- *Corresponding Author:
- Chunbo Zou
Department of Nephrology, Taizhou School of Clinical Medicine, Nanjing Medical University, Taizhou, Jiangsu 225300, China
E-mail: zcb318182@163.com
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “81-90” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Long non-coding RNA taurine upregulated gene 1 plays an important role in the progression of diabetic nephropathy. Studies have shown that there is a complex and precise interaction between taurine upregulated gene 1 and microRNA-22-3p. MicroRNA-22-3p is an important microRNA whose expression level is closely associated with the development and progression of diabetic nephropathy. Through analysis, research has shown that the expression level of microRNA-22-3p is significantly downregulated in patients with diabetic nephropathy, while the high expression of taurine upregulated gene 1 is closely related to the clinical course of diabetic nephropathy. Further studies revealed a negative regulatory mechanism between taurine upregulated gene 1 and microRNA-22-3p. Taurine upregulated gene 1 was able to promote microRNA-22-3p degradation, thereby affecting the biological function of microRNA-22-3p. One of the main intracellular targets of microRNA-22-3p is thioredoxin. Thioredoxin has important physiological functions in redox processes, while its expression level is often regulated in diabetic nephropathy. MicroRNA-22-3p has been shown to inhibit the expression of thioredoxin by directly binding to the 3’ untranslated region of thioredoxin, thereby affecting redox balance and promoting apoptosis and dysfunction in cells. Further studies revealed the regulatory mechanism of taurine upregulated gene 1/microRNA-22-3p/CSE axis in diabetic nephropathy. Taurine upregulated gene 1 modulates the control of thioredoxin by microRNA-22-3p through its influence on microRNA-22-3p expression levels. Elevated taurine upregulated gene 1 expression results in reduced microRNA-22-3p levels, subsequently increasing thioredoxin expression. This cascade helps maintain cellular redox balance, mitigating apoptosis and functional deficits. Overall, long non-coding RNA taurine upregulated gene 1 is involved in important regulatory mechanisms during diabetic nephropathy development by regulating the activity of the microRNA-22-3p/thioredoxin axis. This finding not only contributes to an in-depth understanding of the pathophysiological mechanisms of diabetic nephropathy, but also lays the groundwork for future therapeutic strategies by identifying new theoretical bases and potential therapeutic targets.
Keywords
Thioredoxin, microRNA-22-3p, taurine upregulated gene 1, diabetic nephropathy, podocyte apoptosis
Diabetic Nephropathy (DN) is a prevalent chronic microvascular complication of diabetes and ranks among the primary causes of end-stage renal disease[1]. The general treatment strategy for DN is intensive glycemic and blood pressure control, but for most of the cases, DN will develop into end stage renal disease[2]. Therefore, new therapeutic targets for DN are urgently needed.
According to former research, Long non-coding RNA (LncRNA) could participate in Diabetic Nephropathy (DN) regulation[3]. Taurine Upregulated Gene 1 (TUG1) is an lncRNA initially discovered for its involvement in photoreceptor development and its crucial role in retinal formation[4]. Recent research indicates that TUG1 not only participates in the regulation of various tumor processes[5], but is also closely related to the development and progression of DN. TUG1 is involved in the pathogenesis of DN through multiple pathways. Firstly, TUG1 can affect the fibrosis process in the glomeruli and interstitium of diabetic patients, promoting damage to kidney structure and function. Secondly, TUG1 can also regulate apoptosis and inflammation in kidney cells of diabetic patients, accelerating the deterioration of kidney function. Additionally, TUG1 participates in the onset and progression of microvascular lesions in the kidneys of diabetic patients, further exacerbating the condition of DN. Research has shown that lncRNA TUG1 regulates mitochondrial bioenergetics and mitigates Extracellular Matrix (ECM) accumulation in DN by targeting microRNA (miR)-377, thereby influencing Peroxisome Proliferator-Activated Receptor Gamma (PPAR-γ) [6]. Moreover, elevated levels of TUG1 are linked to endothelial cell apoptosis, and aberrant TUG1 expression is correlated with the onset of numerous neurological disorders[7]. Nevertheless, the precise involvement of lncRNA TUG1 in podocyte apoptosis in DN remains uncertain.
Unlike lncRNAs, microRNAs (miRNAs) are small non-coding RNA molecules that play a crucial role in gene regulation[3]. They are typically about 21-25 nucleotides in length and are involved in post-transcriptional regulation of gene expression. miRNAs can bind to specific messenger Ribonucleic Acid (mRNA) molecules and either inhibit their translation into proteins or promote their degradation, thereby influencing the level of proteins produced by the cell. These molecules are important in various biological processes such as development, differentiation, apoptosis, and disease pathways, including cancer and metabolic disorders like diabetes[8]. The expression level of miR-22-3p is significantly downregulated, leading to a series of molecular dysregulations including promoting cell apoptosis, increasing inflammation response, and injury repair processes. Additionally, miR-22-3p may also participate in renal pathological processes by regulating inflammatory mediators and cell signaling pathways, affecting the development and progression of DN[9]. Secondly, miR-22-3p is also involved in the fibrosis process in the glomeruli and interstitium of diabetic patients, further worsening the condition of DN[10]. Additionally, miR-22-3p can affect the function of kidney microvasculature in diabetic patients, influencing renal microcirculation and exacerbating the progression of DN[11], while the potential mechanism still need to be clarified.
