- *Corresponding Author:
- Li Yin
National Health Commission of the People's Republic of China (NHC) Key Laboratory of Control of Tropical Diseases, School of Tropical Medicine, Hainan Medical University, Haikou, Hainan Province 571199, China
E-mail: hyinli@163.com
| Date of Received | 22 December 2022 |
| Date of Revision | 10 July 2023 |
| Date of Acceptance | 25 January 2024 |
| Indian J Pharm Sci 2024;86(1):336-345 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Breast cancer is one of the most common cancers in women worldwide. Therefore, it is important to understand the mechanism of breast cancer occurrence and development. In this study, we aimed to explore the potential use of chromosome 19 open reading frame 48 as a predictive biomarker by examining its expression patterns and how they respond to various treatments. Additionally, we conducted molecular docking simulations to determine the likelihood of drugs binding to this protein. We analyzed the expression of chromosome 19 open reading frame 48 in breast cancer using online tools. The relationship between its expression and survival times was explored too. Receiver operating characteristic analysis and molecular docking was used to explore the possibility of chromosome 19 open reading frame 48 as a biomarker. Chromosome 19 open reading frame 48 is highly expressed in breast cancer, and the higher the expression, the shorter the patients survival. Chromosome 19 open reading frame 48 plays a crucial role in the development of breast cancer by participating in cell-cycle-related biological processes. Through receiver operating characteristic analysis and molecular docking simulations, it is likely that it is also a promising predictive biomarker. Chromosome 19 open reading frame 48 could be a prognostic biomarker and predictive biomarkers for predicting therapy response in breast cancer. This provides a theoretical basis for the research and treatment of breast cancer.
Keywords
Breast cancer, chromosome 19 open reading frame 48, predictive biomarker, molecular docking, morbidity, metastasis
Breast cancer, one of the most commonly diagnosed cancers among women worldwide, has been extensively studied[1]. According to statistics from the American Cancer Society (ACS), there were over 2.3 billion new breast cancer cases and 685 000 breast cancer deaths globally in 2020[2]. Breast cancer is considered a heterogeneous disease, with genetic and epigenetic changes having a significant impact on its development, progression, and metastasis. Therefore, it is essential to further understand the mechanisms underlying breast cancer development and progression, which may provide a new approach for early diagnosis and prognosis prediction. Early detection and easier treatment at an earlier stage can significantly reduce the morbidity and mortality associated with breast cancer[3].
Currently, the most popular ways for breast cancer clinical treatment are surgery and subsequent chemotherapy, targeted therapy, or radiation therapy. Studies have shown that surgery combined with drug therapy can effectively prevent recurrence and metastasis. However, frequent drug resistance has been a major challenge in the treatment of breast cancer. Long-term continuous chemo radiotherapy can lead to certain toxic side effects and reduce patient resistance, and its effectiveness in killing cancer cells is also poor.
Breast cancer can be divided into several categories; ductal carcinoma in situ, invasive ductal carcinoma, Triple Negative Breast Cancer (TNBC), inflammatory breast cancer, metastatic breast cancer, and others[4]. Among all breast cancer types, the TNBC subgroup is negative for Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal Growth Factor Receptor 2 (HER2)[5]. Therefore, TNBC lacks specific therapeutic targets. In addition, TNBC is highly invasive and leads to a poor prognosis. Therefore, the management of TNBC remains a huge clinical challenge. There is an urgent need to explore predictive biomarkers and therapeutic targets for TNBC patients. As a multifactorial disease, breast cancer is closely related to obesity and smoking[6,7]. The interplay between obesity and breast cancer risk largely hinges on the menopausal status of women. An ideal screening method should be accurate, painless, non-invasive and safe. However, discomfort, radiation exposure, false positive results and overtreatment are unavoidable when mammography is used[8,9]. Therefore, it is essential to investigate more precise and reactive biomarkers in breast cancer screening.
The development of breast cancer involves mutations in oncogenes and tumor suppressor genes, as well as other chromosomal abnormalities.
Gene special deletions or copy number changes in breast cancer may change the hyperplasia to ductal carcinoma in situ. Furthermore, breast cancer may also be triggered when transcription factors are dysregulated. Series of studies have indicated that oncogenic transcription factors are related to abnormal apoptosis control and cell cycle as well as cell invasion. Recent studies have confirmed that breast cancer may also be driven by epigenetic changes, including Deoxyribonucleic Acid (DNA) methylation, histone modifications and so on. Understanding the molecular mechanisms connected with breast cancer development and progression will provide strategies for identifying novel diagnostic and prognostic biomarkers while providing better prevention and treatment for breast cancer patients. Increasing evidence suggests that loss of tumor suppressor leads to increased expression of oncogenes, and increased expression of oncogenes can inhibit expression of tumor suppressor. Carcinogenesis is a multi-step phenomenon that often appears as changes in genetic levels and controls the major intracellular pathways of cell growth and development. Increased evidence suggests that oncogenes are involved in the breast cancer introduction. Modulation of oncogenes results in a cell function that signals and contributes to a tumorigenic phenotype. Therefore, at the same time, these signals produce stupendous amounts of protein, leading to cell growth and inhibition of apoptosis. The expression of specific genes is associated with ER, PR expression and other key characteristics such as cell proliferation, invasion and metastasis[10]. Therefore, specific genes have become candidate biomarkers for their pivotal role in breast cancer regulatory networks. Therefore, it is necessary to find potential biomarkers in high- risk breast cancer as a driving factor for early tumorigenesis and development.
