- *Corresponding Author:
- Jingfang Zhu
Department of Obstetrics, People’s Hospital of Dongxihu, Wuhan, Hubei 430063, China
E-mail: Zhujingfang800531@163.com
| Date of Received | 17 January 2021 |
| Date of Revision | 08 November 2022 |
| Date of Acceptance | 06 May 2023 |
| Indian J Pharm Sci 2023;85(3):591-602 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Baicalein has anti-tumor effect and participates in multi-channel regulation mechanism. However, the mechanism of baicalein action has not been fully clarified in cervical cancer. Tumor protein D52, circ_0007364 and microRNA-665 expression were determined by quantitative reverse transcriptionpolymerase chain reaction. Tumor protein D52 and apoptosis-related proteins were detected by Western blot. Cell proliferation was measured via methyl thiazolyl tetrazolium, 5-ethynyl-2'-deoxyuridine and colony formation assays. The associations among microRNA-655, circ_0007364 and tumor protein D52 were determined by dual luciferase report assay. Cell invasion and apoptosis were assessed by Transwell assay and flow cytometry. The function experiments were carried out in vivo and in vitro. Baicalein significantly decreased circ_0007364 and tumor protein D52 expression and increased microRNA-665 expression in cervical cancer cell samples. Circ_0007364 or tumor protein D52 introduction and microRNA-665 down regulation weakened the suppressive effects of baicalein in cervical cancer cells. Moreover, circ_0007364 functioned by binding to microRNA-665 to regulate tumor protein D52 and affect cellular process in cervical cancer. Our founding showed that baicalein inhibited cervical cancer cell growth via regulating circ_0007364/microRNA-665/tumor protein D52 axis, further understanding the anticancer mechanism of baicalein and contributing to find more effective anticancer drug components for cervical cancer.
Keywords
Baicalein, circ_0007364, microRNA-665, tumor protein D52, cervical cancer
Cervical Cancer (CC) mainly occurs in the cervix and poses a serious threat to women’s life, constituting a leading cause of morbidity[1,2]. Moreover, most CCs are caused by Human Papillomavirus (HPV) [3]. The treatments of CC are mainly surgery and chemoradiotherapy[2]. Thus, exploring the treatment methods of CC and understanding the molecular mechanism will contribute to develop new strategies to hind CC.
Baicalein is a bioactive flavonoid used as the main active component of traditional Chinese medicine and its application effect in tumor treatment is very significant[4]. Various studies have proved that baicalein has anti-tumor effect and participates in multi-channel regulation mechanism in cancers, including Hepatocellular Carcinoma (HCC), breast cancer and CC[5-7]. For example, Wu et al.[8] pointed out that baicalein suppressed mammalian Target of Rapamycin Complex 1 (mTORC1) inhibitorinduced autophagy and could selectively promote the chemical sensitivity of Tumor-Initiating Cells (TICs) and HCC without affecting mouse and human primary hepatocytes. In addition, baicalein regulated breast cancer apoptosis and autophagy, so as to achieve anticancer effect[6]. Moreover, baicalein affected circ Hippocampus Abundant Transcript 1 (circHIAT1)/ microRNA (miR)-19a-3p pathway to inhibit cell growth in CC[9]. Nevertheless, how baicalein acts in CC remains to be explored. Understanding the anticancer molecular mechanism of baicalein will help to improve the treatment efficiency and treatment methods in CC.
Circular Ribonucleic Acid (circRNA) is a noncoding RNA that plays an indispensable role in cancers[10-12]. Interestingly, circRNA does not play a role alone in cells, but combines with miRNA to affect messenger RNA (mRNA) transcription, thus affecting the development and progress of cells[13,14]. With the deepening of research, circRNA was found to regulate cell development and disease development, including breast cancer, HCC, colorectal cancer, immune diseases and CC[15-19]. For example, circ_100367 bound to miR-217 to regulate Wnt3, further modulating esophageal squamous cell carcinomas cell growth[20]. Circ_101996 promoted CC cell proliferation by inducing miR-8075- dependent Targeting Protein for Xenopus Kinesinlike Protein 2 (TPX2) expression[21]. Previous work showed a high circ_0007364 expression in CC[22]. Interestingly, in our study, baicalein significantly decreased circ_0007364 expression, but the related regulatory pathway in CC has not been fully clarified. In this paper, we found that circ_0007364 content was reduced in baicalein-treated CC cells. Through further prediction and verification, we determine that baicalein inhibited cell growth and invasion through circ_0007364/miR-665/Tumor Protein D52 (TPD52) axis in CC.
Materials and Methods
Samples collection:
Twenty seven pairs of CC tissues and matched normal cervical tissue were collected from patients who have undergone a surgical resection in Wuchang Hospital Affiliated to Wuhan University of Science and Technology. The experiment has been approved by the ethics committee of Wuchang Hospital Affiliated to Wuhan University of Science and Technology, and written informed consent has been obtained from all patients. Collected tissues were stored in a refrigerator.
Cell culture, treatment and transfection:
SiHa, HeLa and Ect1/E6E7 cells were purchased from EK-Bioscience (Shanghai, China) and cultured in Dulbecco's Modified Eagle's medium (DMEM) contained 10 % Fetal Bovine Serum (FBS) and streptomycin/penicillin with 5 % Carbon dioxide (CO2) at 37° for followed experiments.
Circ_0007364 mimic (circ_0007364), small interfering RNA (siRNA) against circ_0007364 (sicirc_ 0007364), plasmid cloning Deoxyribonucleic Acid (pcDNA)-TPD52 and negative controls were obtained from GenePharma (Shanghai, China). MiR-665 mimic, miR-665 inhibitor and negative controls were purchased from Ribo Bio (Guangzhou, China). When the confluence of cells reached 70 %~90 %, cells were transfected into CC cells via Lipofectamine 2000 (Invitrogen, Carlsbad, California, United States of America (USA)). SiHa and HeLa cells were treated with baicalein (0, 10, 20, and 30 μg/ml) for 48 h and then were collected for the next experiments. Finally, 20 μg/ml baicalein was used to function experiments.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR):
Collected cell and tissue RNA were extracted with TRIzol (Invitrogen). After that, RNA quality was assessed through the NanoDrop™ 2000c. According to the detection concentration, the volume required for RNA was calculated and 1 μg RNA was reversely transcribed into complementary Deoxyribonucleic Acid (cDNA) utilizing highcapacity reverse transcription reagents (Qiagen, Hilden, Germany). Then, the circRNA and mRNA expression was detected SYBR Premix Ex TaqTM II (Invitrogen). TaqMan miRNA assay was applied to detect miR-655. U6 was used as the control for miR-665. Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) was used as the control for circ_0007364, Protein Tyrosine Phosphatase 4A2 (PTP4A2) and TPD52. The primers are shown in Table 1. Gene expression was measured through calculating with 2−ΔΔCt method.
| Name | Primers for PCR (5’-3’) |
|---|---|
| hsa_circ_0007364 | F-GTGACGACTTTGGTTCGAGT |
| R-CTCACTTGTCAGCGAAAATGC | |
| TPD52 | F-GCAAGACGTGACAGCAACAT |
| R-TTGCAGGTCCCATCTGTGTAAG | |
| miR-665 | F-GCGTATGAACCAGGAGGCTGAG |
| R-CTCAACTGGTGTCGTGGAG | |
| GAPDH | F-GACAGTCAGCCGCATCTTCT |
| R-GCGCCCAATACGACCAAATC | |
| U6 | F-CTCGCTTCGGCAGCACA |
| R-AACGCTTCACGAATTTGCGT | |
| PTP4A2 | F-GGAGTGACGACTTTGGTTCG |
| R-TCAAAGCAAGTGCAACCAGC |
Table 1: Primer Sequences used for QRT-PCR
RNase R treatment:
After RNA extraction, added 3 U/μg Ribonuclease R (RNase R) (Sigma-Aldrich, St. Louis, Missouri, USA) and incubated at 37° for 15 min. The control sample of RNA was incubated without RNase R.