A recent study has found that Cystathionine-γ-lyase (CSE) could be involved in miR-21 related with vascular resistance in growth-restricted pregnancies. A direct interaction between CSE and miR-22 was also revealed[12]. Carboxylesterases (CES) can catalyze the production of Hydrogen Sulfide (H2S) in vivo and is highly expressed in the cardiovascular system and kidney. The upregulated CES expression level is the characteristic of cardiovascular disease, neurodegenerative diseases and metabolic diseases, such as diabetes. Many studies revealed that CSE can both modulate the process of epigenome remodeling and regulate cellular miRNA signatures. A growing body of research indicates that the control of CSE expression is influenced by redox-dependent mechanisms and is enhanced under conditions of cellular oxidative stress. Consequently, the introduction of external Hydrogen peroxide (H2O2) has been demonstrated to increase CSE gene expression in fibroblasts[13]. Moreover, recent studies have shown that oxidative stress is involved in DN, and CSE alters oxidative stress in vivo[14]. The underlined mechanism for CSE mediated DN disease progression need to be further studied.
In our study, we confirmed the regulatory role of the lncRNA TUG1/miR-22-3p/CSE axis in controlling cell apoptosis in High Glucose (HG)-stimulated podocytes. Our research results indicate that this regulatory axis plays a crucial role in modulating podocyte’s response to HG environments, providing important clues for further understanding its mechanism of action in diseases like DN.
Materials and Methods
All experiments were conducted following appropriate guidelines and regulations, and they received approval from the Ethics Committee of Taizhou People’s Hospital (Approval Number KY: 202003001). All DN patients and corresponding healthy controls involved in this study at Taizhou People’s Hospital provided informed consent.
Microarray analysis:
66 individuals from Taizhou People’s Hospital were categorized into two cohorts (normal and diabetic nephrotic proteinuria groups) based on their Albumin Creatinine Rate (ACR). The normal group comprised 33 patients with an ACR of 30 mg/g or lower, while the diabetic nephrotic proteinuria group included 33 patients with an ACR ranging from 30-300 mg/g. Peripheral blood samples were collected from all participants for high-throughput microarray analysis. Using microarray data from the gene expression omnibus database (http://www.ncbi.nlm.nih.gov/geo/), we identified lncRNAs that are differentially expressed in podocyte cells and are related to DN.
Weighted Gene Co-expression Network Analysis (WGCNA):
The starting point of WGCNA is to construct a network based on systematic gene expression levels with the aim of showing co-expression relationships between genes. We conducted sample clustering to illustrate the association between expression profiles and clinical characteristics. Following initial raw data processing, we applied the WGCNA algorithm, as described previously, to pinpoint significant gene modules linked to DN.
Quantitative real-time Polymerase Chain Reaction (qPCR) assay:
Total RNA was isolated from cultured podocytes using TRIzol reagent. Take 1 mg of RNA and reverse transcribe it into complementary Deoxyribonucleic Acid (cDNA) using a reagent kit. Perform RT-qPCR using the ABI-7300 PCR instrument (Applied Biosystems, United State of America (USA)) following the experimental protocol outlined in the product manual.
Cells and transfection:
The human podocyte cell line AB8/13 was obtained from American type culture collection (American Type Culture Collection (ATCC), Manassas, Virginia). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640. For differentiation induction, AB8/13 cells were incubated in RPMI 1640 at 37°, until the confluence of the podocytes reached 80 %. The cells were treated with glucose concentrations of 5 mol/l or 30 mol/l (Sigma, St. Louis, Missouri, USA) for 48 h. Overexpression vectors were constructed and transfected into cells.
Flow cytometry apoptosis assay:
We collected AB8/13 podocyte cells and analyzed them using a cell apoptosis detection kit. By staining the cells with fluorescent dyes or probes, we were able to distinguish between live cells, early apoptotic cells, late apoptotic/necrotic cells and dead cells. Subsequently, we quantified the stained cells using BD Biosciences FACSCalibur flow cytometer and CellQuest software.
Western blot analysis:
Protein extraction from podocyte cells was carried out using the Bradford protein kit from Beyotime (Shanghai, China). The extracted proteins were subjected to polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes by electro blotting. Immunoblot analysis was performed using a monoclonal CSE antibody.
Dual luciferase reporter assay:
The cDNAs encoding TUG1 and CSE were synthesized and inserted into the luciferase reporter vector pmirGLO to create the recombinant vectors TUG1-Wild Type (WT) and CSE-WT, respectively. Similarly, mutated versions of the TUG1 and CSE 3’-Untranslated Regions (3’UTRs) were generated and named TUG1-Mutant (MT) and CSE-MT, respectively. WT vectors, MT vectors or empty reporter vectors were co-transfected with miR-22-3p or miR-NC into 293T cells using Lipofectamine 2000.
Determination of Superoxide Dismutase (SOD), Reactive Oxygen Species (ROS) and Malondialdehyde (MDA) content:
The SOD, ROS, and MDA levels in podocyte cells were assessed following previously established protocols with slight adjustments. Podocyte cells were subjected to trypsinization, followed by two washes in 50 mM phosphate buffer (pH 7.8) at 4°, and subsequent homogenization. The homogenate underwent centrifugation at 13 000×g for 10 min, yielding a supernatant used for measuring SOD activity, ROS levels and MDA content.
Statistical analysis:
Statistical analysis was conducted using GraphPad Prism 9.0 software. Experimental data underwent analysis employing either Student’s t-test or one-way Analysis of Variance (ANOVA) test.