Chromosome 19 open reading frame 48 (C19orf48), is located on chromosome 19q13.33. C19orf48-encoded peptides are widely expressed in kidney tumors and other histological solid tumors[11,12]. It has been found that the donor T cell response of HLA-A*0201 restricted secondary H antigen encoded by C19orf48 may contribute to the regression of renal cell carcinoma. In addition, C19orf48 has been shown to be an androgen response element that may be involved in protein synthesis and trafficking, oxidative stress, transcription, proliferation, apoptosis, and differentiation of prostate cancer[13]. Therefore, C19orf48 may be a potential biomarker for prostate cancer. In normal breast tissue, as an age-related DNA methylation (aDNAm), C19orf48 expression is decreased, which is inversely proportional to the methylation level. Because of the similar findings of breast tissue in healthy women and breast tumors, aDNAm might be a way to increase breast cancer risk with age[14].
Prognostic biomarkers can predict survival, as well as treatment response. In our study, we investigated the potential of C19orf48 as a predictive biomarker by analysing its expression and how it responds to different treatments. Additionally, we simulated the binding possibilities between drugs and this protein through molecular docking.
In our study, we analyzed the messenger Ribonucleic Acid (mRNA) expression of C19orf48 in several types of human cancers. We also explored the relationship between its expression and survival time of breast cancer patients using online databases. We selected genes co-expressed with C19orf48 and performed enrichment analysis on biological processes and pathways.
Materials and Methods
The mRNA expression of C19orf48 in cancers:
Expression of mRNA in adenocarcinomas: We utilized the Gene Expression Profiling Interactive Analysis (GEPIA) online tool (http://gepia.cancer-pku.cn/), a robust data mining platform based on The Cancer Genome Atlas (TCGA) data, to analyse the expression of the C19orf48 gene in nine types of adenocarcinomas. In our study, the parameters were set as follows; |Log2FC|=1, and p=0.01.
Enrichment analysis of the genes co-expressed with C19orf48: CBio Cancer Genomic Portal (http://cbioportal.org) is a comprehensive online web database. Cancer genomics data, including mutation, expression, and Copy Number Variation (CNV), can be explored[15]. The genes co-expressed with C19orf48 in breast cancer were downloaded from cBioPortal. The genes whose absolute correlation coefficient was >0.4 were selected for the enrichment analysis using Metascape (http://metascape.org). Using this tool, we identified the enriched ontology terms using the standard accumulative hypergeometric statistical test[16].
In our study, the enriched Gene Ontology (GO) biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were selected with the threshold of p=0.01[17-19]. At the same time, the relationship between the mRNA expression and CNV was explored.
The survival analysis using Kaplan-Meier plotter:
The Kaplan-Meier plotter (www.kmplot.com) is an online tool that is used for conducting a meta-analysis based on biomarker assessment. The effect of 54 675 genes on survival using 18 674 cancer samples including breast cancer, ovarian cancer, lung cancer, gastric cancer and pan-cancer was assessed[20]. In our study, Kaplan-Meier survival analysis was performed to explore the relationship between the expression of C19orf48 and the survival time of different type cancers patients. The logrank p value and the hazard ratio were calculated.
Receiver Operating Characteristic (ROC) analysis:
The predictive biomarker has the potential to aid in selecting a specific treatment over another. The ROC plotter (https://www.rocplot.org/) was employed to analyse the correlation between gene expression and the response to therapy, utilizing transcriptome data from breast and ovarian patients[21]. In our study, ROC analysis was performed to find the relationship between the expression of C19orf48 and the response to different therapy. The pathological response was selected.
Molecular docking:
Molecular docking is a powerful tool that has revolutionized our understanding of drug discovery and therapy. By computationally predicting the binding affinity of small molecules with their target proteins, molecular docking has enabled researchers to identify novel drug targets, develop more effective treatments, and improve patient outcomes. The relationship between the expression of C19orf48 and the response to different therapy has been performed using ROC analysis. To explore the possible combination forms of drugs used to treat breast cancer and C19orf48, we conducted molecular docking using Autodock Vina[22,23] and visualized by PyMol molecular graphics system (http://www.pymol.org). The computed structure model of C19orf48 was download from AlphaFold protein structure database[24]. The simile’s file of ligand obtained from PubMed[25] was converted to Protein Data Bank (PDB) format using the online tool NovoPro (https://www.novopro.cn/tools/ smiles2pdb).