Western blot assay:
Total protein extraction was performed using Radioimmunoprecipitation Assay (RIPA) lysis buffer and then Bicinchoninic Acid (BCA) kits (Thermo Fishe, Carlsbad, California, USA) were utilized for protein concentration analysis. Subsequently, Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDSPAGE) was performed to separate protein on the midi-cell electrophoresis system and then the protein was transferred to nitrocellulose membrane. The membranes were incubated with anti-GAPDH (1:1000 dilution; Cat. No: AF0006; Beyotime, Shanghai, China), anti-cleavedcaspase- 3 (1:1000; Cat. No: AB2302, Abcam, Cambridge, Massachusetts, USA), anti- Matrix Metalloprotease-9 (MMP-9) (1:1000; cat. no. ab219372, Abcam) and anti-TPD52 (1:1000 dilution; Cat. No: AB182578, Abcam) at 4° overnight. Then the membrane was incubated with anti-rabbit or anti-mouse Immunoglobulin G (IgG) (1:1000 dilution; Cat. No: AB228414, Thermo Fisher). Finally, proteins blot was detected using an Electrochemiluminescence (ECL) system (Thermo Fisher).
Colony formation assay:
Transfected cells were seeded into six-well petri dishes added with DMEM and then went through 2 w incubation. Then cells were fixed with 4 % paraformaldehyde (Phygene, Fuzhou, China), followed by the staining with crystal violet (Sigma). Finally, cell colonies were assessed under microscope.
Methyl Thiazolyl Tetrazolium (MTT) assay:
Transfected cells were seeded into 96-well petri dishes added with DMEM and then went through 72 h incubation at 37°. Then, each well was added with 20 μl MTT solution (Sigma-Aldrich) and incubated for 4 h. After each well was added with 200 μl dimethyl sulfoxide (Sigma), each sample was measured at 490 nm..
Transwell invasion assay:
Transfected cells grew in the upper chamber of Transwell (six-well plate, Corning Costar, Lowell, Massachusetts, USA), and the Transwell inserts were coated with Matrigel. Complete DMEM medium was added into the bottom chamber. After incubation for 48 h, the invasion cells were fixed and stained with 0.1 % crystal violet (Sigma- Aldrich) solution and then counted on microscope.
Hematoxylin and Eosin (HE) Staining:
The tissue was fixed with 4 % formalin and dehydrated with gradient concentration ethanol. Subsequently, the tissue was paraffin-embedded and the tissue section was cut into 4 μm sections. Subsequently, the sections were stained with 3,3-Diaminobenzidine (DAB) and then subjected to counterstaining with hematoxylin. Finally, a microscope was used for observation.
5-Ethynyl-2'-Deoxyuridine (EdU) incorporation assay:
Transfected CC cells were incubated with 20 μM EdU reagent (RiboBio, Guangzhou, China) in 96-well plates for 2 h. Subsequently, cells were fixed with 4 % paraformaldehyde (Phygene) and stained in 5 μg/ml 4,6-Diamidino-2-Phenylindole (DAPI). After incubation for 30 min in the dark, EdU-positive cells were observed by fluorescence microscope.
Dual luciferase report assay:
Circ_0007364 and TPD52 3′ Untranslated Regions (3′ UTRs) contained binding sites of miR-665 were synthesized and inserted into the psiCHECK-2 vector (Promega, Madison, Wisconsin, USA), named WT-circ_0007364or WT-TPD52 3′UTR. In addition, the circ_0007364 or TPD52 3′UTR contained miR-665 mutated sites were synthesized and then were inserted into the psiCHECK-2 vector (Promega), named Mutant-Type (MUT)- circ_0007364 or MUT-TPD52 3′UTR. These vectors were co-transfected with miR-NC or miR- 665 mimic (miR-665) into cells via Lipofectamine 2000 (Invitrogen). At 48 h after culture, the firefly and Renilla luciferase activities acted as internal control.
Flow cytometry analysis:
Cell apoptosis was assessed by the FITC apoptosis detection kit (BD Biosciences, Franklin Lakes, New Jersey, USA). 2×105 transfected CC cells were seeded into well and incubated with 5 μl Annexin V-Fluorescein Isothiocyanate (FITC) and 5 μl Propidium Iodide (PI) in the dark. Eventually, cell apoptosis was measured using FACScan® flow cytometry (BD Biosciences).
Xenografts experiments:
This animal experiment was approved by the Ethics Committee of Wuchang Hospital Affiliated to Wuhan University of Science and Technology. HeLa cells (2×105) transfected with sh-circ_0007364 or sh-NC were subcutaneously injected into the right back of Bagg and Albino (BALB/c) female nude mice (4 w-5 w, n=24). The experiment was divided into four groups. After 8 d, baicalein (45 mg/kg) was given every 3 d and tumor volume was observed and measured every 3 d. 23 d later, the tumor was taken out, the tumor growth curve and tumor weight were obtained.
Statistical analysis:
Statistical analysis was displayed via using GraphPad Prism 7.0 software (GraphPad, Bethesda, Maryland, USA). Pearson correlation analysis, Student’s t-test or one-way analysis of variance was used for comparison. Statistical significance was indicated when p<0.05.
Results and Discussion
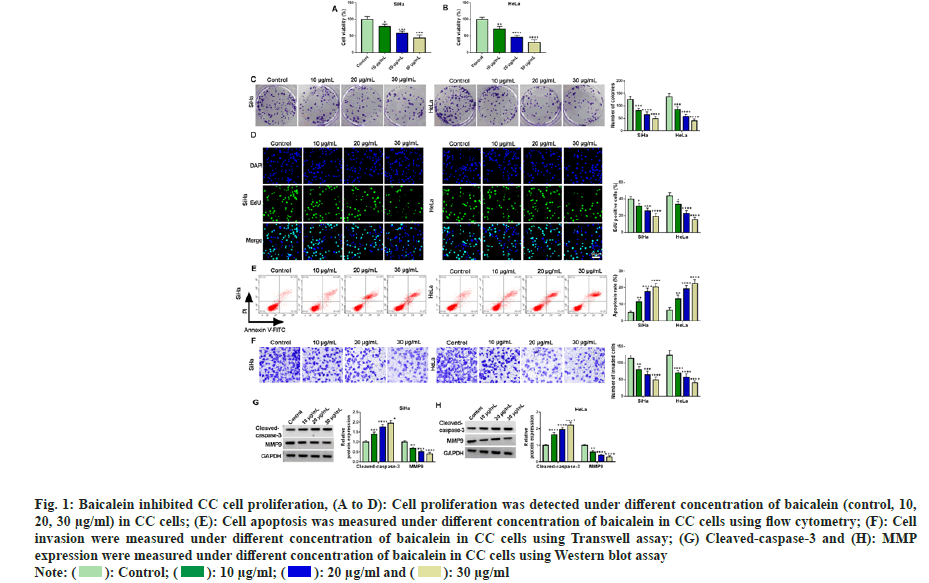
To understand whether baicalein showed inhibitory effects on CC cells, we selected two CC cell lines routinely used in CC research and treated them with different concentrations of baicalein (10, 20, 30 μg/ml). Then, the physiological indexes of cells were measured. The results of fig. 1A-fig. 1D showed that the cell viability and cell colony number and EdU positive cells were obviously decreased with the increase of baicalein concentration, indicating that baicalein inhibited CC cell proliferation. Subsequently, CC cell apoptosis was gradually increased with increasing baicalein concentration (fig. 1E). Results of Transwell showed that the higher the concentration of baicalein, the greater the inhibitory effect on CC cell invasion (fig. 1F). Baicalein significantly induced cleaved-caspase-3 and obviously inhibited MMP-9 production in CC cells (fig. 1G and fig. 1H). In conclusion, baicalein inhibited the growth of CC cells at different concentrations, indicating that it could be applied for the treatment of CC.