Results and Discussion
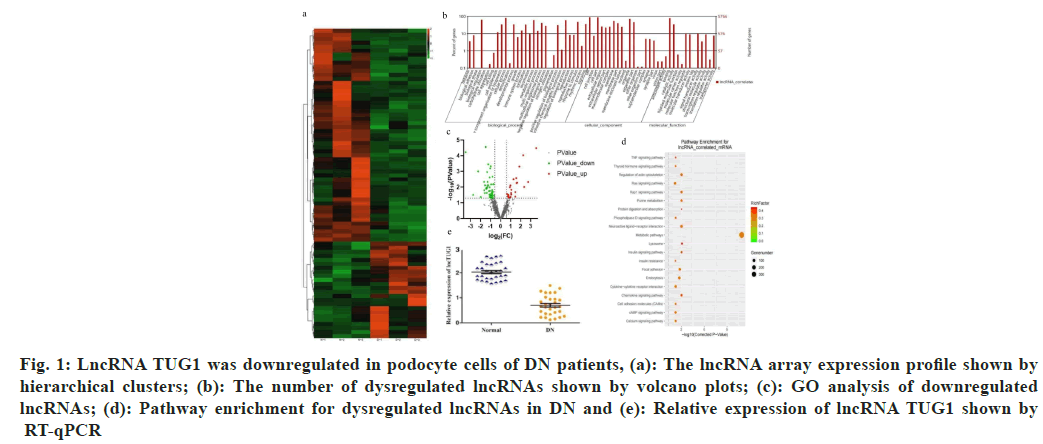
To investigate the potential role of lncRNAs in DN, a high-throughput microarray assay for lncRNAs was conducted with three paired DN samples and normal samples. As shown in (fig. 1a).
Fig. 1: LncRNA TUG1 was downregulated in podocyte cells of DN patients, (a): The lncRNA array expression profile shown by hierarchical clusters; (b): The number of dysregulated lncRNAs shown by volcano plots; (c): GO analysis of downregulated lncRNAs; (d): Pathway enrichment for dysregulated lncRNAs in DN and (e): Relative expression of lncRNA TUG1 shown by RT-qPCR
77 differently expressed lncRNAs were screened, including 24 upregulated lncRNAs and 53 downregulated lncRNAs (due to the large number of genes in the heat map, the gene ID cannot be clearly seen, so it is hidden). Variations in lncRNA expression are shown in the volcano plot (fig. 1b). Gene Ontology (GO) analysis revealed that the predicted target lncRNAs were among the top 58 enriched GO categories (fig. 1c).
Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation indicated that dysregulated lncRNA genes were notably enriched in 20 canonical pathways (fig. 1d). Moreover, the expression of lncRNA TUG1 was markedly reduced in the DN groups (fig. 1e).
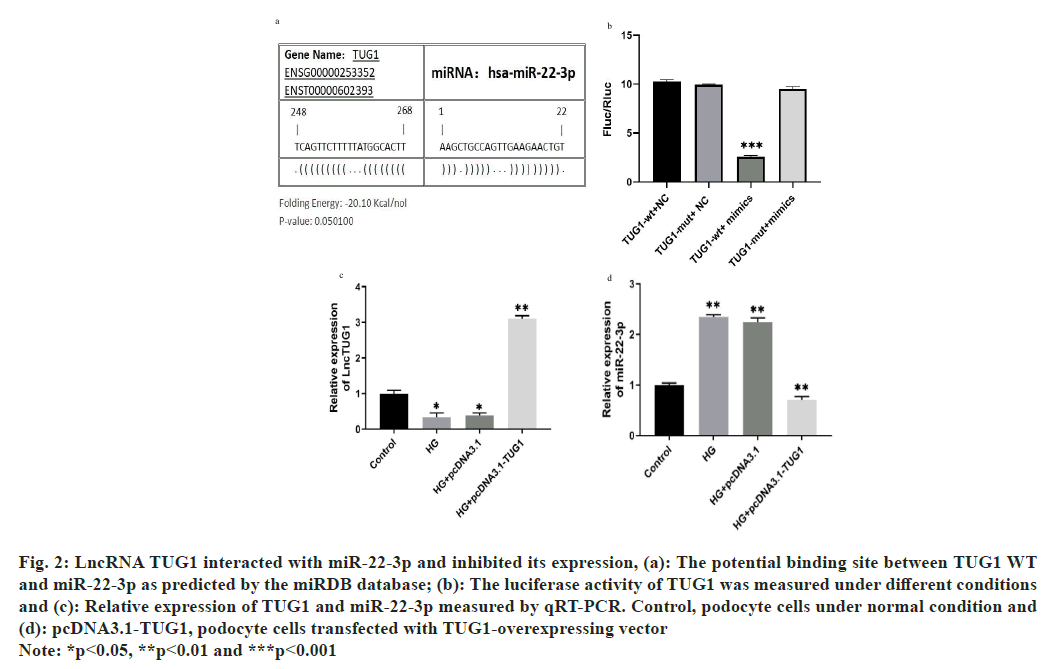
To enhance the understanding of the regulatory pathway involving lncRNA TUG1 in DN, we performed prediction analysis using the miR database, which revealed an interaction between lncRNA TUG1 and miR-22-3p (fig. 2a). To further investigate the relationship between them, we conducted co-transfection experiments for validation. The dual-luciferase reporter analysis revealed that cells transfected with miR-22-3p exhibited significantly reduced luciferase activity of TUG1-WT (fig. 2b). Additionally, furthermore, to explore the impact of TUG1 on the expression of miR-22-3p under HG conditions, we conducted the experiment shown in fig. 2c and fig. 2d. The study indicates that overexpression of TUG1 significantly suppresses the expression of miR-22-3p.