Results and Discussion
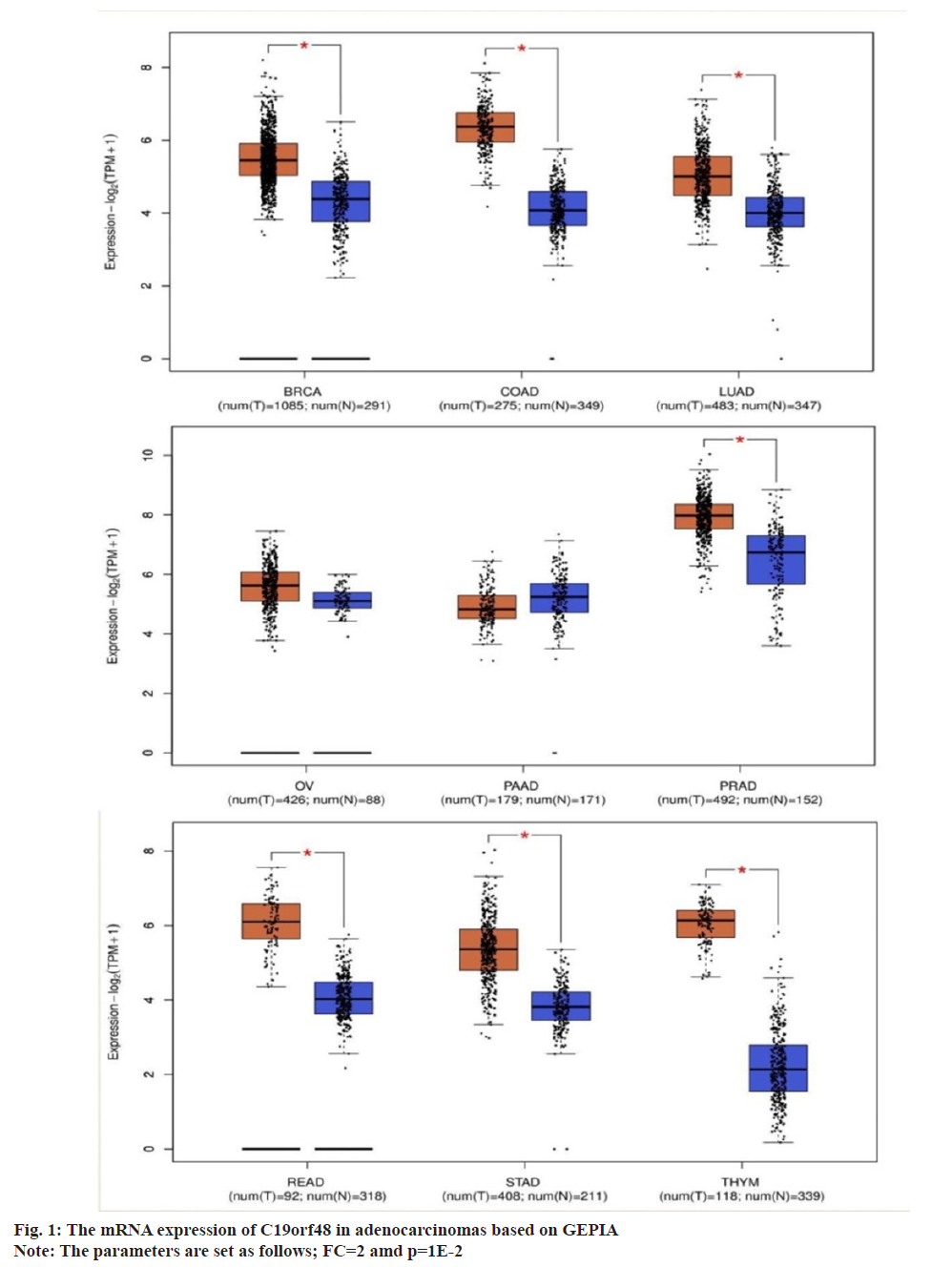
According to GEPIA results, it was found that C19orf48 was over-expressed in all adenocarcinomas including breast cancer compared to the normal samples which was shown in fig. 1.
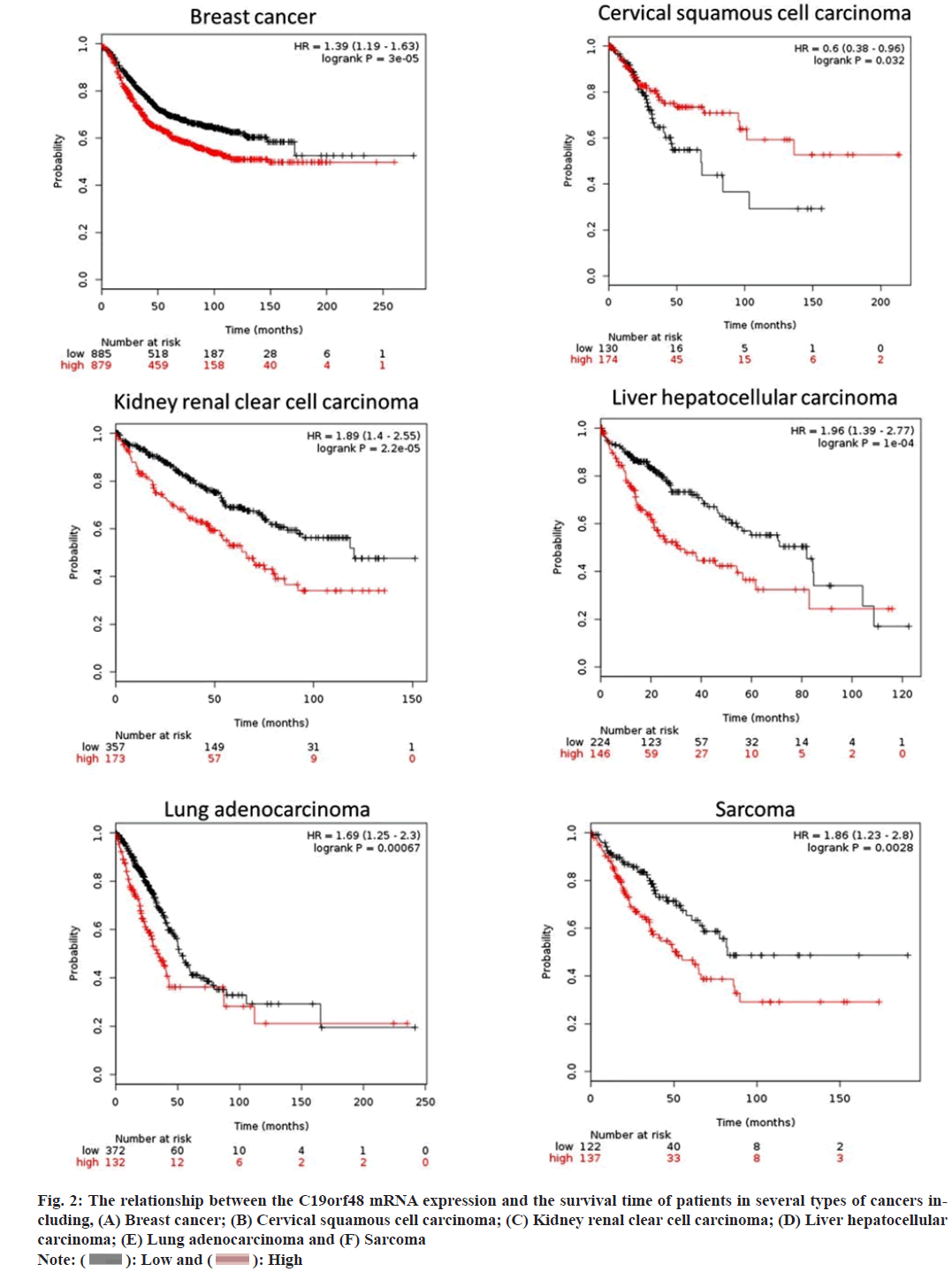
The Kaplan-Meir plotter was employed to identify the relationship between the expression of C19orf48 and the overall survival time. It showed that the elevated expression of C19orf48 contributed to the short overall survival time in several types of cancers including breast cancer, cervical squamous cell carcinoma, kidney renal clear cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma and sarcoma (fig. 2). All the above suggest that the C19orf48 is an unfavorable prognostic biomarker in cancer development.