Fig. 1: Baicalein inhibited CC cell proliferation, (A to D): Cell proliferation was detected under different concentration of baicalein (control, 10,
20, 30 μg/ml) in CC cells; (E): Cell apoptosis was measured under different concentration of baicalein in CC cells using flow cytometry; (F): Cell
invasion were measured under different concentration of baicalein in CC cells using Transwell assay; (G) Cleaved-caspase-3 and (H): MMP
expression were measured under different concentration of baicalein in CC cells using Western blot assay
Note: ( ): Control; (
): Control; ( ): 10 μg/ml; (
): 10 μg/ml; ( ): 20 μg/ml and (
): 20 μg/ml and ( ): 30 μg/ml
): 30 μg/ml
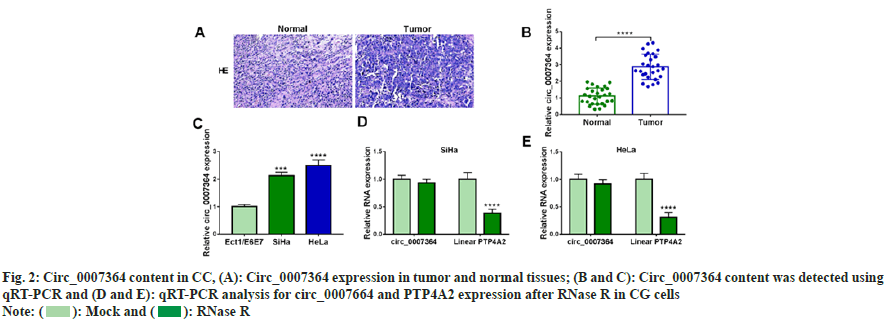
To determine circ_0007364 expression property in CC, we collected 27 pairs of CC tissues and their adjacent tissues. The staining in tumor tissues was deepened in fig. 2A. Circ_0007364 content was elevated in CC tissues and cells (fig. 2B and fig. 2C). As shown in fig. 2D and fig. 2E, after RNase R treatment, circ_0007364 was more stable than its linear mRNA (PTP4A2). Taken together, circ_0007364 was highly expressed in CC.
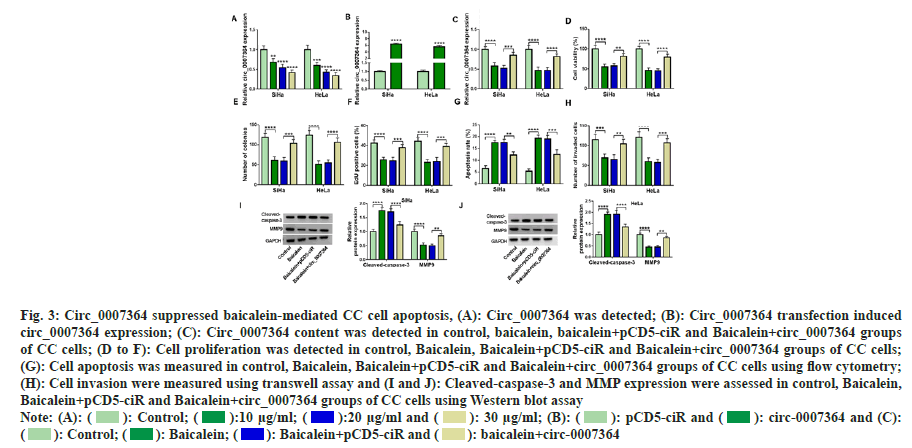
Combined with the above research, we speculated that circ_0007364 was related to baicalein in CC cells growth. Therefore, we chose baicalein to treat CC cells and observe whether circ_0007364 expression was changed. As shown in fig. 3A, with the increase of baicalein treatment concentration, circ_0007364 content was gradually reduced. Therefore, we speculated that baicalein decreased circ_0007364 expression to inhibit the growth of CC. To prove this idea, we built the overexpression cell line of circ_0007364 (circ_0007364) and control cell line (pCD5-ciR). The expression of circ_0007364 increased significantly (fig. 3B). Next, 20 μg/ml baicalein was used to treat transfected or non-transfected CC cells and baicalein inhibited circ_0007364 expression, but circ_0007364 transfection attenuated the inhibitory effect (fig. 3C). Consistent with previous studies[22], baicalein inhibited the proliferation of CC cells (fig. 3D-fig. 3F), promoted apoptosis (fig. 3G), weakened the ability of cell invasion and metastasis (fig. 3H), enhanced cleavedcapase- 3 production and reduced MMP9 protein expression (fig. 3I and fig. 3J). However, enhanced circ_0007364 expression reverses the suppression effects of baicalein on CC cell progression (fig. 3C to fig. 3J). Therefore, promotion of circ_0007364 significantly attenuated the inhibitory influence of baicalein in CC cell lines.