Fig. 2: LncRNA TUG1 interacted with miR-22-3p and inhibited its expression, (a): The potential binding site between TUG1 WT
and miR-22-3p as predicted by the miRDB database; (b): The luciferase activity of TUG1 was measured under different conditions
and (c): Relative expression of TUG1 and miR-22-3p measured by qRT-PCR. Control, podocyte cells under normal condition and
(d): pcDNA3.1-TUG1, podocyte cells transfected with TUG1-overexpressing vector
Note: *p<0.05, **p<0.01 and ***p<0.001
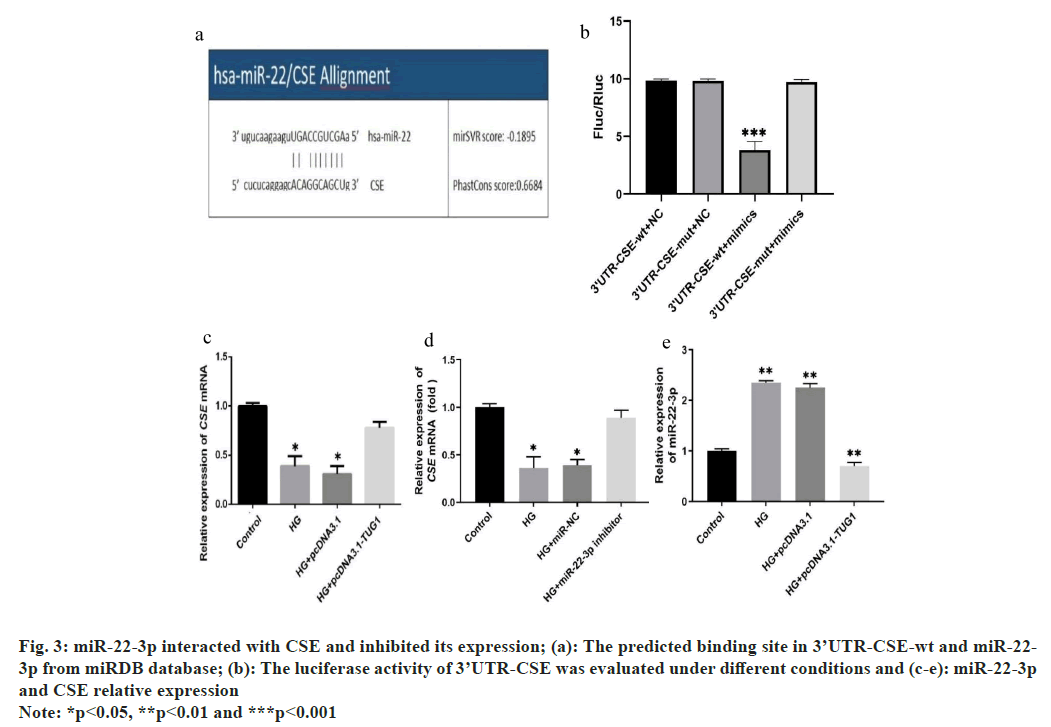
Previous studies have indicated that CSE acts as a downstream gene regulated by miR-22 in response to oxidative stress[15-17]. According to predictions from the miRDB database, miR-22-3p was identified as directly interacting with the 3’UTRs of CSE (fig. 3a). After preparing constructs containing the WT 3’UTR or Mut 3’UTR, they were co-transfected into 293T cells with miR-22-3p mimics. Subsequently, the dual luciferase reporter analysis revealed a substantial decrease in luciferase activity linked to the WT 3’UTR, demonstrating the regulatory influence exerted by miR-22-3p on this specific region of the target gene. These findings underscore the role of miR-22-3p in modulating gene expression through post-transcriptional mechanisms (fig. 3b). Moreover, under HG conditions, CSE expression significantly decreased but notably increased after transfection with TUG1 or a miR-22-3p inhibitor (fig. 3c and fig. 3d) Additionally, TUG1 overexpression resulted in decreased miR-22-3p expression (fig. 3e).
Fig. 3: miR-22-3p interacted with CSE and inhibited its expression; (a): The predicted binding site in 3’UTR-CSE-wt and miR-22-3p from miRDB database; (b): The luciferase activity of 3’UTR-CSE was evaluated under different conditions and (c-e): miR-22-3p and CSE relative expression
Note: *p<0.05, **p<0.01 and ***p<0.001
These findings suggest that miR-22-3p inhibits CSE expression through targeted regulation, with TUG1 potentially playing a crucial role in this regulatory mechanism.
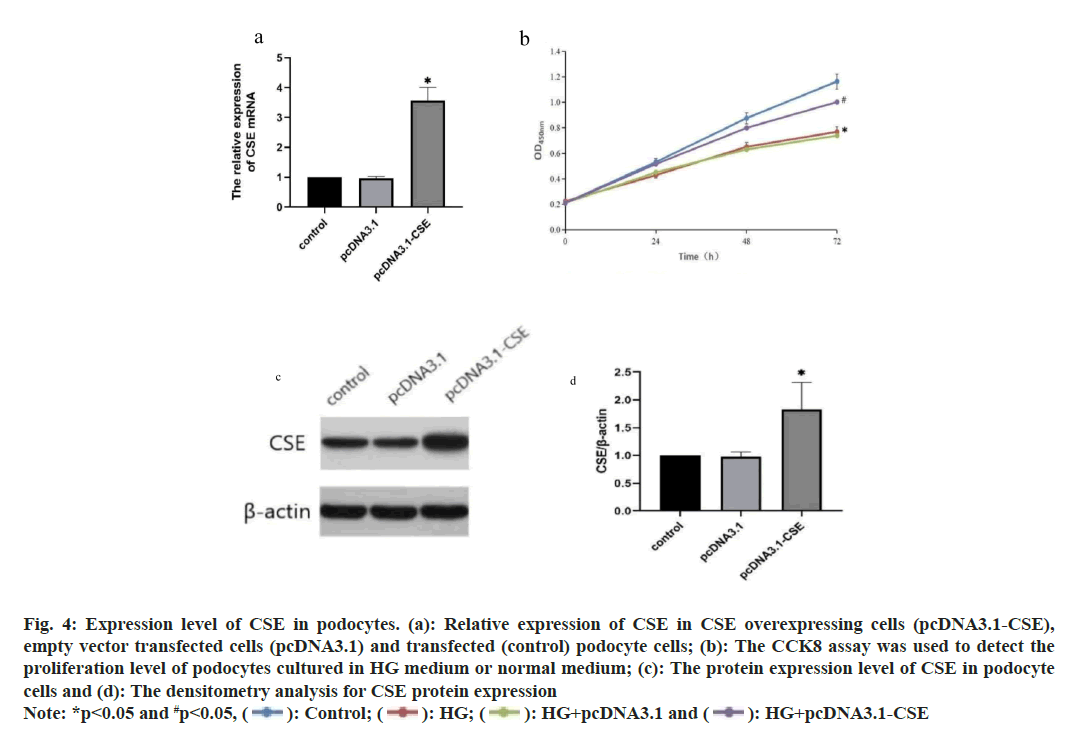
To investigate the function of CSE, we conducted an overexpression of CSE in podocytes. The outcomes demonstrated a notable increase in CSE expression in podocytes with CSE overexpression (fig. 4a). Furthermore, the Cell Counting Kit-8 (CCK8) assay indicated that exposure to HG notably decreased the viability of podocytes (fig. 4b). However, the CSE overexpressing cells exhibited increased cell proliferation compared to cells transfected with an empty vector and transfected cells under HG treatment (fig. 4b). The expression of CSE protein were further confirmed by Western blot and only the CSE overexpressing cells showed increased CSE protein expression (fig. 4c). The densitometry analysis for Western blot results was also shown in fig. 4d.