Fig. 2: The relationship between the C19orf48 mRNA expression and the survival time of patients in several types of cancers including, (A) Breast cancer; (B) Cervical squamous cell carcinoma; (C) Kidney renal clear cell carcinoma; (D) Liver hepatocellular carcinoma; (E) Lung adenocarcinoma and (F) Sarcoma
Note:  High
High
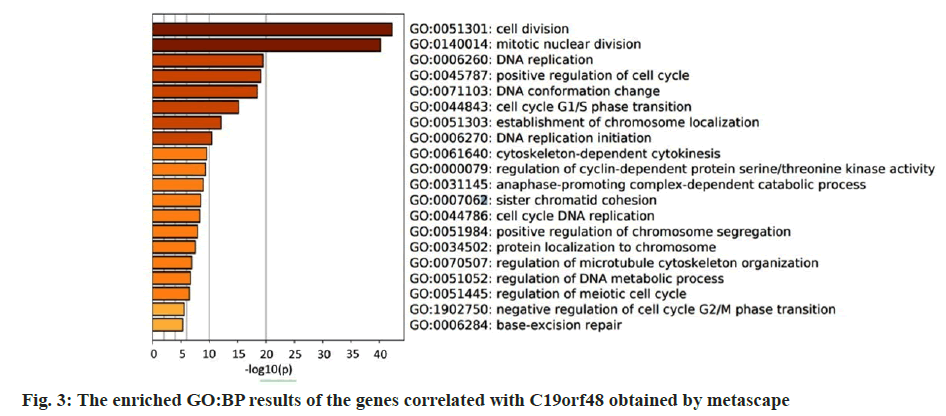
As a relatively new gene, the detailed role of C19orf48 in breast cancer is not known yet. We constructed the Protein-Protein Interaction (PPI) with the genes which co-expressed with C19orf48 using the breast cancer dataset. In together, 141 genes correlated with C19orf48 whose absolute Spearman's correlation coefficient score ≥0.4 were selected to construct the PPI network. It showed that the enriched GO:Base Pair (BP) terms include cell cycle related biological process which illustrated that the potential role of C1orf48 in cell proliferation as shown in fig. 3. The enriched KEGG pathways comprise mainly cell cycle, Human T-Lymphotropic Virus Type-1 (HTLV-I) infection etc., which was shown in Table 1.