Fig. 3: Circ_0007364 suppressed baicalein-mediated CC cell apoptosis, (A): Circ_0007364 was detected; (B): Circ_0007364 transfection induced
circ_0007364 expression; (C): Circ_0007364 content was detected in control, baicalein, baicalein+pCD5-ciR and Baicalein+circ_0007364 groups
of CC cells; (D to F): Cell proliferation was detected in control, Baicalein, Baicalein+pCD5-ciR and Baicalein+circ_0007364 groups of CC cells;
(G): Cell apoptosis was measured in control, Baicalein, Baicalein+pCD5-ciR and Baicalein+circ_0007364 groups of CC cells using flow cytometry;
(H): Cell invasion were measured using transwell assay and (I and J): Cleaved-caspase-3 and MMP expression were assessed in control, Baicalein,
Baicalein+pCD5-ciR and Baicalein+circ_0007364 groups of CC cells using Western blot assay
Note: (A): ( ): Control; (
): Control; ( ):10 μg/ml; (
):10 μg/ml; ( ):20 μg/ml and (
):20 μg/ml and ( ): 30 μg/ml; (B): (
): 30 μg/ml; (B): ( ): pCD5-ciR and (
): pCD5-ciR and ( ): circ-0007364 and (C):
(
): circ-0007364 and (C):
( ): Control; (
): Control; ( ): Baicalein; (
): Baicalein; ( ): Baicalein+pCD5-ciR and (
): Baicalein+pCD5-ciR and ( ): baicalein+circ-0007364
): baicalein+circ-0007364
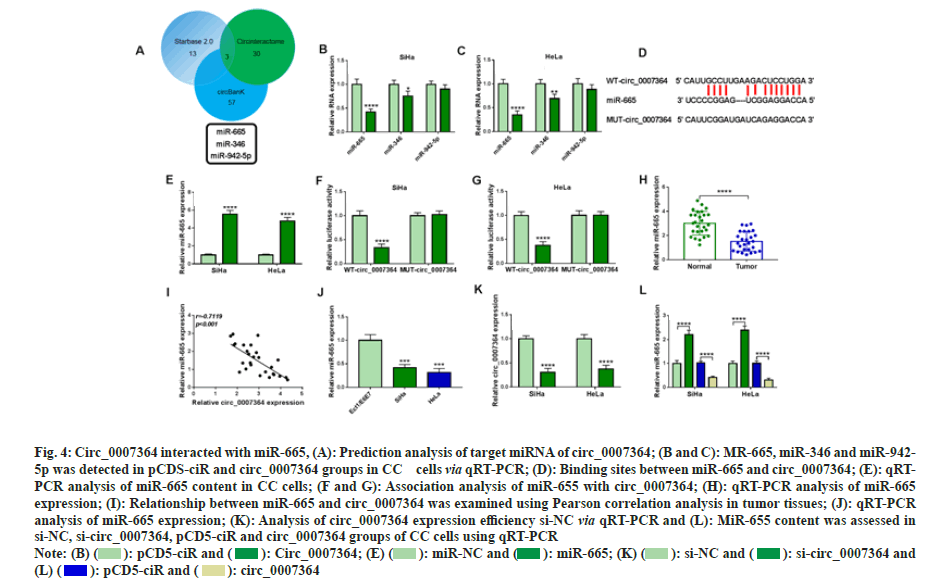
After the prediction of Star base 2.0, circBank and Circinteractome, we found three candidate miRNAs of circ_0007364, including miR-665/- 346/-942-5p (fig. 4A). In addition, circ_0007364 transfection could significantly decreased miR- 665 and miR-356, but did not affect miR-942-5p (fig. 4B and fig. 4C). Therefore, we chose miR- 665 as circ_0007364 target miRNA for further study. We constructed wild type circ_0007364 containing complementary sites of miR-655 (WTcirc_ 0007364) and mutant circ_0007364 containing mutation sites of miR-655 (MUT-circ_0007364) (fig. 4D) and overexpressing vector of miR-665 (fig. 4E) was also obtained. The results showed that when WT-circ_0007364 combined with miR-655, relative luciferase activity decreased significantly, while MUT-circ_0007364 and miR- 655 were transfected into cells had no significant effect on relative luciferase activity (fig. 4F and fig. 4G). Moreover, miR-655 was significantly down regulated in CC tissues and cells (fig. 4H and fig. 4I), and had negative correlation with circ_0007364 in CC tissues (fig. 4J). In addition, inhibition of circ_0007364 sharply decreased miR- 655 expression in CC cells (fig. 4K). Circ_0007364 silencing reduced miR-655 expression in CC cells (fig. 4L). Moreover, miR-655 expression in circ_0007364 groups was significantly lower in circ_0007364 groups compared with that in pCD5- miR group. Therefore, circ_0007364 bound to miR-655 and regulated its expression in CC cells.
Fig. 4: Circ_0007364 interacted with miR-665, (A): Prediction analysis of target miRNA of circ_0007364; (B and C): MR-665, miR-346 and miR-942-
5p was detected in pCDS-ciR and circ_0007364 groups in CC cells via qRT-PCR; (D): Binding sites between miR-665 and circ_0007364; (E): qRTPCR
analysis of miR-665 content in CC cells; (F and G): Association analysis of miR-655 with circ_0007364; (H): qRT-PCR analysis of miR-665
expression; (I): Relationship between miR-665 and circ_0007364 was examined using Pearson correlation analysis in tumor tissues; (J): qRT-PCR
analysis of miR-665 expression; (K): Analysis of circ_0007364 expression efficiency si-NC via qRT-PCR and (L): MiR-655 content was assessed in
si-NC, si-circ_0007364, pCD5-ciR and circ_0007364 groups of CC cells using qRT-PCR
Note: (B) ( ): pCD5-ciR and (
): pCD5-ciR and ( ): Circ_0007364; (E) (
): Circ_0007364; (E) ( ): miR-NC and (
): miR-NC and ( ): miR-665; (K) (
): miR-665; (K) ( ): si-NC and (
): si-NC and ( ): si-circ_0007364 and
(L) (
): si-circ_0007364 and
(L) ( ): pCD5-ciR and (
): pCD5-ciR and ( ): circ_0007364
): circ_0007364
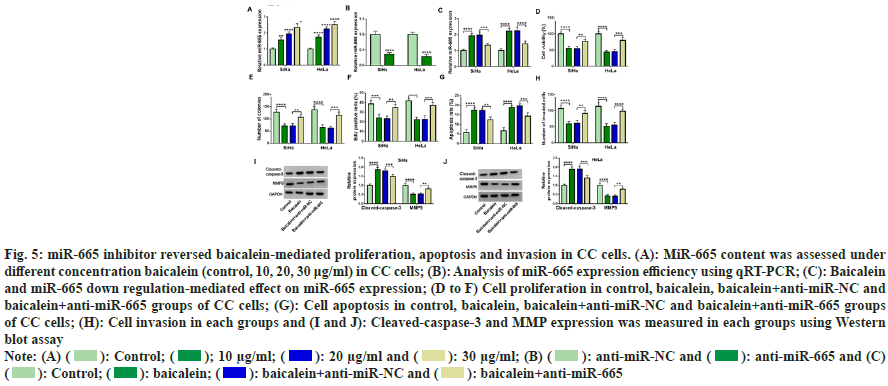
We measured the expression of miR-665 in CC cells under different concentrations to understand the relationship between miR-665 and baicalein. MiR-655 was gradually increased with the increase of baicalein concentration (fig. 5A). Therefore, we transfected the oligos of anti-miR-665 into CC cells and anti-miR-665 transfection inhibited the expression of miR-665 (fig. 5B). Less than 20 μg baicalein, inhibition of miR-665 could weaken miR-665 expression induced by baicalein (fig. 5C). Moreover, inhibiting the expression of miR- 665 weakened baicalein-mediated inhibition in cell viability (fig. 5D), cell clone number (fig. 5E), EdU positive cells (fig. 5F) and invasion (fig. 5G) and the promotion of apoptosis (fig. 5H). Additionally, baicalein increased cleaved-caspase-3 production and decreased MMP9 expression in CC cells and these phenomena were reversed through down regulating miR-665 (fig. 5I and fig. 5J). Therefore, inhibiting miR-665 expression could weaken baicalein-induced effects in CC cells.