Fig. 4: Expression level of CSE in podocytes. (a): Relative expression of CSE in CSE overexpressing cells (pcDNA3.1-CSE),
empty vector transfected cells (pcDNA3.1) and transfected (control) podocyte cells; (b): The CCK8 assay was used to detect the proliferation level of podocytes cultured in HG medium or normal medium; (c): The protein expression level of CSE in podocyte cells and (d): The densitometry analysis for CSE protein expression
Note: *p<0.05 and #p<0.05,  HG+pcDNA3.1-CSE
HG+pcDNA3.1-CSE
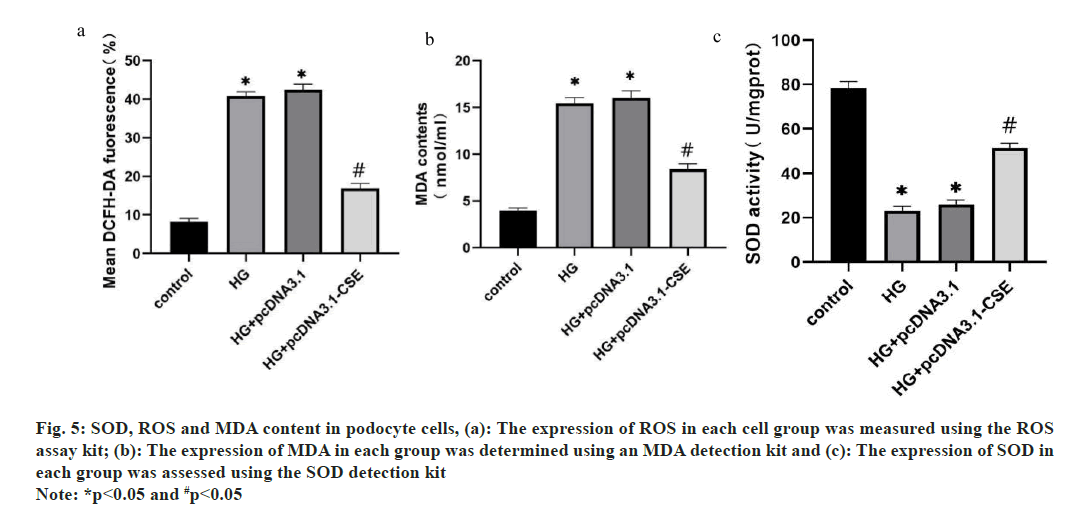
As cells oxidative stress is closely related to cell apoptosis, the ROS activity, MDA and SOD content were measured in podocyte cells transfected with CSE or negative control. These results demonstrated that overexpression of CSE increased the cell SOD content, while it deceased the level of MDA. The ROC content exhibited a notable decrease in the HG group (fig. 5).
Fig. 5: SOD, ROS and MDA content in podocyte cells, (a): The expression of ROS in each cell group was measured using the ROS
assay kit; (b): The expression of MDA in each group was determined using an MDA detection kit and (c): The expression of SOD in
each group was assessed using the SOD detection kit
Note: *p<0.05 and #p<0.05
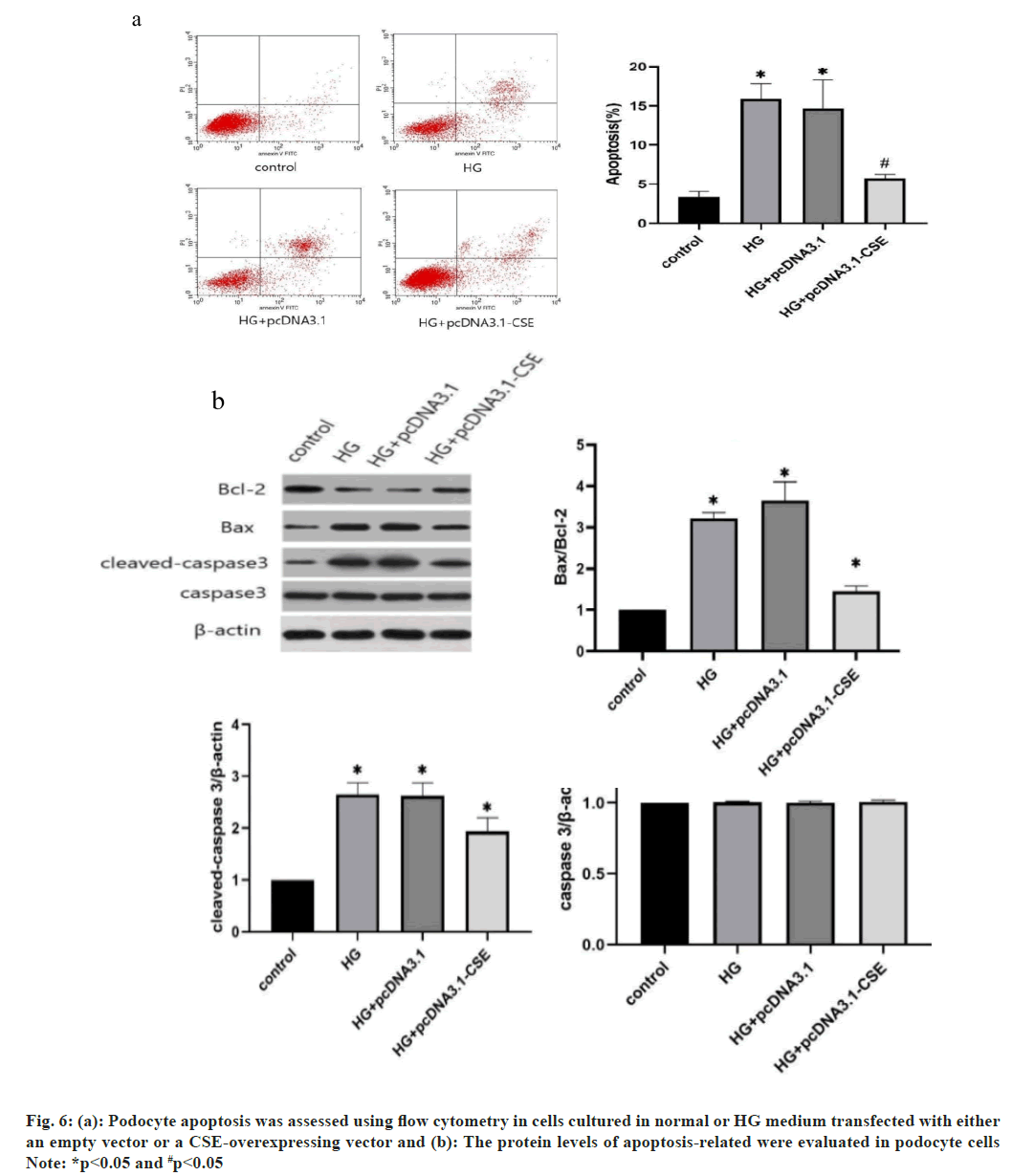
The apoptotic rates were also determined by flow cytometry assay. As depicted in fig. 6a and fig. 6b, the apoptotic rate of podocyte cells notably increased with HG treatment. However, podocytes overexpressing CSE showed a significantly lower apoptotic rate compared to other cells (fig. 6a). Moreover, elevated CSE levels resulted in increased B-cell lymphoma 2 (Bcl-2) expression. Conversely, the HG environment led to a reduction in the expression of pro-apoptotic proteins like Bcl-2 Associated X-protein (BAX) and cleaved-asparaginase 3 (fig. 6b). Notably, the overexpression of CSE did not alter the expression of the apoptosis-related protein caspase 3 in podocytes (fig. 6b). These findings suggest that CSE overexpression promotes cell viability, reduces HG-induced cell apoptosis, and enhances the resistance of podocyte cells to HG conditions.
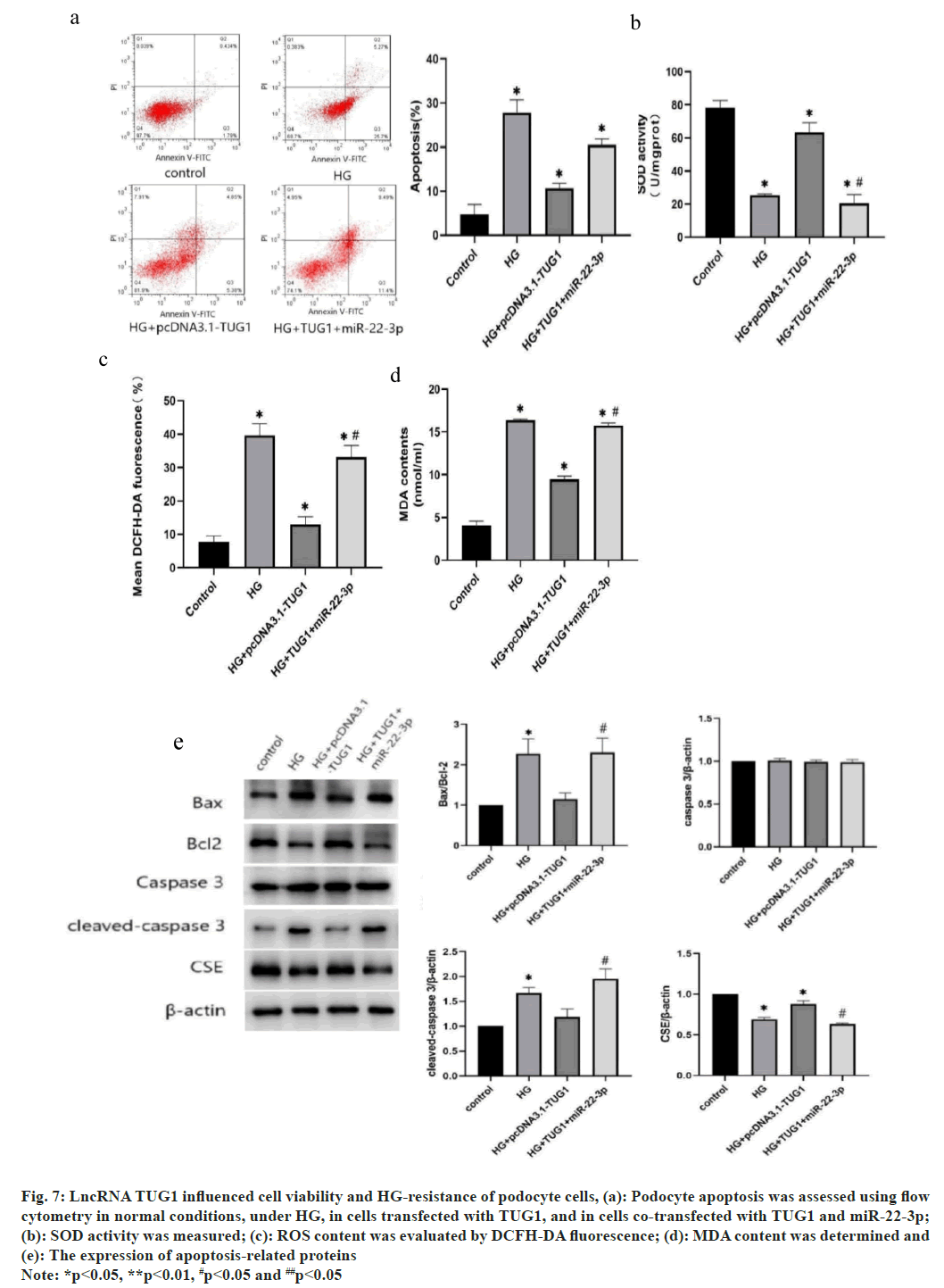