| Term ID | Description | LogP |
|---|---|---|
| hsa04110 | Cell cycle | -27.611 |
| hsa04114 | Oocyte meiosis | -8.51 |
| hsa05166 | HTLV-I infection | -7.432 |
| hsa03030 | DNA replication | -7.222 |
| hsa05222 | Small cell lung cancer | -2.825 |
| hsa03440 | Homologous recombination | -2.753 |
Table 1: The Enriched KEGG Results of the Genes Co-Expressed with C19orf48 Obtained by Metascape
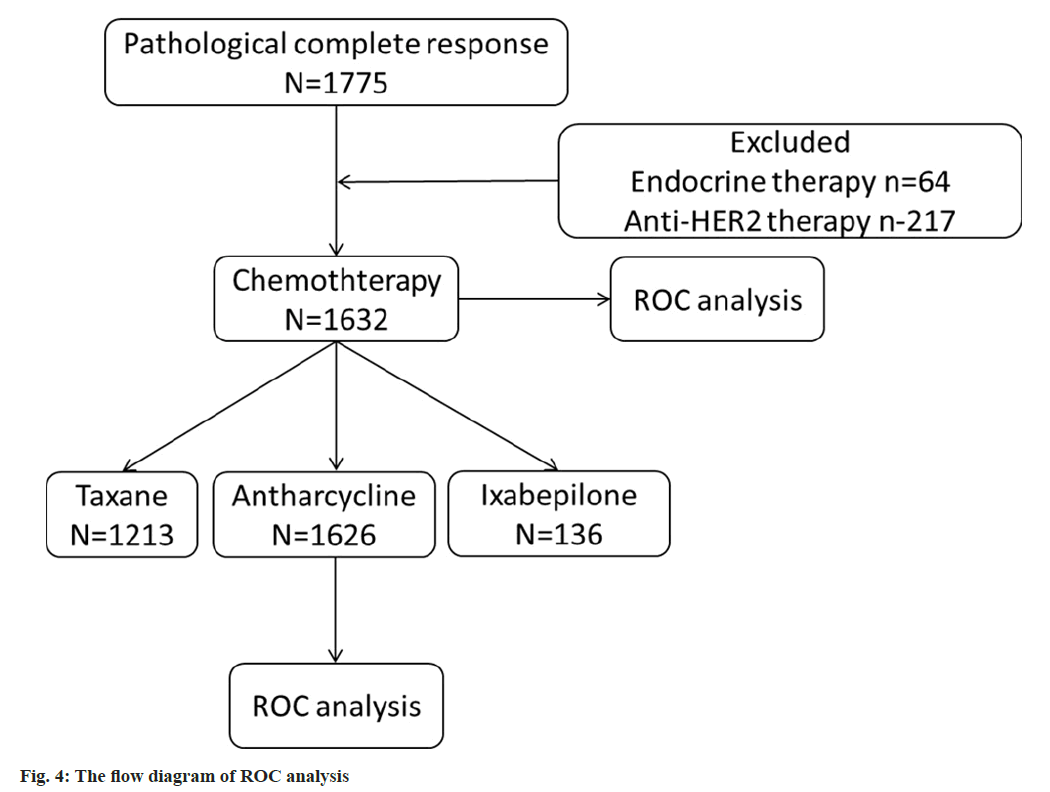
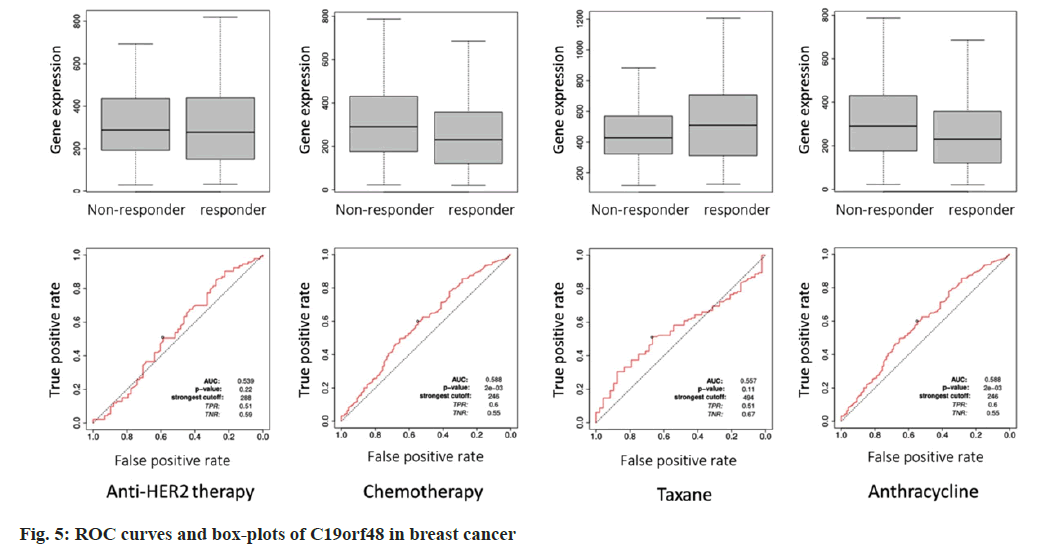
We analyzed C19orf48 for anti-HER2 therapy (p=0.22, Area Under the Curve (AUC)=0.539) and chemotherapy in breast cancer (p=2e-03, AUC=0.588) (fig. 4 and fig. 5). In general, C19orf48 has strong correlation to resistance especially chemotherapy (any) in all samples (n=1632). In order to explore the relation between C19orf48 expression and specific drugs, the ROC analysis was performed including taxane (p=0.11 and AUC=0.557) and anthracycline (p=0.588 and AUC=0.588). The ROC plots and the mean plots for C19orf48 are presented in fig. 5. It can be inferred that C19orf48 could be a target of anthracycline.