Fig. 5: miR-665 inhibitor reversed baicalein-mediated proliferation, apoptosis and invasion in CC cells. (A): MiR-665 content was assessed under
different concentration baicalein (control, 10, 20, 30 μg/ml) in CC cells; (B): Analysis of miR-665 expression efficiency using qRT-PCR; (C): Baicalein
and miR-665 down regulation-mediated effect on miR-665 expression; (D to F) Cell proliferation in control, baicalein, baicalein+anti-miR-NC and
baicalein+anti-miR-665 groups of CC cells; (G): Cell apoptosis in control, baicalein, baicalein+anti-miR-NC and baicalein+anti-miR-665 groups
of CC cells; (H): Cell invasion in each groups and (I and J): Cleaved-caspase-3 and MMP expression was measured in each groups using Western
blot assay
Note: (A) ( ): Control; (
): Control; ( ); 10 μg/ml; (
); 10 μg/ml; ( ): 20 μg/ml and (
): 20 μg/ml and ( ): 30 μg/ml; (B) (
): 30 μg/ml; (B) ( ): anti-miR-NC and (
): anti-miR-NC and ( ): anti-miR-665 and (C)
(
): anti-miR-665 and (C)
( ): Control; (
): Control; ( ): baicalein; (
): baicalein; ( ): baicalein+anti-miR-NC and (
): baicalein+anti-miR-NC and ( ): baicalein+anti-miR-665
): baicalein+anti-miR-665
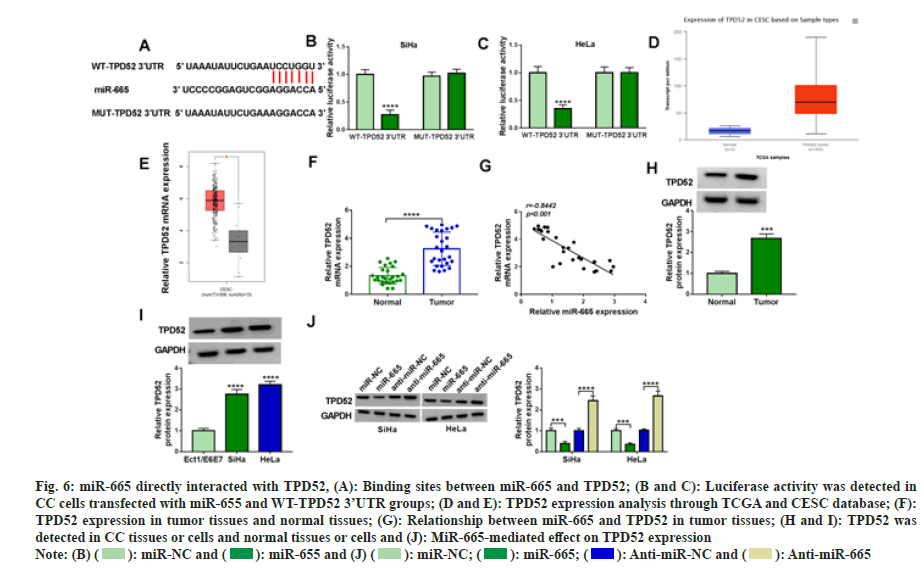
The target gene of miR-665 through Star base 2.0. TPD52 contained binding sites with miR-665 (fig. 6A). In addition, when WT-TPD52 3 'UTR rather than MUT-TPD52 3' UTR was transfected with miR-665, the luciferase activity decreased significantly (fig. 6B and fig. 6C). Moreover, the network database TCGA showed TPD52 was highly expressed in CC tissues (fig. 6D and fig. 6E). In our experiment, we also found a high TPD52 expression in CC tissues and cells (fig. 6F-fig. 6I) and TPD52 was negatively correlated with miR-665 (fig. 6J). Additionally, miR-665 negatively regulated TPD52 expression. Therefore, these results suggested miR-665 directly bound to TPD52 in CC cells.
Fig. 6: miR-665 directly interacted with TPD52, (A): Binding sites between miR-665 and TPD52; (B and C): Luciferase activity was detected in
CC cells transfected with miR-655 and WT-TPD52 3’UTR groups; (D and E): TPD52 expression analysis through TCGA and CESC database; (F):
TPD52 expression in tumor tissues and normal tissues; (G): Relationship between miR-665 and TPD52 in tumor tissues; (H and I): TPD52 was
detected in CC tissues or cells and normal tissues or cells and (J): MiR-665-mediated effect on TPD52 expression
Note: (B) ( ): miR-NC and (
): miR-NC and ( ): miR-655 and (J) (
): miR-655 and (J) ( ): miR-NC; (
): miR-NC; ( ): miR-665; (
): miR-665; ( ): Anti-miR-NC and (
): Anti-miR-NC and ( ): Anti-miR-665
): Anti-miR-665
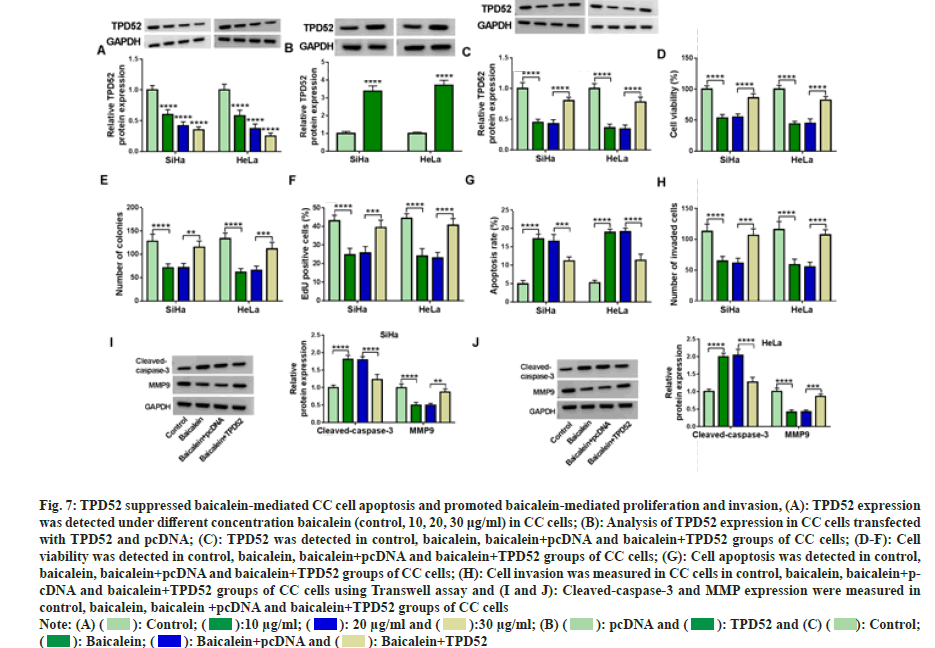
Next, TPD52 content was assessed in CC cells after baicalein treatment. The results showed that TPD52 level in cancer cells was decreased with increasing baicalein concentration (fig. 7A). We transfected the overexpression vector of TPD52 into CC cells to clarify the relationship between TPD52 and baicalein (fig. 7B). Increasing the expression of TPD52 can restore the inhibitory effect of baicalein on TPD52 expression (fig. 7C). Moreover, baicalein significantly inhibited cell proliferation (fig. 7D-fig. 7F) and invasion (fig. 7G), induced apoptosis (fig. 7H), while overexpression of TPD52 attenuated these effects of baicalein on the progression of CC cells. Increasing TPD52 expression reversed the expression of cleavedcaspase- 3 and MMP9 affected by baicalein (fig. 7I and fig. 7J). Therefore, overexpression of TPD52 weakened baicalein induced effects in CC cells.