To delve deeper into the impact of TUG1 on podocyte cell resistance to HG, the TUG1 overexpressing podocyte cells were constructed. Compared with HG group, the apoptotic rate of podocyte cells decreased significantly when transfected with TUG1. However, the podocyte cells showed an increased apoptotic rate when miR-22-3p was transfected into the TUG1 overexpressing podocyte cells (fig. 7a). As shown in fig. 7b, SOD activity was decreased under HG condition, overexpressing TUG1 retrieved SOD activity to control level, but co-transfection of TUG1 and miR-22-3p decreased SOD activity to HG level. Similarly, overexpression of TUG1 significantly decreased ROS and MDA content under HG condition, but co-transfection of TUG1 and miR-22-3p retrieved the ROS and MDA content (fig. 7c and fig. 7d). In cells transfected with TUG1 and miR-22-3p, deletion at the TUG1 gene inhibited the increase in Bcl-2. In addition, the reductions of BAX and cleaved asparaginase-3 were restored to levels similar to those in HG conditions (fig. 7e). These findings suggest that the lncRNA TUG1 impedes cell viability and reduces HG-induced apoptosis and that miR-22-3p act downstream of TUG1.
Fig. 7: LncRNA TUG1 influenced cell viability and HG-resistance of podocyte cells, (a): Podocyte apoptosis was assessed using flow cytometry in normal conditions, under HG, in cells transfected with TUG1, and in cells co-transfected with TUG1 and miR-22-3p;
(b): SOD activity was measured; (c): ROS content was evaluated by DCFH-DA fluorescence; (d): MDA content was determined and (e): The expression of apoptosis-related proteins
Note: *p<0.05, **p<0.01, #p<0.05 and ##p<0.05
DN is one of the most serious complications of diabetes mellitus with a high risk of multi-organ failure and death[13]. Research indicates that podocytopathy is likely implicated in the advancement of DN. Additionally, podocyte apoptosis serves as a direct marker for DN[15]. In this study, a high-throughput microarray assay for lncRNAs was conducted and the expression of lncRNA TUG1 was found to be down-regulated in DN samples and HG-stimulated podocyte cells. Here, TUG1 overexpressing podocyte cells also exhibited lower cell apoptotic rate and higher HG resistance (fig. 7).