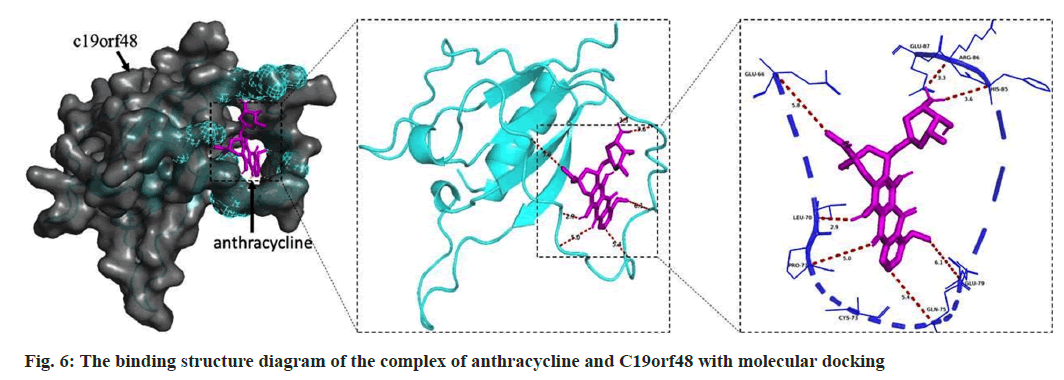
It is generally believed that the lower the binding energy between a ligand and a receptor, the more stable it is. According to the results of molecular docking, the binding energy is -7.7 kJ/mol, indicating that anthracycline has a high binding activity with C19orf48 (fig. 6). Based on this, it can be speculated that C19orf48 may be the binding site of anthracycline.
Breast cancer poses a great threat to the health of the world and is one of cancer death reasons among women[26]. Breast cancer mortality has declined due to early detection and advanced treatment. However, the complex mechanisms of breast cancer development and progression remain a significant barrier to the treatment of this disease. At present, effective tools for evaluating the treatment effects and predicting prognosis in breast cancer patients are always wanted. The Tumor, Node and Metastasis (TNM) staging system can accurately assess tumor size, lymphatic involvement and distant metastasis[27,28]. A more precise system for indicating the molecular characteristics of breast cancer is needed for personalization and precise treatment. The study of breast cancer biomarkers will fill this gap. Early diagnosis is critical for the prognosis. However, the median size of clinically diagnosed breast tumors is currently 2 to 2.5 cm, which may be advanced (stage III) breast tumors that have metastasized from tissue to axillary lymph nodes. At present, there is a lack of highly accurate breast cancer diagnostic tests. For example, the sensitivity of standard mammography tests is only 54 % to 77 %. Ultrasound, Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) have higher sensitivity but higher cost[29]. Therefore, a more precise, cost-effective and non-invasive alternatives for diagnosis in breast cancer is what urgently needed currently. In oncology, biomarkers are regarded can be used to predict the malignant potential, prognosis, or response to treatment of a tumor. Therefore, a more effective marker is urgently needed. Conventional histopathology is difficult to predict the breast cancer patient prognosis. Therefore, biomarkers indicating intrinsic features of tumors at the molecular level have become a hotspot in biomedical research. Many biomarkers are widely used in current clinical practice[30]. For example, hormone receptors for breast cancer subtype classification and genes associated with genome maintenance can be used to predict breast cancer susceptibility[31]. ER, PR and HER2 have been identified as prognostic biomarkers and therapeutic targets for breast cancer[32,33]. However, many biomarkers require further investigation in order to find better biomarkers and drug development goals[34]. In addition, many single- or multi-gene biomarkers have been allowed in clinical application of breast cancer patient prognosis and to promote the breast cancer diagnosis and management, especially in the early detection, accurate prediction of metastasis and treatment options. However, most of them are still in the preclinical stage. Therefore, it is necessary to explore more sensitive markers in clinical development to improve the breast cancer patient diagnosis, prognosis and therapy. Despite significant advances in the prevention, diagnosis, and therapy, metastatic breast cancer is still a disease can`t be cure. At the molecular level, breast cancer, one of the heterogeneous diseases, whose development and progression is regulated by genes connected with cell growth, proliferation and differentiation. With the advent of the era of targeted therapy, increasing molecularly targeted drugs are available for clinical practice. However, the main challenge currently remains to identify predictive biomarkers for selecting the best treatment to protect patients with breast cancer from treatment-related side effects and to minimize treatment costs.
In our study, the GEPIA database was employed to identify the expression of C19orf48 in different type of cancers. GEPIA has the most comprehensive spectrum of cancer mutations, gene expression data and related clinical information, which facilitates the discovery of new biomarkers or potential therapeutic targets. In addition, cBioPortal integrates data from 126 oncology genome research projects, including TCGA, the International Cancer Genome Consortium and other large oncology research projects, in addition, it includes data from 28 000 cases[35]. In recent years, ever-increasing amounts and quality of data have caused to the unit of biological networks, with the final goal of revealing potential cellular processes. Therefore, PPIs have become one of the most vital and researched networks. The PPI network is a useful tool for understanding cell function, disease mechanisms, and drug design or repositioning. It helps to study the molecular mechanisms of disease from a systems perspective and to discover new drug targets.