Fig. 7: TPD52 suppressed baicalein-mediated CC cell apoptosis and promoted baicalein-mediated proliferation and invasion, (A): TPD52 expression
was detected under different concentration baicalein (control, 10, 20, 30 μg/ml) in CC cells; (B): Analysis of TPD52 expression in CC cells transfected
with TPD52 and pcDNA; (C): TPD52 was detected in control, baicalein, baicalein+pcDNA and baicalein+TPD52 groups of CC cells; (D-F): Cell
viability was detected in control, baicalein, baicalein+pcDNA and baicalein+TPD52 groups of CC cells; (G): Cell apoptosis was detected in control,
baicalein, baicalein+pcDNA and baicalein+TPD52 groups of CC cells; (H): Cell invasion was measured in CC cells in control, baicalein, baicalein+pcDNA
and baicalein+TPD52 groups of CC cells using Transwell assay and (I and J): Cleaved-caspase-3 and MMP expression were measured in
control, baicalein, baicalein +pcDNA and baicalein+TPD52 groups of CC cells
Note: (A) ( ): Control; (
): Control; ( ):10 μg/ml; (
):10 μg/ml; ( ): 20 μg/ml and (
): 20 μg/ml and ( ):30 μg/ml; (B) (
):30 μg/ml; (B) ( ): pcDNA and (
): pcDNA and ( ): TPD52 and (C) (
): TPD52 and (C) ( ): Control;
(
): Control;
( ): Baicalein; (
): Baicalein; ( ): Baicalein+pcDNA and (
): Baicalein+pcDNA and ( ): Baicalein+TPD52
): Baicalein+TPD52
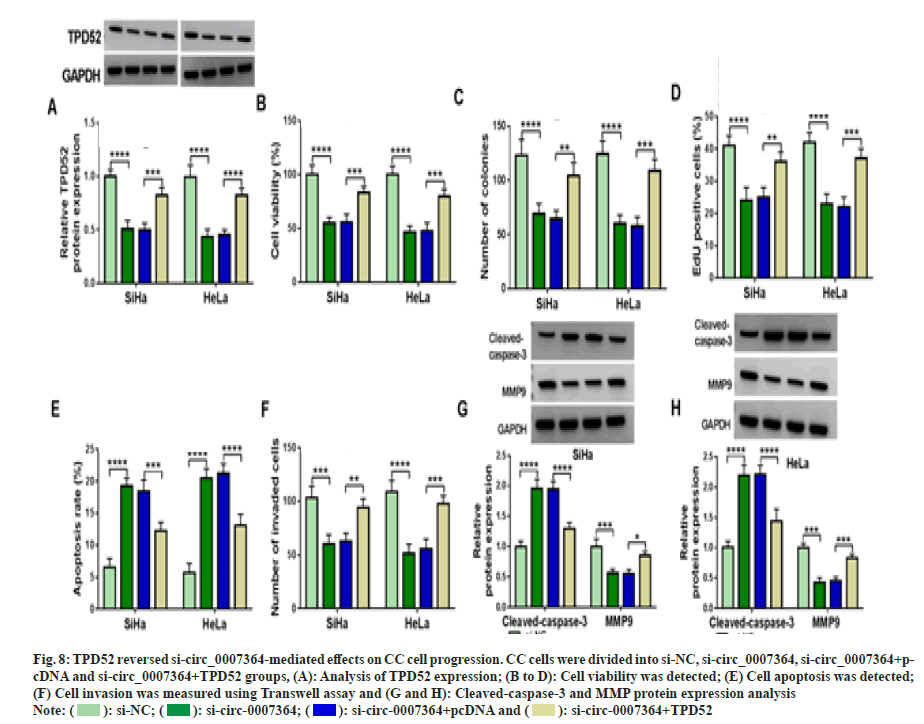
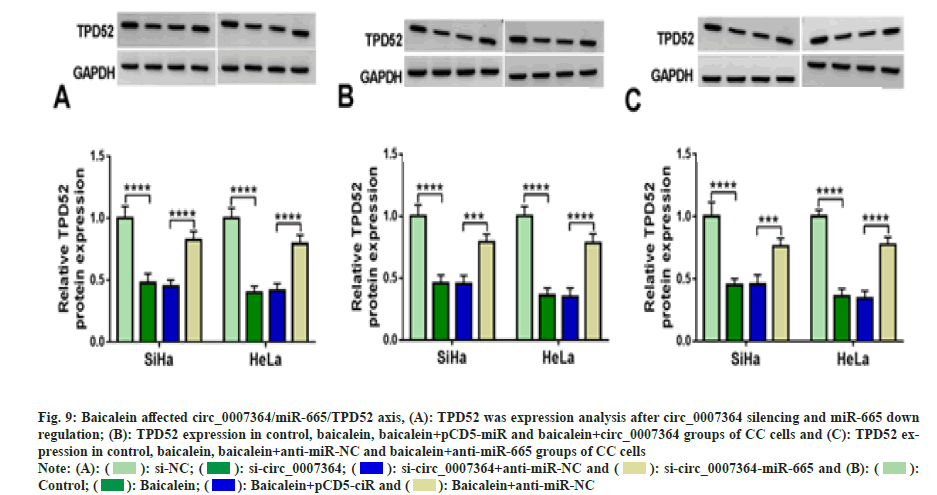
Next, we found that si-circ_0007364 transfection not only inhibited the expression of TPD52 (fig. 8A), cell proliferation (fig. 8B-fig. 8D), cell invasion (fig. 8E) and the protein expression of MMP9 (fig. 8F and fig. 8G), but also induced apoptosis (fig. 8H) and cleavedcaspase- 3 production (fig. 8G and fig. 8H), while overexpression of TPD52 reversed these effects. Additionally, inhibition of miR-665 improved TPD52 expression decreased by si-circ_0007364 transfection (fig. 9A). Therefore, circ_0007364 regulated the expression of TPD52 by binding miR-665 and affected cell proliferation invasion and apoptosis in CC cells. Moreover, under baicalein treatment, promotion of circ_0007364 or inhibition of miR-665 significantly induced TPD52 expression that was inhibited by baicalein (fig. 9B and fig. 9C). In conclusion, baicalein inhibited the growth of CC cells by affecting circ_0007364/ miR-665/TPD52 axis.