It has been previously demonstrated that lncRNA TUG1 promotes the expression of Tissue Inhibitor of Metalloproteinase 3 (TIMP3) by inhibiting miR-21, thereby improving the condition of DN[16]. In this study, an in-depth investigation into the downstream regulators of the TUG1 pathway was undertaken, utilizing predictive analysis from the miRDB database. The direct interaction between TUG1 and miR-22-3p, as revealed by the miRDB database, was subsequently confirmed through luciferase reporter assays.
Moreover, the expression of miR-22-3p was found to be significantly suppressed in TUG1-overexpressing podocyte cells. In summary, miR-22-3p emerges as a potential direct downstream regulatory factor of lncRNA TUG1 and holds promise as a novel therapeutic target for the treatment of DN. Modulating the expression levels of both miR-22-3p and lncRNA TUG1 may offer therapeutic avenues to control and manages the progression of DN effectively.
As reported, CSE catalyzes the production of H2S in vivo and is highly expressed in the cardiovascular system and kidneys[12]. Geng et al.[17] found that H S can improve the ROS produced under high-glucose conditions. Szostak et al.[18] reported that mesangial cells cultured in vitro under HG conditions produced ROS, which affected collagen IV synthesis through oxidative stress effects. Apoptosis is a basic process of cells that is necessary to maintain normal tissue homeostasis under physiological conditions. Previous reports had shown that podocyte injury and apoptosis often occur in diabetes mellitus and its complications[19]. According to Wang et al. CSE is the downstream gene for miR-22 against oxidative stress[8]. The miR-22-3p inhibited CSE expression by direct targeting regulation was also revealed. Here, it shows that miR-22-3p could inhibit CSE expression through direct targeted regulation, and when lncRNA TUG1 was overexpressed, CSE expression was significantly up-regulated, leading to reduced pod cellular damage and enhanced cell resistance to oxidative stress under HG conditions. This indicates CSE is a downstream gene of the lncRNA TUG1/miR-22-3p pathway, which are jointly involved in podocyte injury and apoptosis. To further confirm the lncRNA TUG1-miR-22-3p-CSE regulatory chain, HG resistance was tested in both TUG1 overexpressing cells and TUG1+miR-22-3p co-overexpressed cells. The expression of miR-22-3p in TUG1 overexpressing cells retrieved both cell HG resistance and CSE expression. This indicates an lncRNA TUG1-miR-22-3p-CSE regulatory chain for controlling podocyte apoptosis in DN.