C19orf48 encodes a minor histocompatibility antigen. In renal cell carcinoma patients, this antigen is found being identified by CD8+ cytotoxic T cells. After Major Histocompatibility Complex (MHC)-matched allergic nonogene ablative allogeneic Hematopoietic Cell Transplantation (HCT), induce T cell responses to HLA-A*0201 restricted secondary H antigen encoded by C19orf48 may contribute to regression of renal cell carcinoma. C19orf48 is up-regulated in prostate cancer. Prostate development and maintenance are dependent on androgen and androgen receptors. The androgen pathway remains important in prostate cancer. One study evaluated the transcriptome results of androgen-resistant prostate cancer cells using a Long Serial Analysis of Gene Expression (LongSAGE) library of gene expression showed that C19orf48 is upregulated in the androgen response. The research indicated that C19orf48 is vital in cell-cycle related processes.
Breast cancer incidence is positively correlated with age, and breast cancer gene methylation is also associated with aging. The research on aDNAm in women’s normal breast tissue without evidence of cancer or breast benign disease can provide insight into the naturally occurring conditions in the breast and promote the treatment of breast cancer. Previous study identified 1214 aDNAm in all autosomes, most of which are methylated and associated with increased age, and are commonly detected in CpG islands and non-enhancers. Among them, there is an obvious negative connection between C19orf48 gene expression and methylation level. Furthermore, based on the independent TCGA- breast cancer dataset, compared to adjacent normal tissue and normal breast tissue, C19orf48 verified in women breast tissue without breast cancer history is highly methylated in breast tumors. Compared to other subtypes, C19orf48 has a lower level of methylation in basal tumors. Therefore, C19orf48 could contribute to the development of breast cancer.
In this study, we evaluated the potential of C19orf48 as a predictive biomarker by analysing its expression levels in different cancer tissues and comparing them to patient outcomes. We also simulated the binding possibilities between drugs and C19orf48 through molecular docking, which could help identify potential drug targets.
The results of the study showed that C19orf48 expression was significantly higher in cancer tissues compared to normal tissues, and it was associated with poor survival outcomes and resistance to treatment. Molecular docking simulations also suggested that C19orf48 may be a promising target for developing new anti-cancer drugs.
Overall, these findings highlight the potential of C19orf48 as a prognostic and predictive biomarker. According to our molecular docking results, C19orf48 may be a functional site of anthracycline.
C19orf48 may be a promising prognostic biomarker. At the same time, through ROC analysis and molecular docking simulations, it is likely that it is also a promising predictive biomarker. To the best of our knowledge, this is the first report describing the expression of C19orf48, its potential driving factors leading to this expression and its effects.
Funding:
This study was supported by Chunhui Project Foundation of the Education Department of China (GGH200002), Hainan Provincial Natural Science Foundation of China (820RC652, 820QN270), Hainan Medical University Program for Talents Introduction (XRC190036).
Authors' contributions:
The study conception and design by Li Yin; the analysis and interpretation of the data by Li Yin, Haoxiu, Na He, and Qinghui Sun; and the drafting of the paper by Li Yin. All authors reviewed the results and approved the final version of the manuscript.
Conflict of interests:
The authors declared no conflict of interests.
References
- Spronk I, Schellevis FG, Burgers JS, de Bock GH. Incidence of isolated local breast cancer recurrence and contralateral breast cancer: A systematic review. Breast 2018;39:70-9.
[Crossref] [Google Scholar] [PubMed]
- Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022;66:15-23.
[Crossref] [Google Scholar] [PubMed]
- de Munck L, Fracheboud J, de Bock GH, den Heeten GJ, Siesling S, Broeders MJ. Is the incidence of advanced-stage breast cancer affected by whether women attend a steady-state screening program? Int J cancer 2018;143(4):842-50.
[Crossref] [Google Scholar] [PubMed]
- Gonzalez-Alonso P, Cristobal I, Zazo S, Martin-Aparicio E, Chamizo C, Madoz-Gurpide J, et al. Recent insights into the development of preclinical trastuzumab-resistant HER2+ breast cancer models. Curr Med Chem 2018;25(17):1976-98.
[Crossref] [Google Scholar] [PubMed]
- Saliou A, Bidard FC, Lantz O, Stern MH, Vincent-Salomon A, Proudhon C, et al. Circulating tumor DNA for triple-negative breast cancer diagnosis and treatment decisions. Expert Rev Mol Diagn 2016;16(1):39-50.
[Crossref] [Google Scholar] [PubMed]
- Kispert S, Schwartz T, McHowat J. Cigarette smoke regulates calcium-independent phospholipase A2 metabolic pathways in breast cancer. Am J Pathol 2017;87(8):1855-66.
[Crossref] [Google Scholar] [PubMed]
- Liu YL, Saraf A, Catanese B, Lee SM, Zhang Y, Connolly EP, et al. Obesity and survival in the neoadjuvant breast cancer setting: Role of tumor subtype in an ethnically diverse population. Breast Cancer Res Treat 2018;167(1):277-88.
[Crossref] [Google Scholar] [PubMed]
- Eriksson L, Bergh J, Humphreys K, Wärnberg F, Törnberg S, Czene K. Time from breast cancer diagnosis to therapeutic surgery and breast cancer prognosis: A population-based cohort study. Int J Cancer 2018;143(5):1093-104.
[Crossref] [Google Scholar] [PubMed]
- Phillips M, Cataneo RN, Cruz-Ramos JA, Huston J, Ornelas O, Pappas N, et al. Prediction of breast cancer risk with volatile biomarkers in breath. Breast Cancer Res Treat 2018;170(2):343-50.
