMed]
- Abbas A, Shah AN, Shah AA, Nadeem MA, Alsaleh A, Javed T, et al. Genome-wide analysis of invertase gene family and expression profiling under abiotic stress conditions in potato. Biology 2022;11(4):539.
[Crossref] [Google scholar] [PubMed]
- Wang X, Chen Y, Jiang S, Xu F, Wang H, Wei Y, et al. PpINH1, an invertase inhibitor, interacts with vacuolar invertase PpVIN2 in regulating the chilling tolerance of peach fruit. Hortic Res 2020;7:1-14.
[Crossref] [Google scholar] [PubMed]

 tertiary structure for DcNI4
tertiary structure for DcNI4