In this investigation, lncRNA TUG1 was found to target miR-22, thereby modulating CSE expression. Cells overexpressing TUG1 showed decreased miR-22 levels and increased CSE expression. Furthermore, lncRNA TUG1 was able to regulate cell apoptosis induced by DN, offering insights into the regulatory pathways involved in DN. These findings suggest that lncRNAs could potentially serve as novel drug targets, opening new avenues for the treatment of DN. In conclusion, our results suggest that lncRNA TUG1 is downregulated in renal tissues of DN patients as well as in HG-stimulated podocytes. Dual luciferase assays showed that miR-22-3p is a target gene of lncRNA TUG1, which in turn is a target gene of the downstream CSE gene. The increased expression of CSE can inhibit cell apoptosis and oxidative stress in podocyte cells stimulated by HG. These results lay a potential theoretical foundation for future DN treatments involving lncRNA modulation.
Conflict of interests:
The authors declared no conflict of interests.
References
- Selby NM, Taal MW. An updated overview of diabetic nephropathy: Diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes Metab 2020;22:3-15.
[Crossref] [Google Scholar] [PubMed]
- Tseng CH, Shah KM, Chiu IJ, Hsiao LL. The role of autophagy in type 2 diabetic kidney disease management. Cells 2023;12(23):2691.
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nature Rev Mol Cell Biol 2020;21(8):475-90.
[Crossref] [Google Scholar] [PubMed]
- Kohler B, Dubovik S, Horterer E, Wilk U, Stockl JB, Tekarslan-Sahin H, et al. Combating drug resistance by exploiting miRNA-200c-controlled phase II detoxification. Cancers 2022;14(22):5554.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Cao J, Hu X, Cheng H. Long noncoding RNA TUG1 upregulates VEGFA to enhance malignant behaviors in stomach adenocarcinoma by sponging miR-29c-3p. J Clin Lab Anal 2021;35(12):e24106.
[Crossref] [Google Scholar] [PubMed]
- Sun L, Ding M, Chen F, Zhu D, Xie X. Long non-coding RNA L13Rik promotes high glucose-induced mesangial cell hypertrophy and matrix protein expression by regulating miR-2861/CDKN1B axis. PeerJ 2023;11:e16170.
[Crossref] [Google Scholar] [PubMed]
- Tan X, Liu Y, Liu Y, Zhang T, Cong S. Dysregulation of long non-coding RNAs and their mechanisms in Huntington's disease. J Neurosci Res 2021;99(9):2074-90.
[Crossref] [Google Scholar] [PubMed]
- Zhao Y, Qin F, Han S, Li S, Zhao Y, Wang H, et al. MicroRNAs in drug addiction: Current status and future perspectives. Pharmacol Ther 2022;236:108215.
[Crossref] [Google Scholar] [PubMed]
- Golmakani H, Azimian A, Golmakani E. Newly discovered functions of miRNAs in neuropathic pain: Transitioning from recent discoveries to innovative underlying mechanisms. Mol Pain 2024;20:17448069231225845.
- Qiu D, Zhao N, Chen Q, Wang M. Knockdown of circ_CDYL contributes to inhibit angiotensin II–induced podocytes apoptosis in membranous nephropathy via the miR-149-5p/TNFSF11 pathway. J Cardiovasc Pharmacol 2022;79(6):887-95.
[Crossref] [Google Scholar] [PubMed]
- Hamdy MM, Abdel-Rahman MS, Badary DM, Sabra MS. Effects of furosemide and tadalafil in both conventional and nanoforms against adenine-induced chronic renal failure in rats. Eur J Med Res 2022;27(1):117.
[Crossref] [Google Scholar] [PubMed]
- Youness RA, Habashy DA, Khater N, Elsayed K, Dawoud A, Hakim S, et al. Role of hydrogen sulfide in oncological and non-oncological disorders and its regulation by non-coding RNAs: A comprehensive review. Noncoding RNA 2024;10(1):7.
[Crossref] [Google Scholar] [PubMed]
- Barrow K, Wang Y, Yu R, Zhu J, Yang G. H2S protects from oxidative stress-driven ACE2 expression and cardiac aging. Mol Cell Biochem 2022;477(5):1393-403.
- Hu HC, Lei YH, Zhang WH, Luo XQ. Antioxidant and anti-inflammatory properties of resveratrol in diabetic nephropathy: A systematic review and meta-analysis of animal studies. Front Pharmacol 2022;13:841818.
[Crossref] [Google Scholar] [PubMed]
- Hongbo M, Yanjiao D, Shuo W, Kun S, Yanjie L, Mengmeng L. Podocyte RNF166 deficiency alleviates diabetic nephropathy by mitigating mitochondria impairment and apoptosis via regulation of CYLD signal. Biochem Biophys Res Commun 2021;545:46-53.
[Crossref] [Google Scholar] [PubMed]
- Marques IS, Tavares V, Savva-Bordalo J, Rei M, Liz-Pimenta J, de Melo IG, et al. Long non-coding RNAs: Bridging cancer-associated thrombosis and clinical outcome of ovarian cancer patients. Int J Mol Sci 2023;25(1):140.
[Crossref] [Google Scholar] [PubMed]
- Geng M, Liu W, Li J, Yang G, Tian Y, Jiang X, et al. LncRNA as a regulator in the development of diabetic complications. Front Endocrinol 2024;15:1324393.
[Crossref] [Google Scholar] [PubMed]
- Szostak J, Gorący A, Durys D, Dec P, Modrzejewski A, Pawlik A. The role of microRNA in the pathogenesis of diabetic nephropathy. Int J Mol Sci 2023;24(7):6214.
- Qu G, Li X, Jin R, Guan D, Ji J, Li S, et al. MicroRNA-26a alleviates tubulointerstitial fibrosis in diabetic kidney disease by targeting PAR4. J Cell Mol Med 2024;28(3):e18099.
[Crossref] [Google Scholar] [PubMed]