[Crossref] [Google Scholar] [PubMed]
- Shidfar A, Costa FF, Scholtens D, Bischof JM, Sullivan ME, Ivancic DZ, et al. Expression of miR-18a and miR-210 in normal breast tissue as candidate biomarkers of breast cancer risk. Cancer Prev Res 2017;10(1):89-97.
[Crossref] [Google Scholar] [PubMed]
- Tykodi SS, Fujii N, Vigneron N, Lu SM, Mito JK, Miranda MX, et al. C19orf48 encodes a minor histocompatibility antigen recognized by CD8+ cytotoxic T cells from renal cell carcinoma patients. Clin Cancer Res 2008;14(16):5260-9.
[Crossref] [Google Scholar] [PubMed]
- Tykodi SS, Warren EH, Thompson JA, Riddell SR, Childs RW, Otterud BE, et al. Allogeneic hematopoietic cell transplantation for metastatic renal cell carcinoma after nonmyeloablative conditioning: Toxicity, clinical response, and immunological response to minor histocompatibility antigens. Clin Cancer Res 2004;10(23):7799-811.
[Crossref] [Google Scholar] [PubMed]
- Song MA, Brasky TM, Weng DY, McElroy JP, Marian C, Higgins MJ, et al. Landscape of genome-wide age-related DNA methylation in breast tissue. Oncotarget 2017;8(70):114648-62.
[Crossref] [Google Scholar] [PubMed]
- Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi PW, et al. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. J Exp Clin Cancer Res 2018;37:298.
[Crossref] [Google Scholar] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):11.
[Crossref] [Google Scholar] [PubMed]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta-and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe 2015;18(6):723-35.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2018;46(D1):D649-55.
[Crossref] [Google Scholar] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res 2015;43(D1):D1049-56.
[Crossref] [Google Scholar] [PubMed]
- Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, et al. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160(3):439-46.
[Crossref] [Google Scholar] [PubMed]
- Fekete JT, Gyorffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3104 breast cancer patients. Int J Cancer 2019;145(11):3140-51.
[Crossref] [Google Scholar] [PubMed]
- Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock vina 1.2.0: New docking methods, expanded force field, and python bindings. J Chem Inf Model 2021;61(8):3891-8.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. Alpha fold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022;50(1):439-44.
[Crossref] [Google Scholar] [PubMed]
- Fiorini N, Canese K, Bryzgunov R, Radetska I, Gindulyte A, Latterner M, et al. PubMed labs: An experimental system for improving biomedical literature search. Database (Oxford) 2018;2018:94.
[Crossref] [Google Scholar] [PubMed]
- Li X, Zhang Y, Meisel J, Jiang R, Behera M, Peng L. Validation of the newly proposed American Joint Committee on Cancer (AJCC) breast cancer prognostic staging group and proposing a new staging system using the national cancer database. Breast Cancer Res Treat 2018;171(2):303-13.
[Crossref] [Google Scholar] [PubMed]
- Chavez‐MacGregor M, Mittendorf EA, Clarke CA, Lichtensztajn DY, Hunt KK, Giordano SH. Incorporating tumor characteristics to the American joint committee on cancer breast cancer staging system. Oncologist 2017;22(11):1292-300.
[Crossref] [Google Scholar] [PubMed]
- Huang S, Chong N, Lewis NE, Jia W, Xie G, Garmire LX. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med 2016;8(1):34.
[Crossref] [Google Scholar] [PubMed]
- Kazarian A, Blyuss O, Metodieva G, Gentry-Maharaj A, Ryan A, Kiseleva EM, et al. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br J Cancer 2017;116(4):501-8.
[Crossref] [Google Scholar] [PubMed]
- Li G, Hu J, Hu G. Biomarker studies in early detection and prognosis of breast cancer. Adv Exp Med Biol 2017;1026:27-39.
[Crossref] [Google Scholar] [PubMed]
- Bagaria SP, Ray PS, Sim MS, Ye X, Shamonki JM, Cui X, et al. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg 2014;149(2):125-9.
[Crossref] [Google Scholar] [PubMed]
- Mudduwa L, Peiris H, Gunasekara S, Abeysiriwardhana D, Liyanage N, Rayala SK, et al. KIBRA; a novel biomarker predicting recurrence free survival of breast cancer patients receiving adjuvant therapy. BMC Cancer 2018;18(1):589.
[Crossref] [Google Scholar] [PubMed]
- Orucevic A, Chen J, McLoughlin JM, Heidel RE, Panella T, Bell J. Is the TNM staging system for breast cancer still relevant in the era of biomarkers and emerging personalized medicine for breast cancer-An institution's 10-year experience. Breast J 2015;21(2):147-54.
[Crossref] [Google Scholar] [PubMed]
- Vella D, Marini S, Vitali F, di Silvestre D, Mauri G, Bellazzi R. MTGO: PPI network analysis via topological and functional module identification. Sci Rep 2018;8(1):5499.
[Crossref] [Google Scholar] [PubMed]
- Deng Y, He R, Zhang R, Gan B, Zhang Y, Chen G, et al. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: A study based on the cancer genome atlas, oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol Lett 2018;15(6):8556-72.
[Crossref] [Google Scholar] [PubMed]