Fig. 8: TPD52 reversed si-circ_0007364-mediated effects on CC cell progression. CC cells were divided into si-NC, si-circ_0007364, si-circ_0007364+pcDNA
and si-circ_0007364+TPD52 groups, (A): Analysis of TPD52 expression; (B to D): Cell viability was detected; (E) Cell apoptosis was detected;
(F) Cell invasion was measured using Transwell assay and (G and H): Cleaved-caspase-3 and MMP protein expression analysis
Note: ( ): si-NC; (
): si-NC; ( ): si-circ-0007364; (
): si-circ-0007364; ( ): si-circ-0007364+pcDNA and (
): si-circ-0007364+pcDNA and ( ): si-circ-0007364+TPD52
): si-circ-0007364+TPD52
Fig. 9: Baicalein affected circ_0007364/miR-665/TPD52 axis, (A): TPD52 was expression analysis after circ_0007364 silencing and miR-665 down
regulation; (B): TPD52 expression in control, baicalein, baicalein+pCD5-miR and baicalein+circ_0007364 groups of CC cells and (C): TPD52 expression
in control, baicalein, baicalein+anti-miR-NC and baicalein+anti-miR-665 groups of CC cells
Note: (A): ( ): si-NC; (
): si-NC; ( ): si-circ_0007364; (
): si-circ_0007364; ( ): si-circ_0007364+anti-miR-NC and (
): si-circ_0007364+anti-miR-NC and ( ): si-circ_0007364-miR-665 and (B): (
): si-circ_0007364-miR-665 and (B): ( ):
Control; (
):
Control; ( ): Baicalein; (
): Baicalein; ( ): Baicalein+pCD5-ciR and (
): Baicalein+pCD5-ciR and ( ): Baicalein+anti-miR-NC
): Baicalein+anti-miR-NC
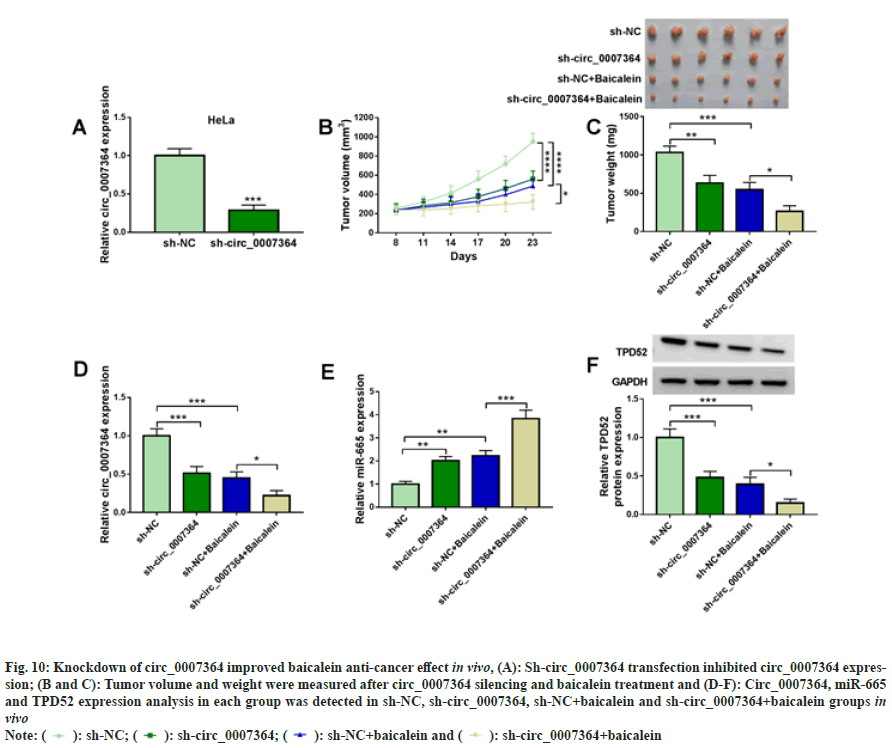
Mouse model assay was used to determine circ_0007364 in vivo and the results showed that circ_0007364 expression was sharply decreased in sh-circ_0007364 group (fig. 10A). After 8 d, mice were injected with baicalein (45 mg/kg) with sh-NC or sh-circ_0007364, once every 3 d. As shown in fig. 10B and fig. 10C, circ_0007364 silencing inhibited tumor volume and weight, and sh-circ_0007364 combined with baicalein could further inhibit tumor growth. In addition, baicalein also induced miR-665 expression that was promoted by sh-circ_0007364 and inhibited circ_0007364 and TPD52 expression that was suppressed by sh-circ_0007364 (fig. 10D to fig. 10F). Therefore, knockdown of circ_0007364 increased baicalein anti-cancer effect in vivo.
Fig. 10: Knockdown of circ_0007364 improved baicalein anti-cancer effect in vivo, (A): Sh-circ_0007364 transfection inhibited circ_0007364 expression;
(B and C): Tumor volume and weight were measured after circ_0007364 silencing and baicalein treatment and (D-F): Circ_0007364, miR-665
and TPD52 expression analysis in each group was detected in sh-NC, sh-circ_0007364, sh-NC+baicalein and sh-irc_0007364+baicalein groups in
vivo
Note: ( ): sh-NC; (
): sh-NC; ( ): sh-circ_0007364; (
): sh-circ_0007364; ( ): sh-NC+baicalein and (
): sh-NC+baicalein and ( ): sh-circ_0007364+baicalein
): sh-circ_0007364+baicalein
Nowadays, disease-related databases are constantly improving with the development of technology, providing high-quality resources for the study of disease mechanism. Traditional Chinese medicine is remarkably effective in treating diseases[23-26]. Baicalein is the main active component of Scutellaria baicalensis and has inhibitory effects on cancer progression[27,28]. However, the specific regulation mechanism is not perfect.
In CC, circRNA/miRNA/mRNA is one of the important regulatory pathways. Circ_0000515/ miR-326/ELK1 axis was involved in CC progression[29]. In addition, circ_Nuclear Receptor- Interacting Protein 1 (NRIP1) enhanced cell invasion through competitively combining miR- 629-3p to regulate PTP4A1/ERK1/2 pathway in CC[30]. CircRNA is also involved in the inhibitory mechanism of baicalein in CC development. For example, baicalein inhibited CC cells growth through regulation of circHIAT1/miR-19a-3p pathway[9]. In our study, circ_0007364 was up regulated in CC tissue samples and baicalein inhibited circ_0007364 expression. Thus, we speculated that circ_0007364 participated in the inhibition mechanism of baicalein in CC. Otherwise, circ_0007364 is derived from PTP4A2, which encodes PTP4A2 protein and belongs to class of protein tyrosine phosphatase family[31]. PTP4A2 can promote the production of breast tumor cells[32,33]. Previous articles have studied that circ_0007364 was highly expressed in CC. Moreover, the increased circ_0007364 expression affected CC progression via adjusting miR-101- 5p/MAT2A axis[22]. Our study showed upregulation of circ_0007364 could suppress baicalein inhibited cell progression in CC cells.
MiRNA is a noncoding small RNA, which mediates the biological process of cells[34,35]. Moreover, miRNA negatively regulates gene expression by directly degrading target mRNA[36]. In this paper, miR-665 was the target miRNA of circ_0007364. In previous studies, miR-665 can inhibit or promote cancer progression in many cancers, like ovarian cancer, HCC, pancreatic cancer and CC[37-40]. In this paper, miR-665 expression was down regulated in CC tissues and cell lines, and baicalein promoted miR-665 expression. Function experiment showed that baicalein regulated CC progression though regulating circ_0007364 to sponge miR-665 in CC.
Moreover, TPD52 was determined to bind to miR-665 in CC and was associated with the regulatory pathway of tumor inhibited by baicalein through dual luciferase report assay and function experiment. Various study demonstrated that TPD52, act as cancer-oncogene, participated in cancer development, which are upregulated in many cancer types, including CC[41-44]. Consistent with previous study[44], TPD52 was up regulated in CC cells and was related to cell progression in CC in the present work. Furthermore, baicalein reduced TPD52 expression in CC cells. Thus, we determined that baicalein inhibited CC development through modulating circ_0007364/ miR-665/TPD52 axis.
This study described how baicalein inhibited the regulatory pathway of CC cell development. Baicalein regulated the growth of CC cells by affecting circ_0007364/miR-665/TPD52 axis, providing a new molecular mechanism of baicalein in CC and finding the key regulatory marker. This exploration could further improve the medication of baicalein and played a better anti-cancer effect. At the same time, this investigation could also be used as a research idea to find the molecular mechanism of other anticancer drugs, and find more effective anticancer drug components and contribute to cancer treatment.
Author’s contributions:
Xuefang Liu and Jing Xie have contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet 2007;370(9590):890-907.
[Crossref] [Google Scholar] [PubMed]
- Hu Z, Ma D. The precision prevention and therapy of HPV-related cervical cancer: New concepts and clinical implications. Cancer Med 2018;7(10):5217-36.
[Crossref] [Google Scholar] [PubMed]
- Olusola P, Banerjee HN, Philley JV, Dasgupta S. Human papilloma virus-associated cervical cancer and health disparities. Cells 2019;8(6):622.
[Crossref] [Google Scholar] [PubMed]
- Sowndhararajan K, Deepa P, Kim M, Park SJ, Kim S. Baicalein as a potent neuroprotective agent: A review. Biomed Pharmacother 2017;95:1021-32.
[Crossref] [Google Scholar] [PubMed]
- Bie B, Sun J, Guo Y, Li J, Jiang W, Yang J, et al. Baicalein: A review of its anti-cancer effects and mechanisms in hepatocellular carcinoma. Biomed Pharmacother 2017;93:1285-91.
[Crossref] [Google Scholar] [PubMed]
- Yan W, Ma X, Zhao X, Zhang S. Baicalein induces apoptosis and autophagy of breast cancer cells via inhibiting PI3K/AKT pathway in vivo and in vitro. Drug Des Devel Ther 2018;12:3961-72.
[Crossref] [Google Scholar] [PubMed]
- Yu X, Liu Y, Wang Y, Mao X, Zhang Y, Xia J. Baicalein induces cervical cancer apoptosis through the NF-κB signaling pathway. Mol Med Rep 2018;17(4):5088-94.
[Crossref] [Google Scholar] [PubMed]
- Wu R, Murali R, Kabe Y, French SW, Chiang YM, Liu S, et al. Baicalein targets GTPase-mediated autophagy to eliminate liver tumor–initiating stem cell–like cells resistant to mTORC1 inhibition. Hepatology 2018;68(5):1726-40.
[Crossref] [Google Scholar] [PubMed]
- Hu J, Wang R, Liu Y, Zhou J, Shen K, Dai Y. Baicalein represses cervical cancer cell growth, cell cycle progression and promotes apoptosis via blocking AKT/mTOR pathway by the regulation of circHIAT1/miR-19a-3p axis. Onco Targets Ther 2021;14:905-16.
[Crossref] [Google Scholar] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56(1):55-66.
[Crossref] [Google Scholar] [PubMed]
- Lei B, Tian Z, Fan W, Ni B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int J Med Sci 2019;16(2):292-301.
[Crossref] [Google Scholar] [PubMed]
- Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer 2020;19(1):117.
[Crossref] [Google Scholar] [PubMed]
- Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ, et al. A circRNA–miRNA–mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med 2018;16(1):1-21.
- Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, et al. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Prac 2017;213(5):453-6.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 2019;10(2):55.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer 2019;18(1):119.
- Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 2020;19(1):13.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in immune responses and immune diseases. Theranostics 2019;9(2):588-607.
- Chen RX, Liu HL, Yang LL, Kang FH, Xin LP, Huang LR, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci 2019;23(20):8771-8.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY) 2019;11(24):12412-27.
[Crossref] [Google Scholar] [PubMed]
- Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol 2019;234(8):14296-305.
[Crossref] [Google Scholar] [PubMed]
- Chen H, Gu B, Zhao X, Zhao Y, Huo S, Liu X, et al. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II alpha (MAT2A) expression by restraining microRNA-101-5p. Bioengineered 2020;11(1):1269-79.
[Crossref] [Google Scholar] [PubMed]
- Chan HH, Ng T. Traditional Chinese Medicine (TCM) and allergic diseases. Curr Allergy Asthma Rep 2020;20(11):67.
[Crossref] [Google Scholar] [PubMed]
- Yu-Jie DA, Shi-Yao W, Shuai-Shuai G, Jin-Cheng L, Fang LI, Jun-Ping K. Recent advances of traditional Chinese medicine on the prevention and treatment of COVID-19. Chin J Nat Med 2020;18(12):881-9.
[Crossref] [Google Scholar] [PubMed]
- Xiang Y, Guo Z, Zhu P, Chen J, Huang Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med 2019;8(5):1958-75.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Qi F, Wang Z, Zhang Z, Pan N, Huai L, et al. A review of traditional Chinese medicine for treatment of glioblastoma. Biosci Trends 2019;13(6):476-87.
[Crossref] [Google Scholar] [PubMed]
- Liu H, Dong Y, Gao Y, Du Z, Wang Y, Cheng P, et al. The fascinating effects of baicalein on cancer: A review. Int J Mol Sci 2016;17(10):1681.
[Crossref] [Google Scholar] [PubMed]
- Sun X, Cui X, Chen X, Jiang X. Baicalein alleviated TGF β1-induced type I collagen production in lung fibroblasts via down regulation of connective tissue growth factor. Biomed Pharmacother 2020;131:110744.
[Crossref] [Google Scholar] [PubMed]
- Tang Q, Chen Z, Zhao L, Xu H. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY) 2019;11(22):9982-99.
- Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis 2020;11(5):399.
[Crossref] [Google Scholar] [PubMed]
- Andres SA, Wittliff JL, Cheng A. Protein tyrosine phosphatase 4A2 expression predicts overall and disease-free survival of human breast cancer and is associated with estrogen and progestin receptor status. Horm Cancer 2013;4(4):208-21.
[Crossref] [Google Scholar] [PubMed]
- Zhao D, Guo L, Neves H, Yuen HF, Zhang SD, McCrudden CM, et al. The prognostic significance of protein tyrosine phosphatase 4A2 in breast cancer. Onco Targets Ther 2015:1707-17.
[Crossref] [Google Scholar] [PubMed]
- Hardy S, Wong NN, Muller WJ, Park M, Tremblay ML. Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression role of PRL-2 in breast cancer. Cancer Res 2010;70(21):8959-67.
[Crossref] [Google Scholar] [PubMed]
- Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: An overview. Methods Mol Biolv 2017;1509:1-10.
[Crossref] [Google Scholar] [PubMed]
- Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer 2011;72(1):92-9.
[Crossref] [Google Scholar] [PubMed]
- Zhang W, Edwards A, Fan W, Flemington EK, Zhang K. miRNA-mRNA correlation-network modules in human prostate cancer and the differences between primary and metastatic tumor subtypes. PloS One 2012;7(6):e40130.
[Crossref] [Google Scholar] [PubMed]
- Zhou P, Xiong T, Yao L, Yuan J. MicroRNA-665 promotes the proliferation of ovarian cancer cells by targeting SRCIN1. Exp Ther Med 2020;19(2):1112-20.
[Crossref] [Google Scholar] [PubMed]
- Bai N, Peng E, Xia F, Wang D, Li X, Li X. CircABCC2 regulates hepatocellular cancer progression by decoying miR-665. J Cancer 2019;10(17):3893.
[Crossref] [Google Scholar] [PubMed]
- Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis 2018;9(6):706.
[Crossref] [Google Scholar] [PubMed]
- Cao L, Jin H, Zheng Y, Mao Y, Fu Z, Li X, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci 2019;110(3):913-25.
[Crossref] [Google Scholar] [PubMed]
- Byrne JA, Frost S, Chen Y, Bright RK. Tumor protein D52 (TPD52) and cancer—oncogene understudy or understudied oncogene? Tumor Biol 2014;35(8):7369-82.
[Crossref] [Google Scholar] [PubMed]
- Fu M, Chen CW, Yang LQ, Yang WW, Du ZH, Li YR, et al. MicroRNA-103a-3p promotes metastasis by targeting TPD52 in salivary adenoid cystic carcinoma. Int J Oncol 2020;57(2):574-86.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Xi X. LINC01133 contribute to epithelial ovarian cancer metastasis by regulating miR-495-3p/TPD52 axis. Biochem Biophys Res Commun 2020;533(4):1088-94.
[Crossref] [Google Scholar] [PubMed]
- Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S. Hsa_circ_0084927 regulates cervical cancer advancement via regulation of the miR-634/TPD52 Axis. Cancer Manag Res 2020;12:9435-48.
[Crossref] [Google Scholar] [PubMed]